Abstract
Nuclear bodies including nucleoli, Cajal bodies, nuclear speckles, Polycomb bodies, and paraspeckles are membrane-less subnuclear organelles. They are steady-state structures that dynamically respond to basic physiological processes as well as various forms of stress, altered metabolic conditions and alterations in cellular signaling. The formation of specific nuclear bodies has been suggested to follow stochastic and ordered assembly models. In addition, a seeding mechanism has been proposed to assemble, maintain, and regulate particular nuclear bodies. In coordination with noncoding RNAs, chromatin modifiers and other machineries, various nuclear bodies have been shown to sequester and modify proteins, process RNAs and assemble ribonucleoprotein complexes, as well as epigenetically regulate gene expression. Understanding the functional relationships between the three-dimensional organization of the genome and nuclear bodies is essential to fully uncover the regulation of gene expression and its implications in human diseases.
The cell nucleus is highly organized and harbors many nuclear bodies
The nucleus contains the vast majority of the cell's genetic material, organized as multiple chromosomes. Individual chromosomes reside in limited and nonrandom regions of the interphase nucleus, known as chromosome territories (Box 1) [1, 2]. Studies of how the nucleus is physically and functionally organized within the interchromatin space have revealed that the nucleus is very organized and highly dynamic. One prominent feature of the nuclear landscape is the ability to harbor a variety of discrete subnuclear organelles, collectively referred to as nuclear bodies [3, 4] (Figure 1). Structures of nuclear bodies have been identified as distinct nuclear foci at both light and electron microscopic levels. Numerous nuclear bodies have been characterized thus far including nucleoli, Cajal bodies (CBs), nuclear speckles, paraspeckles, Polycomb bodies, etc. (Table 1).
Box 1. Chromosome Territories.
During interphase, individual chromosomes occupy distinct regions known as chromosome territories. Neighboring chromosome territories have varying levels of overlap and chromatin can loop out from one territory to another territory. The interior of chromosome territories contains spaces, allowing access of gene regulatory factors [1] (Figure 1). The intranuclear radical distribution of chromosome territories seems to be nonrandom, with an evolutionarily conserved feature that gene-poor chromosomes tend to be oriented to the nuclear periphery and gene-rich chromosomes tend to localize in more internal nuclear regions [2]. However, these structures and distributions are not static. The mobility of chromatin allows dynamic interactions between genomic loci and loci with other nuclear structures. It has been shown that in some cases the repositioning of specific loci with respect to nuclear domains is correlated with their transcriptional activity [111]. Whether these specific interactions provide means to regulate gene expression or are the consequences of changes in transcriptional activity remains to be determined. The higher order organization of chromosome territories and its relationship with the regulation of gene expression merit further investigation with a higher spatial and temporal resolution.
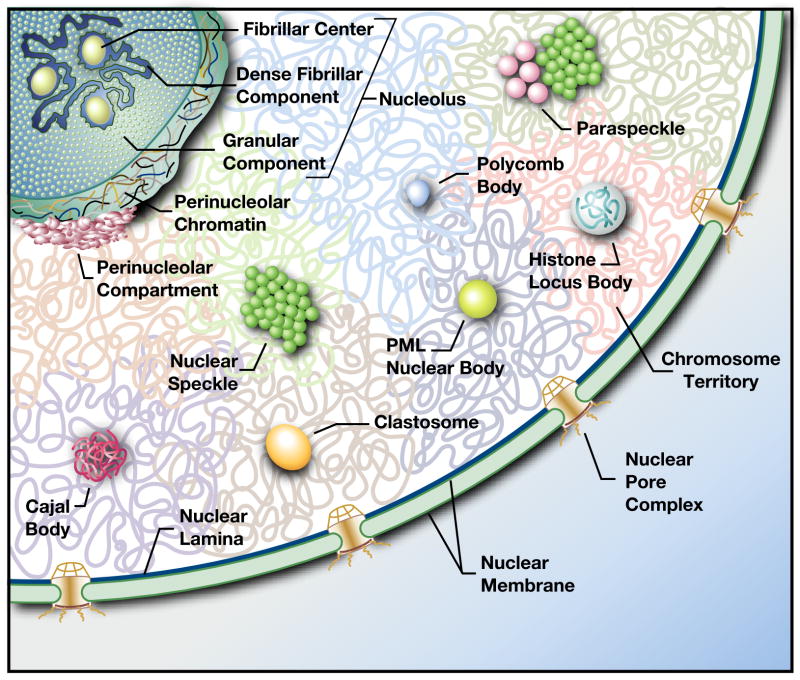
Figure 1.
Diversity of nuclear bodies. The cartoon represents the landscape of an interphase mammalian cell nucleus. The nucleus is enclosed by a double membrane structure called the nuclear envelope, which is contiguous with the rough endoplasmic reticulum and serve as a physical barrier to separate nuclear contents from cytoplasm. The nuclear envelope is interrupted in places by nuclear pore complexes controlling the nucleocytoplasmic transport. Under the inner face of the nuclear envelope, the nuclear lamina provides mechanical support and participates in chromatin organization. The nucleus contains the vast majority of the cell's genetic material organized as multiple chromosomes. Interphase chromosomes occupy distinct chromosome territories. Chromatin fibers and loops from the same chromosome territory and from neighboring chromosome territories can make contact and intermingle in cis and trans. The interchromatin space is very organized, highly dynamic, and harbors multiple nuclear bodies, such as Cajal bodies, clastosomes, histone locus bodies, nuclear speckles, nucleoli, paraspeckles, perinucleolar compartments, PML-nuclear bodies, and Polycomb bodies. The nucleolus is composed of fibrillar centers, dense fibrillar component and granular components and is surrounded by perinucleolar heterochromatin.
Table 1. Basic characteristics of several nuclear bodies.
| Body Name | No./Cell | Typical Size (μm) | Defining components | (Putative) Functions |
|---|---|---|---|---|
| Cajal body | 0-10 | 0.1-2.0 | coilin, SMN | Involved in snRNAs and snoRNAs modification, and assembly and trafficking of snRNPs and snoRNPs. Also plays a role in telomerase assembly and telomere length regulation. |
| Clastosome | 0-3 | 0.2-1.2 | 19S, 20S proteasome | Contains 20S and 19S proteasomes, ubiquitin conjugates, and protein substrates of the proteasome. Forms in response to stimuli that activate proteasome-dependent proteolysis. |
| Histone locus body | 2-4 | 0.2-1.2 | NPAT, FLASH | Involved in the transcription and processing of histone pre-mRNAs. |
| Nuclear speckle | 25-50 | 0.8-1.8 | SRSF2, SRSF1, Malat1 | Involved in the storage, assembly, and modification of pre-mRNA splicing factors. |
| Nuclear stress body | 2-10 | 0.3-3.0 | HSF1, HAP | Contains satellite III ncRNAs and is a part of the general response to stress. Precise function not yet determined. |
| Nucleolus | 1-4 | 0.5-8.0 | RNA Pol I machinery | Involved in the transcription and processing of rRNA and the assembly of ribosomal subunits. Plays roles in the modification and assembly of other nuclear RNAs and RNPs. Regulates cell cycle progression by sequestering and modifying many proteins. |
| Paraspeckle | 10-20 | 0.5 | PSP1, p54nrb, Men ε/β (Neat1) | Involved in nuclear retention of some A-to-I hyperedited mRNAs. |
| Perinucleolar compartment | 1-4 | 0.2-1.0 | PTB, CUGBP | Precise functions are unknown but its prevalence positively correlates with metastatic capacity. |
| PML-nuclear body | 10-30 | 0.3-1.0 | PML | Involved in response to many forms of stress, viral defense, and genome stability by the sequestration, modification, and degradation of many partner proteins. |
| Polycomb body | 12-16 | 0.3-1.0 | Bmi1, Pc2 | Involved in Polycomb proteins-mediated gene paring and silencing in Drosophila. Precise function in mammalian cells remains to be determined. |
Nuclear bodies spatially compartmentalize the nuclear environment and create distinct sites within a confined volume, thereby concentrating reactants and substrates to potentially facilitate more efficient biological reactions. This compartmentalization paradigm is similar to that which occurs in the cytoplasm, where intracellular organelles carry out various metabolic processes in isolated areas demarcated by membranes to achieve specificity and efficiency. However, unlike cytoplasmic organelles, nuclear bodies lack a defining membrane to separate them from their surroundings. Recent studies are beginning to elucidate the molecular mechanisms responsible for the assembly and maintenance of several nuclear bodies [5-7].
In this review, we highlight recent advances in our understanding of the biogenesis and organization of nuclear bodies, focusing primarily on mechanisms of the assembly process. We emphasize the functional implications of several nuclear bodies during cellular differentiation and development. The significance of some nuclear bodies in response to stress and their connections with human diseases are also discussed.
Nuclear bodies are highly organized and dynamically regulated
Although not membrane-bound, nuclear bodies maintain their structural integrity at steady-state, implicating a biogenesis and maintenance mechanism that differs from membrane-bound cytoplasmic organelles [8, 9]. Elucidating the mechanism(s) of how cells assemble, maintain, and regulate nuclear bodies in fine molecular details is critical to understanding their biological functions in different metabolic conditions and during development, and their potential contribution to human diseases.
Nuclear bodies are dynamic steady-state structures
The absence of membranes around nuclear bodies allows their components to exchange more freely with the surrounding nucleoplasm. Most protein components of nuclear bodies are also diffusely distributed in the interchromatin spaces at lower concentrations. Fluorescence recovery after photobleaching (FRAP) studies have demonstrated the rapid exchange of many components between a nuclear body and the nucleoplasm. For example, all three paraspeckle core protein components (PSP1, p54nrb and PSF) show a similar mobile fraction (60-70%) within the paraspeckle. The recovery half life after photobleaching (t1/2) of these mobile populations, which is on the order of seconds, is larger than their nucleoplasmic populations [6]. The delayed mobility of proteins within paraspeckles as compared to the nucleoplasmic population is similar to what has been shown for SRSF1 (SF2/ASF) in speckles and fibrillarin in nucleoli [10, 11], reflecting the intricate protein-protein and protein-RNA interactions within paraspeckles. These protein-protein/RNA interactions account for the structural integrity of the paraspeckle. In fact, paraspeckle proteins form homo- or hetero-oligomers by coiled-coil domains and associate with RNAs by RNA recognition motifs, and the deletion of these interacting motifs results in the loss of paraspeckles [12]. These self-association properties have also been shown for many other components in most other nuclear bodies [8]. Thus, the rapid and free exchange of individual components and their self-association properties determine that nuclear bodies are steady-state structures formed by dynamic interactions of protein-protein and/or protein-RNA components in the nucleoplasm.
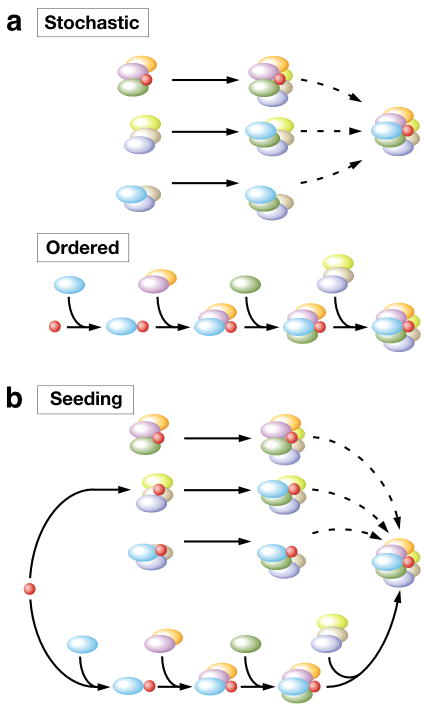
Three models have been proposed to explain how dynamic interactions of individual components in the nucleoplasm results in the assembly of nuclear bodies [6, 8, 9] (Figure 2). In the stochastic assembly model, the structure of nuclear bodies builds up by essentially stochastic interactions of individual components. The assembly process is largely random and with no strict hierarchical order. Each component is equal in terms of assembly order; so multiple pathways can be followed to assemble a nuclear body (Figure 2a). Alternatively, the ordered assembly model posits that the assembly steps follow a tightly controlled sequential order. Individual components are hierarchically different; therefore only one or limited numbers of pathways can lead to the assembly of a nuclear body (Figure 2a). Both models represent extreme theoretical scenarios and it is not yet clear if they are strictly followed in actual biological settings. Alternatively, nuclear body assembly may follow a compromised assembly process. For example, a seeding assembly model has been proposed in which a single or subset of components act hierarchically as a seed to initiate nuclear body formation (Figure 2b).
Figure 2.
Mechanisms of nuclear body assembly. (a) The formation of a nuclear body can occur via a stochastic assembly mechanism in which multiple pathways can be followed to assemble a nuclear body (top), or a hierarchically ordered mechanism in which only one or limited number of pathways are followed to assemble a nuclear body (bottom). (b) The seeding model of nuclear body assembly. RNAs or proteins serve as seeds (shown in red) to nucleate the formation of a nuclear body. The subsequent steps after the initial seeding event could be either stochastic or ordered. If they are completely random (top), the seeding model still differs from the stochastic model because the seeding component is hierarchically different from the other components. If they are tightly ordered (bottom), the seeding model becomes an ordered mechanism.
Stochastic assembly model
An earlier study using a bacterial Lac operator/repressor (LacO/LacI) tethering system, in which LacI fusion protein can be artificially tethered to the integration site of the LacO array within the mammalian cell genome, has provided evidence that the CB might be de novo assembled via stochastic interactions [5]. In this system, individual CB components were tethered to a genomic locus in HeLa cells. The immobilization of any given component was sufficient to initiate the recruitment of most, if not all, of the other CB components. The newly formed structures had a similar size, shape, and composition to endogenous CBs and their components also had comparable dissociation kinetics. Thus, these de novo formed structures were regarded as “bona fide” CBs. However, it is possible that the CBs observed at the tethering sites are endogenous CBs that have recruited the LacO array, similar to what has been shown using a LacI-Lamin B fusion protein to tether a locus to the nuclear lamina [13]. Live cell imaging experiments are required to distinguish between these scenarios. Since many components can initiate the formation of new CBs, there should be no strict requirement for a sequential order of assembly, supporting the conclusion that the assembly of CBs follows the stochastic model. These findings represent a major step forward for understanding the mechanism of nuclear body assembly. However, it remains to be determined if these structures are actually functional CBs, whether this assembly reflects the native in vivo situation, and if so, what nuclear elements serve as the endogenous “tethers”. Nevertheless, this experimental system allows the testing of models for nuclear body organization and provides potential ways to probe nuclear body function.
Seeding assembly model
A recent study has adopted a similar strategy to tether individual paraspeckle proteins in C2C12 myoblasts [6]. In this case, protein tethering did not efficiently recruit other paraspeckle protein components or Men ε/β (also known as Neat1) noncoding RNAs (ncRNAs), important structural components of paraspeckle. However, some paraspeckle protein components were shown to interact in the absence of paraspeckles demonstrating that recruitment of some protein components to the tethered site is an indication of protein-protein interactions rather than paraspeckle assembly. Finally, the protein complex formed at the tethering site showed no capacity to retain adenosine to inosine (A-to-I) hyperedited mRNAs (Box 2), which is a known feature of paraspeckles. Hence, it was concluded that paraspeckles do not form via a stochastic assembly model.
Box 2. Adenosine to Inosine (A-to-I) RNA Editing.
The deamination of adenosine to inosine (A-to-I) in RNA is catalyzed by members of adenosine deaminase that acts on RNA (ADAR) enzyme family [112]. These enzymes are ubiquitously expressed in the nuclei of higher eukaryotes and target double-stranded structures encompassing exons, intron, and 5′ and 3′ untranslated regions (UTRs). A-to-I editing can be site-selective or promiscuous. Since inosines are recognized as guanosines by the translation machinery, site-selective A-to-I editing within coding sequences alters codon meaning therefore diversifying the proteome [112]. Mammalian ADAR2 acts on an intron of its own pre-mRNA creating a new splice site which results in the change of splicing pattern [113]. It has been shown that the presence of inverted repeated Alu elements or short interspersed nuclear elements (SINEs) in the 3′ UTR can induce A-to-I hyperediting in many mRNAs. This event has been proposed to result in nuclear retention of some hyperedited mRNAs by promoting their binding with p54nrb and PSF, two major paraspeckle components [114]. For example, mouse Cat2 transcribed nuclear RNA (CTN-RNA) is transcribed from the mouse cationic amino acid transporter 2 (mCat2) gene loci and the 3′ UTR of CTN-RNA contains SINE elements required for A-to-I hyperediting [115]. Under normal condition, A-to-I hyperedited CTN-RNA is retained in the nucleus, partially within paraspeckles. Under stress, the 3′ UTR of CTN-RNA is post-transcriptionally cleaved to produce mCat2 mRNA which is then released to the cytoplasm [115]. However, many endogenous mRNAs with inosines in their 3′ UTRs have been found on polysomes within the cytoplasm of mammalian cells and Caenorhabditis elegans, suggesting that this nuclear retention phenomenon is not absolute [116]. A-to-I editing alters RNA structure, coding potential, splicing pattern, or cellular distribution, and offers means to regulate gene expression at a variety of post-transcriptional levels.
Studies to understand paraspeckle assembly turned to analyses of Men ε/β ncRNAs, as the depletion of these ncRNAs results in paraspeckle disassembly [14-18]. To address the role of Men ε/β ncRNAs in paraspeckle assembly, a live cell imaging system was developed that allows for the inducible transcription of Men ε/β ncRNAs and the direct visualization of the recruitment of paraspeckle proteins [6]. Upon induction of Men ε/β ncRNAs transcription, all four paraspeckle proteins tested (PSP1, p54nrb, PSF and PSP2) were efficiently recruited to the transcription sites. Multiple criteria were applied to conclude that the newly formed structures are bona fide functional paraspeckles. First, all three core paraspeckle proteins exhibited indistinguishable kinetics in de novo formed and endogenous paraspeckles. Second, de novo formed paraspeckles had similar assembly/disassembly dynamics through the cell cycle compared with their endogenous counterparts. Third, de novo formed paraspeckles were functional as they had the ability to retain A-to-I hyperedited mRNAs. Furthermore, it was demonstrated that it is the active transcription of the Men ε/β gene, together with the ncRNAs, that regulates paraspeckle maintenance and dynamics. Finally, FRAP analyses revealed a significant longer t1/2 of Men ε/β ncRNAs than protein components in paraspeckles, consistent with a mechanism wherein Men ε/β ncRNAs serve as the seeding molecules to assemble paraspeckles.
These results have identified a hierarchical difference between ncRNA and protein components and established that nascent Men ε/β transcripts act as a seed to recruit nucleoplasmic proteins to assemble paraspeckles. The subsequent assembly steps after the initial RNA seeding event could be either stochastic or ordered, which needs to be distinguished by future live cell imaging studies. In this seeding model, one initial component is sufficient and necessary to initiate the nuclear body assembly process (Figure 2b). This concept emphasizes the initial determinant as the pivotal and essential factor, and studies aimed at signaling pathways that control the availability of seeding molecules will expand our understanding of how such nuclear bodies are dynamically regulated.
Nuclear body organization is highly regulated
The RNA seeding model wherein newly transcribed RNAs provide platforms to assemble nuclear bodies sheds light on the regulatory mechanism of controlling the location, size, and number of nuclear bodies. It was found that paraspeckles form and localize in close proximity to the Men ε/β gene loci, corroborating that Men ε/β ncRNAs continuously serve as a nucleator and active transcription is required to maintain paraspeckles. We also found that the size of paraspeckles correlates with the amount of Men ε/β ncRNAs being transcribed. However, there is apparently a size limit and once the size of the paraspeckle exceeds this limitation, fission/splitting/budding events occur to generate a cluster of paraspeckles that remain in the vicinity of the Men ε/β gene locus. Indeed, only two paraspeckles or two clusters of paraspeckles are observed adjacent to two Men ε/β gene loci in normal diploid mammalian tissue (unpublished data), whereas many more are observed next to multiple Men ε/β gene loci in aneuploid cultured cells [6].
The advantage of the seeding model is to provide means to rapidly regulate nuclear bodies by modulating the level of seeds in the nuclei in response to environmental stimuli, differentiation states, and metabolic conditions. For example, human embryonic stem cells (hESCs) lack the expression of Men ε/β ncRNAs and paraspeckle structures [14]. Upon hESC differentiation, the expression of Men ε/β ncRNAs increases, resulting in the formation of paraspeckles which retain A-to-I hyperedited mRNAs in the nuclei of differentiated cells [14]. hESCs lacking paraspeckles efficiently release hyperedited Lin28 mRNAs to the cytoplasm which contributes to pluripotency [19]. The increase of Men ε/β ncRNAs expression has also been demonstrated upon muscle differentiation and viral infection, although the biological functions in these processes are yet to be identified [17, 20].
RNA seeding serves to assemble multiple nuclear bodies
The paradigm of utilizing the RNA seeds to nucleate and organize nuclear bodies also applies to other nuclear bodies. Histone locus bodies (HLBs) are enriched in histone pre-mRNA 3′ end processing components and are known to be involved in processing of histone pre-mRNAs [21]. By artificially tethering histone H2b pre-mRNAs, which are tagged with bacteriophage-derived MS2 stem-loop structures, to a specific engineered site in the HeLa cell genome, NPAT, a HLB component, was recruited to the tethering site implying that histone pre-mRNA seeds the formation of HLBs [7]. However, the assembly process in vivo might be more complex. Histone pre-mRNA transcription and processing are limited to S phase. While in Drosophila, HLBs are found throughout the cell cycle [21]. Similarly, tethering human satellite (Sat) III transcripts can recruit components of nuclear stress bodies (nSBs) indicating the formation of nSBs even in the absence of stress [7]. nSBs are unique nuclear structures which form upon heat shock and other cellular stress. Upon heat shock, HSF1 binds to human pericentric heterochromatin on chromosome 9 q11-12 region composed of long tandem arrays of Sat III repeats [22, 23]. HSF1 redistribution initiates a series of events including the recruitment of CBP/p300, chromatin remodeling, the recruitment of RNA polymerase (Pol) II, and the transcriptional activation of pericentric heterochromatin. Nascent Sat III RNA transcripts then act as seeds to assemble nSBs, which could function as a molecular trap for transcription and splicing factors contributing to global shut down of transcription and splicing alteration. Alternatively, these transcripts may play roles in heterochromatin assembly and maintenance, or affect the organization of the cell nucleus in response to stress [22, 23].
Proteins can also act to seed nuclear bodies
In addition to RNA seeding nuclear bodies, certain proteins could also serve as the seed to assemble some nuclear bodies. The tumor suppressor protein promyelocytic leukemia (PML), a major component of the PML-nuclear body (PML-NB) (a.k.a. nuclear domain 10, ND10), is essential for PML-NB formation since human leukemia and solid tumors cells in which the PML gene is inactivated and pml-/- mouse cells fail to assemble PML-NBs [24]. On the other hand, tethering PML initiates the de novo formation of PML-NBs (as defined morphologically by recruiting another component, SP100) [5]. In addition, PML and other PML-NBs components, such as SP100 and DAXX, can be post-translationally modified by small ubiquitin-like modifier (SUMO). PML and DAXX both contain SUMO-interacting motifs (SIMs) allowing PML to interact with itself and other proteins through sumoylation and SIMs. A PML mutant that cannot be modified by SUMO fails to recruit SP100 and DAXX, and de-sumoylation of PML during mitosis results in the disassembly of PML-NBs [24]. CK2 kinase-mediated phosphorylation within SIM of DAXX has recently been demonstrated to increase its binding affinity to SUMO, therefore facilitating DAXX and PML interaction and DAXX targeting to PML-NBs [25]. Finally, under stress such as viral infection and senescence, PML is upregulated and the size and number of PML-NBs increase [26]. Together, these lines of evidence indicate that PML, presumably the sumoylated form, works as a seed to assemble and regulate PML-NBs.
In summary, the interactions among the protein and RNA components of nuclear bodies determine their structural integrity. However, the rapidly exchanging pools of components and the transient nature of their interactions make these steady-state macromolecular complexes highly dynamic, flexible, and prepared for change in response to various forms of stress and different signaling events. Additional investigations are needed to assess the mechanisms by which other nuclear bodies are formed and maintained. In addition, future studies focusing on how cells regulate the assembly process of nuclear bodies in the context of cell differentiation and human diseases at the tissue level will significantly advance our understanding of the precise physiological functions of nuclear bodies.
Nuclear bodies exert diverse and important biological functions
The genome is physically and functionally organized into complex higher-order structures. Nuclear bodies provide a variety of specific functionally compartmentalized microenvironments within the three-dimensional nuclear volume (Figure 1). By concentrating proteins and/or RNAs required for specific biological processes, nuclear bodies can serve as reaction sites to efficiently facilitate these processes (Figure 3a). In addition, by providing unique compartmentalized microenvironments, nuclear bodies can act as hubs to regulate the expression of recruited gene loci (Figure 3b). Nuclear bodies can also function as storage/modification sites to recycle and modify RNAs and proteins (Figure 3c). Notably, one nuclear body is very likely to combine different themes to execute diverse functions and accommodate different processes at the same time. In the following sections, we focus on the functional properties of four prominent nuclear bodies: nucleoli, CBs, nuclear speckles, and Polycomb bodies. Using these bodies as examples we describe their connections to higher-order genomic organization, epigenetic modifications and gene expression, present their relevance for physiological and pathological processes, and summarize common themes of how nuclear bodies function.
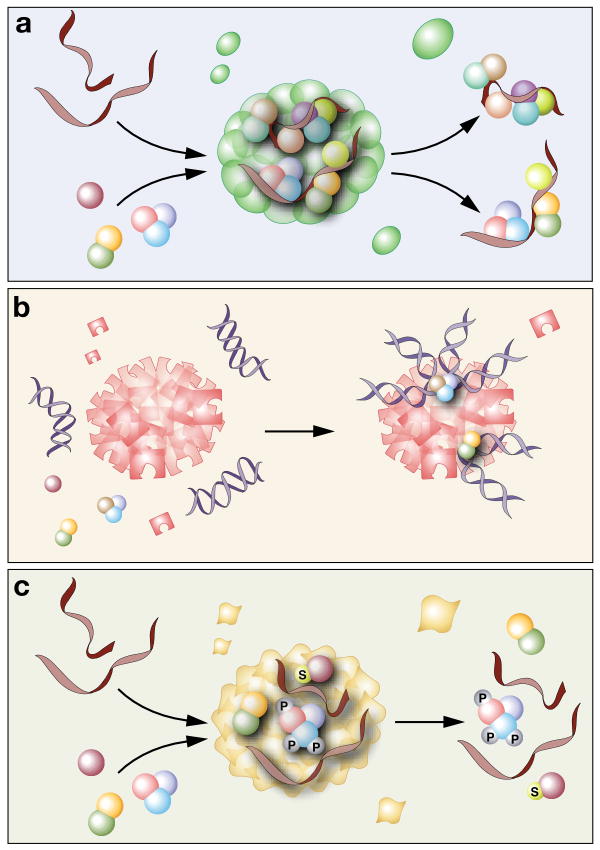
Figure 3.
Conceptual frameworks of how nuclear bodies function. Here, we summarize the common themes of how nuclear bodies may function in the nucleus. However, many nuclear bodies combine multiple mechanisms to perform their diverse cellular functions. (a) Reaction site. Nuclear bodies can concentrate the substrates and enzymes in a confined volume, thereby enhancing the specificity and efficiency of biological reactions. For example, nucleoli serve as pre-rRNA synthesis and processing factories and CBs promote the modification of snRNAs and the assembly of snRNPs, by enriching RNAs and proteins required for these processes. (b) Hub. Nuclear bodies can act as hot spots to activate or repress gene expression by recruiting gene loci. For example, the perinucleolar heterochromatin recruits Xi and imprinted regions to establish and maintain their epigenetic heterochromatin state. Nuclear bodies can also recruit multiple gene loci to stabilize their interactions and subsequently regulate their expression. For example, in Drosophila PcG bodies harbor Hox genes interactions. (c) Storage/modification site. Nuclear bodies can store and recycle proteins and RNAs. For example, paraspeckles retain some A-to-I hyperedited mRNAs. Nuclear bodies can also modify the components being stored. For example, a nuclear speckle-associated kinase can regulate the phosphorylation state of SR proteins and PcG bodies act as sumoylation centers for several nuclear proteins.
Nucleolus
rRNA biogenesis
The nucleolus is organized around the multiple tandemly-arrayed rDNA clusters known as nucleolus organizer regions (NORs), and it is the site of rDNA transcription, pre-rRNA processing and modification, and initial steps of pre-ribosome assembly and maturation [27]. Nucleoli in mammals and birds typically contain three morphologically distinct subregions termed fibrillar centers (FCs), dense fibrillar component (DFC) and granular component (GC) (Figure 1). The tripartite architecture of the nucleolus reflects its function as rDNAs are localized in the FCs; transcription occurs on the border between the FCs and the DFC; fibrillarin and small nucleolar ribonucleoproteins (snoRNPs) are in the DFC where pre-rRNAs are processed; and pre-ribosomal subunits accumulate in the GC where they are assembled [28]. Nucleolar organization changes when ribosome biogenesis is altered. Low doses of actinomycin D which inhibit RNA Pol I transcription, and many forms of stress which affect rDNA transcription have been shown to induce nucleolar reorganization [29, 30].
Mammalian rDNA clusters are characterized by multiple alternating modules of a long intergenic spacer (IGS) of approximately 30 kb and a pre-rRNA coding region of approximately 14 kb. The level of rDNA transcription in eukaryotic cells is tightly regulated according to the protein synthesis requirements of the cell. It has been estimated that approximately half of the several hundred copies of rDNA repeats are transcriptionally active. Generally, transcriptionally active rDNAs exhibit an open euchromatic structure with histone H4 acetylation and histone H3 lysine 4 (H3K4) methylation. Transcriptionally silent rDNA repeats display a more condensed heterochromatic structure with H4 hypoacetylation and H3K9, H3K27, and H4K20 methylation [31].
Recent studies have demonstrated profound and complex roles of long noncoding RNAs (lncRNAs) (Box 3) in regulating rDNA expression. LncRNAs extending 150-250 nucleotides, termed pRNAs, have been found to originate from a RNA Pol I promoter located within the IGS. A specific stem-loop structure found in pRNAs is conserved in several mammalian species and involved in binding with and the recruitment of the nucleolar remodeling complex (NoRC) to rDNA [32]. NoRC, a chromatin remodeling complex, then shifts the positions of rDNA promoter-bound nucleosomes and acts as a scaffold coordinating the activities of other enzymes to rewrite histone modifications, methylate DNA, and establish heterochromatin [33]. pRNA could also form a DNA:RNA triplex structure that is specifically recognized by the DNA methyltransferase DNMT3b to achieve de novo CpG methylation at the rDNA promoter region [34]. Moreover, two nucleolar proteins PHF8, a member of JmjC family of histone demethylases, and KDM2B, a histone demethylase, have also been shown to play roles in regulating the epigenetic state of rDNAs [35-37]. These mechanisms seem to operate at the level of the promoter within each individual rDNA repeat. However, nucleolar dominance (Box 4) has clearly demonstrated that an entire NOR can be repressed as a unit.
Box 3. Long Noncoding RNAs (lncRNAs).
The majority of mammalian and other eukaryotic genomes are transcribed into ncRNAs [117]. LncRNAs are arbitrarily considered longer than 200 nucleotides in length. The expression of some lncRNAs exhibits development- and tissue-specific patterns and the sequence of some lncRNAs is evolutionary constrained, suggesting that they are not merely transcriptional noise [118]. An emerging body of evidence has demonstrated that they play important biological functions, such as in transcriptional interference, epigenetic regulation, dosage compensation, parental imprinting, and nuclear organization, therefore contributing to development and pathogenesis [119-122].
Box 4. Nucleolar Dominance.
Nucleolar dominance is an epigenetic phenomenon observed in interspecific hybrid cells in which the expression of rDNAs is inherited from one parental species due to the silencing of those derived from the other parent. It occurs in both plant and animal kingdoms and provides an excellent system to study how genes are chosen to be and maintained silent epigenetically and how this process is established and regulated [123]. Species–specificity of the Pol I transcription machinery and enhancer–imbalance (the dominant NOR is that containing the greater number of enhancer repeats within the IGS) hypotheses can explain this phenomenon in human–mouse somatic cell hybrids and Xenopus hybrids, respectively [124]. However, the molecular basis of NOR selection and details of its repression remain unclear in plants. Short interfering RNAs (siRNAs) and RNAi pathway components have been shown to be involved in this intriguing process [125].
Other functions of the nucleolus
The nucleolus has been shown to be the site of RNA processing and modifications and assembly of multiple ribonucleoprotein (RNP) complexes, such as the telomerase RNP, RNase P RNA, the signal recognition particle RNA, and microRNAs [27, 38, 39]. The nucleolus can also regulate post-translational modifications such as sumoylation and phosphorylation of some nuclear proteins, and sequester specific proteins to inhibit their activities during the cell cycle to coordinate and regulate multiple aspects of cell cycle progression [27]. In addition, the nucleolus is a target of many viruses. Many virally encoded proteins are detected in the nucleolus and the localization could potentially contribute to virus replication and export of viral RNAs [40]. Finally, the nucleolus plays essential roles coordinating multiple functional aspects in response to many forms of stress. One prominent example is the critical role of the nucleolus in regulating the level of the tumor suppressor protein p53 under stress and its implication to human disease, which has been comprehensively and systematically reviewed [29].
The perinucleolar region
The nucleolus is generally surrounded by a shell of highly compact heterochromatin, known as the perinucleolar heterochromatin (Figure 1). It contains satellite DNA that surrounds NORs, and silent rDNA clusters [4, 31]. Condensation of a part of the rDNA repeats into the perinucleolar heterochromatin could be a general strategy against recombination of these highly repeated rDNAs, thereby preserving rDNA stability [41]. Live cell imaging analysis of chromatin indicated that the perinucleolar association constrains the movement of DNA sequences at different sites in multiple chromosomes [42]. In fact, the perinucleolar region has been demonstrated to function in the maintenance of silencing of non-ribosomal genomic regions. During X-chromosome inactivation, the X-inactivation center targets the inactive X chromosome (Xi) to the perinucleolar region and it has been proposed that the Xi must continuously visit this region during mid-to-late S phase, when the Xi is replicated, to duplicate its epigenetic state and stably repress resident genes. The association of the Xi with the perinucleolar region is dependent on a lncRNA, Xist [43]. By serial-section transmission electron microscopic analysis, a subsequent study confirmed this association and further revealed the extensive contacts of the Xi with the nuclear envelope [44]. A system has recently been developed in ES cells to visualize the X-inactivation center and pairing prior to X-chromosome inactivation [45]. Future studies, including live cell imaging analysis using this system, are essential to determine if, how, and why the Xi continuously visits the perinucleolar region and to characterize the involvement of the nuclear envelope during X chromosome inactivation. Separate studies have found that another lncRNA, Kcnqlot1 antisense ncRNA, mediates the relocalization of an imprinted chromatin region to the perinucleolar region in mid S phase which correlates with subsequent bi-directional imprinting [46, 47]. In Saccharomyces cerevisiae, tRNA-mediated gene silencing, in which tRNA genes transcribed by RNA Pol III can silence nearby Pol II promoters, requires perinucleolar localization as well [48]. Collectively, the temporal and spatial compartmentalization of various chromosomal regions to the perinucleolar region may provide a mechanism to replicate and maintain repressive epigenetic chromatin states of the Xi, imprinted chromatin regions, and other heterochromatin (Figure 3b). Further studies are required to dissect the precise roles that lncRNAs, such as Xist and Kcnqlot1, play in perinucleolar localization and to analyze their functional relationships with histone modifying enzymes, chromatin remodeling complexes, and the DNA replication machinery.
The perinucleolar region also harbors nuclear bodies with unknown functions, such as the perinucleolar compartment (PNC) [49, 50] and the Sam68 nuclear body (SNB) [51]. The PNC contains nascent Pol III transcripts and RNA-binding proteins and the SNB contains various RNA-binding proteins. It seems unlikely that the interaction of Xi or imprinted region occurs in the PNC or SNB since both bodies are primarily found in transformed cells [52]. Notably, PNC prevalence is positively correlated with the metastatic capacity of many types of cancer and has been proposed to serve as a marker for tumor diagnosis and anti-tumor drug selection [53]. However, the specific function of the PNC in malignancy remains to be determined.
In summary, the concept of the nucleolus being “plurifunctional” was first raised in 1998 [38] and is now widely accepted [27, 29]. As the site of rRNA synthesis, the nucleolus provides a perfect system to study Pol I transcription, gene silencing, and heterochromatin metabolism. Moreover, the nucleolus is the modification/assembly site for many other RNAs/RNPs, regulates proteins important for cell cycle progression, and directs cellular responses to many forms of stress. Finally, the perinucleolar region provides a unique compartment for the establishment and maintenance of gene silencing.
Cajal body
snRNPs assembly/modification
The CB contains a variety of proteins and RNAs involved in the assembly and modification of small nuclear RNP (snRNPs) and snoRNPs [21]. For example, spliceosomal snRNAs are first transcribed in the nucleus, exported to the cytoplasm, each assembled into a complex with seven conserved Sm proteins, and hypermethylated at their 5′ end. The newly assembled snRNPs are subsequently imported back into the nucleus. They concentrate first in CBs, later travel to speckles, and eventually move to active genes where they play essential roles in pre-mRNA splicing [54]. CBs have been suggested to play roles in promoting some final steps of snRNP maturation and/or facilitating the interaction of individual snRNPs to form higher-order complexes [21]. However, Arabidopsis thaliana and Drosophila melanogaster lacking CBs, due to the deficiency of a conserved CB protein coilin, are fully viable and develop normally [55, 56]. Coilin knockout mouse lines, which do not form CBs, display semilethality with about 50% dying in the gestation stage. The surviving adults show fertility and fecundity defects [57]. In these three organisms studied, coilin or the CB is not essential for viability. However, a recent study of the function of coilin and the CB during embryogenesis in the zebrafish Danio rerio suggested otherwise [58]. Morpholino-mediated knockdown of coilin resulted in the loss of CBs and the dispersal of snRNPs, and led to developmental arrest at the 15- to 16-somite stage in the embryo, presumably because of increased intron retention and reduced mRNA production. Remarkably, the developmental defect could be rescued by the injection of pre-assembled mature human snRNPs, but not the snRNAs or snRNP proteins alone, suggesting that coilin and possibly CBs are essential for the efficient macromolecular assembly of snRNPs in zebrafish [58].
The CB also harbors a class of CB-specific RNAs (scaRNAs) that are involved in the post-transcriptional modification of snRNAs. Studies in cultured cells demonstrated that scaRNAs in the CB are important for methylation and pseudouridylation of snRNAs [59]. Since all of the snRNAs are correctly modified in coilin-null flies, which lack CBs but have normal levels of scaRNAs, it has been suggested that these scaRNA-mediated snRNA modifications can occur in the absence of CBs [21]. Thus, the role of the CB per se might be locally concentrating the reactants thereby promoting the efficiency of snRNPs assembly and modification (Figure 3a). If any snRNPs maturation step concentrated in the CB becomes rate-limiting due to an altered metabolic requirement (for example, embryogenesis in zebrafish discussed earlier), cells lacking coilin and subsequently CBs might exhibit apparent sensitivity and a notable disadvantage. This presents challenges to identify the appropriate environmental and developmental cues to reveal the physiological functions of the CB. These challenges might apply to other nuclear bodies as well, such as the paraspeckle [18].
Telomerase RNP assembly
The CB also has a role in telomerase assembly and telomere length homeostasis. Telomerase RNP enzyme is composed of telomerase RNA (TR) and telomerase reverse transcriptase (TERT) and is responsible for maintaining telomeric DNA at the ends of eukaryotic chromosomes. The 3′ end of TR is closely related to box H/ACA motif-containing snoRNAs and scaRNAs [60]. TR and TERT have been shown to localize in CBs in human cancer cell lines, suggesting that the CB may function in some aspects of telomerase RNP assembly and maturation [61, 62]. During S phase, when telomeres are elongated, TR-containing CBs were observed to move and make transient associations with telomeres indicating that specific interactions occur between CBs and telomeres [63-65]. Together, these results imply a fascinating hypothesis that the CB mediates the assembly of telomerase RNP and the delivery of telomerase to a subset of telomeres during S phase to regulate telomere elongation [66].
In summary, the CB promotes the efficiency and specificity of snRNPs modification/assembly by enriching the relevant components, and might not be essential under “normal” conditions but become indispensible under certain stress. It is important to determine the physiological or pathological conditions under which the CB is essential. The possibility that CB localization/delivery licenses telomerase RNP for telomere elongation also merits further investigation.
Nuclear speckle
Storage/modification sites of the splicing machinery
Nuclear speckles (a.k.a. interchromatin granule clusters) are generally regarded as the storage/modification compartment of pre-mRNA splicing factors, including snRNPs and serine/arginine-rich (SR) proteins. Several kinases and phosphatases that can phosphorylate and dephosphorylate components of the splicing machinery respectively have been found localized in speckles, supporting its role in regulating the post-translational modification of splicing factors [67] (Figure 3c). Speckles have long been recognized to contain a population of poly(A)+ RNAs [68-70] and a recent report suggested that a lncRNA, Malat1, is part of these long-sought after poly(A)+ RNAs enriched in speckles [71]. Knockdown of Malat1 results in a reduction in the recruitment of SR family splicing factors to the transcription site of a transgene array, suggesting that Malat1 plays an important role in splicing factor dynamics [72]. The contribution of Malat1 to pre-mRNA splicing/processing has also been demonstrated in regard to various endogenous genes [72-74]. A recent study examining alternative splicing patterns after Malat1 knockdown provides some insights into this process [75]. Malat1 depletion in HeLa cells has been shown to significantly decrease the level of phorphorylated SR proteins, which affects the alternative splicing of many genes thereby resulting in aberrant mitosis and increased cell death [75]. However, it remains to be determined if the altered alternative splicing pattern is the direct consequence of Malat1 depletion or a secondary effect because of cell death and mitosis defects, what kinases/phosphatases are responsible for the phosphorylation change of SR proteins, and how these enzymes are modulated by Malat1.
Hubs for gene activation
Previous findings have demonstrated that speckles are in close proximity to many active genes [67]. Multiple specific genes have been shown to cluster around a common nuclear speckle. Therefore, nuclear speckles have been proposed to serve as hubs of enhanced mRNA metabolic activity involved in mRNA maturation and export [76-81]. A recent study has indicated that the association of the Hsp70 gene with speckles is mediated by its promoter and dependent on active transcription, but does not correlate with the level of nascent transcript accumulation [82]. Furthermore, given that the Hsp70 transcript does not contain introns, the functional implication of this association remains to be determined. However, proteins other than pre-mRNA splicing factors, such as some transcription factors, serine 2 phosphorylated RNA Pol II, and components of the transcription elongation complex have also been shown to localize in speckles [67].
Based on these findings, one can envisage the possibility of speckles serving as “hubs” to link active transcription sites (Figure 3b) as has been suggested for other nuclear regions such as “transcription factories” [83, 84]. Indeed, two recent reports have revealed interchromosomal interaction events induced by hormones, which could be potentially mediated by nuclear speckles [85, 86]. In MCF7 breast cancer and human mammary epithelial cells, estradiol triggers the juxtapositioning of two estrogen receptor α (ERα)–responsive genes (TFF1 and GREB1) at nuclear speckles, accompanied by the long-distance movement and interaction of their respective chromosomes 21 and 2 [85]. This association is dependent on ERα and its co-activator CBP/p300. It remains to be determined by live cell imaging studies if the highly active genes move together and recruit a large amount of splicing factors thereby cytologically resembling a speckle or if the genes actually move to an existing speckle [87]. Moreover, this rapid and long-range movement is implied to be mediated by actin and nuclear myosin 1, which have been suggested to play a role in several aspects of transcription [88-90]. Interestingly, nuclear speckles have also been shown to contain actin, a critical actin regulator–phosphatidyinositol 4,5-bisphosphate, and the α isoform of phosphatidylinositol 4-phosphate 5-kinase [91]. The precise roles of actin and myosin in mediating gene movements and the potential function of speckles in this process remain to be deciphered. Depletion of the histone lysine demethylase LSD1, that is essential for the activation of ERα–responsive genes, has no effect on the TFF1 and GREB1 association, but prevents the localization of both loci within speckles and attenuates the transcriptional activation of both genes [85]. These results reveal an essential role of LSD1 in targeting loci to speckles, and the potential function of the speckle in coordinating transcriptional regulation and co-transcriptional processing [85].
Likewise, in LNCaP prostate cancer cells, androgen induces specific intra- and interchromosomal interactions to generate spatial proximity for the androgen receptor (AR)–responsive gene TMPRSS2 and two other genes (ERG and ETV1) at speckles [86]. AR then promotes site-specific DNA double strand breaks (DSBs) by recruiting activation-induced cytidine deaminase and the LINE-1 repeat-encoded ORF2 endonuclease to the interacting loci at speckles. DSBs are later ligated by non-homologous end joining (NHEJ) to create non-random chromosomal translocations in prostate tumors [86]. The potential involvement of speckles in recruiting and assisting the DSB and NHEJ machineries remains to be elucidated.
AR-induced gene loci interactions were also observed in an independent study [92]. However, ER-induced interactions could not be detected in MCF7 and human mammary epithelial cells by a different group [93]. Although the same cell lines were used and the same conditions were followed, such long-range chromosome movement and rapid gene interaction could not be replicated. Moreover, while the initial study suggested a diploid representation of chromosomes 21 and 2 which harbor the TFF1 and GREB1 genes respectively [85], the MCF7 cells used in this study are from hypertriploid to hypotetraploid with 4-6 TFF1 or GREB1 gene loci [93]. Thus, further investigation is required to resolve this discrepancy [94].
In summary, splicing factors are stored and modified at nuclear speckles and are recruited to active genes that are at nuclear sites away from speckles or reside on the periphery of speckles. This proximity to speckles may increase the efficiency of splicing of some genes. Future studies are needed to assess whether speckles may act as hubs to directly influence gene expression.
Polycomb body
Hubs for gene repression
Polycomb group (PcG) proteins are gene-silencing proteins that regulate the expression of a variety of genes. PcG proteins form at least two classes of complexes designated Polycomb repressive complexes 1 and 2 (PRC1 and PRC2) that are thought to collaborate to repress gene transcription [95]. Examination of the localization of PcG proteins has revealed that they are organized into distinct nuclear domains called Polycomb or PcG bodies, which are often localized close to pericentromeric heterochromatin [96]. In Drosophila melanogaster, PcG proteins silence Hox genes through binding to cis-regulatory DNA modules, called Polycomb response elements (PREs) [97]. Drosophila PREs are responsible for inducing long-distance pairing/clustering of multiple endogenous Hox genes, and a transgene containing Fab-7, a well-characterized PRE-containing element. The pairing events occur specifically within PcG bodies [98, 99] (Figure 3b). Intriguingly, three components of the RNA interference (RNAi) machinery, Dicer2, PIWI, and Argonaute1, also localize to distinct nuclear foci and a subset of these foci in Drosophila are frequently colocalized with PcG bodies [98]. After gene pairing is established in PcG bodies, bi-directional transcription in the vicinity of the PRE is stimulated and the RNAi machinery associated with the PcG body cleaves dsRNA to produce siRNA, which is proposed to participate in stabilizing the chromosomal pairing and maintaining gene silencing [98]. In addition, the loss of association of one Hox gene locus with PcG bodies remarkably de-represses the expression of other Hox gene loci, suggesting that specific spatial nuclear organization of Hox gene pairing within PcG bodies reinforces PcG-mediated gene silencing [100]. Strikingly, once long-distance pairing is abolished by removal of endogenous Fab-7, the de-repressed chromatin state induced at the transgene locus can be transmitted through meiosis into a large fraction of the progeny [101].
Sumoylation centers
PcG bodies have also been found in human cells and they appear to directly associate with pericentromeric heterochromatin [102]. Recently, Polycomb-dependent regulatory regions have been identified in vertebrate genomes [103, 104]. However, how PcG bodies function in mammalian systems remains to be determined. The human PcG protein Pc2 has been demonstrated to act as a SUMO E3 ligase by bringing the SUMO E2 (Ubc9) and the substrates (CtBP and CTCF) together, thereby making PcG bodies sumoylation centers [105, 106] (Figure 3c). Moreover, the Caenorhabditis elegans–specific PcG protein SOP-2 is sumoylated, and contains RNA binding motifs which are capable of binding small RNAs and are evolutionarily conserved in vertebrate PcG proteins [107, 108]. Furthermore, recent studies have shown CTCF/cohesin to be implicated in long-range interchromosomal interactions in many systems [109]. Therefore, it is tempting to speculate that PcG-dependent sumoylation may functionally relate to RNAi and CTCF-mediated chromosomal interaction and repression.
In summary, over the past several years, we have gained significant insights into the functions of PcG bodies and PcG-mediated gene pairing, but several important questions remain to be answered. The pairing has been suggested to occur in a locus- and tissue-specific manner [110]. Therefore, identification of tissue-specific factors and characterization of intrinsic properties of specific gene loci are required to fully understand the molecular mechanism underlying this process. How this long-range pairing is accomplished also remains largely unknown. It has been postulated that PRE-containing PcG target genes dynamically localize to PcG bodies, such that genes localized within one PcG body may only linger for a certain time and then leave to incorporate into another PcG body [101]. This PcG body-hopping process prevents PcG target genes from diffusing away randomly in the nucleoplasm, but at the same time, allows these genes to explore parts of the nucleus and to stay in the vicinity of other genes via PcG bodies. Once proximity is achieved, a strong association might be established by regulatory components either on chromatin or from other protein machineries [101]. Live cell imaging with high temporal resolution is crucial to verify the PcG body-hopping hypothesis. PcG target genes are distributed throughout all chromosomes. How the interactions of these genes are re-established after each round of cell division and what roles PcG bodies play during meiotic/mitotic inheritance also warrant further investigation.
Concluding remarks
Nuclear bodies are highly dynamic structures involved in the modulation of numerous nuclear activities. Studies of their biogenesis have provided important mechanistic and molecular insights into their organization and function. While certain nuclear bodies form de novo in a stochastic fashion, others are organized via a seeding model whereby RNA or protein molecules serve as the nucleator to initiate nuclear body assembly. The seeding model provides cells with means to rapidly regulate the localization, number, size, and activity of nuclear bodies by modulating the availability of seeds, under normal physiological conditions or in response to various forms of stress, altered metabolism, and differential signaling. This ability allows for the potential regulation of gene expression at multiple levels.
The conceptual framework of the biological functions of nuclear bodies is just beginning to be uncovered (Figure 3), molecular and mechanistic details still remain largely incomplete. A comprehensive understanding of the three-dimensional organization of the genome in the context of the nuclear body landscape is crucial to fully appreciate the control and regulation of gene expression in normal cellular processes and the implication in human diseases. Future studies combining cell biology, biochemistry, and genetic approaches are likely to provide exciting insights into this rapidly evolving field.
Acknowledgments
We apologize to our colleagues whose work was not cited or discussed in full owing to space limitations. We thank Drs. Joseph G. Gall and Thoru Pederson for critically reading the manuscript and providing helpful insights. Y.S.M. is supported by a National Cancer Center Postdoctoral Fellowship. B.Z. is supported by a Department of Defense Prostate Cancer Research Postdoctoral Fellowship. Research in the Spector laboratory is supported by grants from NIH (NIGMS 42694, EY 18244, and NCI 5PO1CA013106).
Footnotes
Publisher's Disclaimer: This is a PDF file of an unedited manuscript that has been accepted for publication. As a service to our customers we are providing this early version of the manuscript. The manuscript will undergo copyediting, typesetting, and review of the resulting proof before it is published in its final citable form. Please note that during the production process errors may be discovered which could affect the content, and all legal disclaimers that apply to the journal pertain.
References
- 1.Cremer T, Cremer M. Chromosome territories. Cold Spring Harb Perspect Biol. 2010;2:a003889. doi: 10.1101/cshperspect.a003889. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Cremer T, et al. Chromosome territories--a functional nuclear landscape. Curr Opin Cell Biol. 2006;18:307–316. doi: 10.1016/j.ceb.2006.04.007. [DOI] [PubMed] [Google Scholar]
- 3.Handwerger KE, Gall JG. Subnuclear organelles: new insights into form and function. Trends Cell Biol. 2006;16:19–26. doi: 10.1016/j.tcb.2005.11.005. [DOI] [PubMed] [Google Scholar]
- 4.Zhao R, et al. Nuclear neighborhoods and gene expression. Curr Opin Genet Dev. 2009;19:172–179. doi: 10.1016/j.gde.2009.02.007. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Kaiser TE, et al. De novo formation of a subnuclear body. Science. 2008;322:1713–1717. doi: 10.1126/science.1165216. [DOI] [PubMed] [Google Scholar]
- 6.Mao YS, et al. Direct visualization of the co-transcriptional assembly of a nuclear body by noncoding RNAs. Nat Cell Biol. 2011;13:95–101. doi: 10.1038/ncb2140. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Shevtsov SP, Dundr M. Nucleation of nuclear bodies by RNA. Nat Cell Biol. 2011;13:167–173. doi: 10.1038/ncb2157. [DOI] [PubMed] [Google Scholar]
- 8.Dundr M, Misteli T. Biogenesis of nuclear bodies. Cold Spring Harb Perspect Biol. 2010;2:a000711. doi: 10.1101/cshperspect.a000711. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Matera AG, et al. Nuclear bodies: random aggregates of sticky proteins or crucibles of macromolecular assembly? Dev Cell. 2009;17:639–647. doi: 10.1016/j.devcel.2009.10.017. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Phair RD, Misteli T. High mobility of proteins in the mammalian cell nucleus. Nature. 2000;404:604–609. doi: 10.1038/35007077. [DOI] [PubMed] [Google Scholar]
- 11.Chen D, Huang S. Nucleolar components involved in ribosome biogenesis cycle between the nucleolus and nucleoplasm in interphase cells. J Cell Biol. 2001;153:169–176. doi: 10.1083/jcb.153.1.169. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Bond CS, Fox AH. Paraspeckles: nuclear bodies built on long noncoding RNA. J Cell Biol. 2009;186:637–644. doi: 10.1083/jcb.200906113. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Kumaran RI, Spector DL. A genetic locus targeted to the nuclear periphery in living cells maintains its transcriptional competence. J Cell Biol. 2008;180:51–65. doi: 10.1083/jcb.200706060. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Chen LL, Carmichael GG. Altered nuclear retention of mRNAs containing inverted repeats in human embryonic stem cells: functional role of a nuclear noncoding RNA. Mol Cell. 2009;35:467–478. doi: 10.1016/j.molcel.2009.06.027. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Clemson CM, et al. An architectural role for a nuclear noncoding RNA: NEAT1 RNA is essential for the structure of paraspeckles. Mol Cell. 2009;33:717–726. doi: 10.1016/j.molcel.2009.01.026. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Sasaki YT, et al. MENepsilon/beta noncoding RNAs are essential for structural integrity of nuclear paraspeckles. Proc Natl Acad Sci. 2009;106:2525–2530. doi: 10.1073/pnas.0807899106. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Sunwoo H, et al. MEN epsilon/beta nuclear-retained non-coding RNAs are upregulated upon muscle differentiation and are essential components of paraspeckles. Genome Res. 2009;19:347–359. doi: 10.1101/gr.087775.108. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Nakagawa S, et al. Paraspeckles are subpopulation-specific nuclear bodies that are not essential in mice. J Cell Biol. 2011;193:31–39. doi: 10.1083/jcb.201011110. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Viswanathan SR, Daley GQ. Lin28: A microRNA regulator with a macro role. Cell. 2010;140:445–449. doi: 10.1016/j.cell.2010.02.007. [DOI] [PubMed] [Google Scholar]
- 20.Saha S, et al. Identification and characterization of a virus-inducible non-coding RNA in mouse brain. J Gen Virol. 2006;87:1991–1995. doi: 10.1099/vir.0.81768-0. [DOI] [PubMed] [Google Scholar]
- 21.Nizami Z, et al. The Cajal body and histone locus body. Cold Spring Harb Perspect Biol. 2010;2:a000653. doi: 10.1101/cshperspect.a000653. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Biamonti G, Vourc'h C. Nuclear stress bodies. Cold Spring Harb Perspect Biol. 2010;2:a000695. doi: 10.1101/cshperspect.a000695. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Jolly C, Lakhotia SC. Human sat III and Drosophila hsr omega transcripts: a common paradigm for regulation of nuclear RNA processing in stressed cells. Nucleic Acids Res. 2006;34:5508–5514. doi: 10.1093/nar/gkl711. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Bernardi R, Pandolfi PP. Structure, dynamics and functions of promyelocytic leukaemia nuclear bodies. Nat Rev Mol Cell Biol. 2007;8:1006–1016. doi: 10.1038/nrm2277. [DOI] [PubMed] [Google Scholar]
- 25.Chang CC, et al. Structural and Functional Roles of Daxx SIM Phosphorylation in SUMO Paralog-Selective Binding and Apoptosis Modulation. Mol Cell. 2011;42:62–74. doi: 10.1016/j.molcel.2011.02.022. [DOI] [PubMed] [Google Scholar]
- 26.Lallemand-Breitenbach V, de The H. PML nuclear bodies. Cold Spring Harb Perspect Biol. 2010;2:a000661. doi: 10.1101/cshperspect.a000661. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Pederson T. The Nucleolus. Cold Spring Harb Perspect Biol. 2010;3:a000638. doi: 10.1101/cshperspect.a000638. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Boisvert FM, et al. The multifunctional nucleolus. Nat Rev Mol Cell Biol. 2007;8:574–585. doi: 10.1038/nrm2184. [DOI] [PubMed] [Google Scholar]
- 29.Boulon S, et al. The nucleolus under stress. Mol Cell. 2010;40:216–227. doi: 10.1016/j.molcel.2010.09.024. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Dousset T, et al. Initiation of nucleolar assembly is independent of RNA polymerase I transcription. Mol Biol Cell. 2000;11:2705–2717. doi: 10.1091/mbc.11.8.2705. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.McStay B, Grummt I. The epigenetics of rRNA genes: from molecular to chromosome biology. Annu Rev Cell Dev Biol. 2008;24:131–157. doi: 10.1146/annurev.cellbio.24.110707.175259. [DOI] [PubMed] [Google Scholar]
- 32.Mayer C, et al. The structure of NoRC-associated RNA is crucial for targeting the chromatin remodelling complex NoRC to the nucleolus. EMBO Rep. 2008;9:774–780. doi: 10.1038/embor.2008.109. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Zhou Y, et al. Reversible acetylation of the chromatin remodelling complex NoRC is required for non-coding RNA-dependent silencing. Nat Cell Biol. 2009;11:1010–1016. doi: 10.1038/ncb1914. [DOI] [PubMed] [Google Scholar]
- 34.Schmitz KM, et al. Interaction of noncoding RNA with the rDNA promoter mediates recruitment of DNMT3b and silencing of rRNA genes. Genes Dev. 2010;24:2264–2269. doi: 10.1101/gad.590910. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Feng W, et al. PHF8 activates transcription of rRNA genes through H3K4me3 binding and H3K9me1/2 demethylation. Nat Struct Mol Biol. 2010;17:445–450. doi: 10.1038/nsmb.1778. [DOI] [PubMed] [Google Scholar]
- 36.Frescas D, et al. JHDM1B/FBXL10 is a nucleolar protein that represses transcription of ribosomal RNA genes. Nature. 2007;450:309–313. doi: 10.1038/nature06255. [DOI] [PubMed] [Google Scholar]
- 37.Zhu Z, et al. PHF8 is a histone H3K9me2 demethylase regulating rRNA synthesis. Cell Res. 2010;20:794–801. doi: 10.1038/cr.2010.75. [DOI] [PubMed] [Google Scholar]
- 38.Pederson T. The plurifunctional nucleolus. Nucleic Acids Res. 1998;26:3871–3876. doi: 10.1093/nar/26.17.3871. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Gerbi SA, et al. The nucleolus: a site of ribonucleoprotein maturation. Curr Opin Cell Biol. 2003;15:318–325. doi: 10.1016/s0955-0674(03)00049-8. [DOI] [PubMed] [Google Scholar]
- 40.Sirri V, et al. Nucleolus: the fascinating nuclear body. Histochem Cell Biol. 2008;129:13–31. doi: 10.1007/s00418-007-0359-6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Peng JC, Karpen GH. H3K9 methylation and RNA interference regulate nucleolar organization and repeated DNA stability. Nat Cell Biol. 2007;9:25–35. doi: 10.1038/ncb1514. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Chubb JR, et al. Chromatin motion is constrained by association with nuclear compartments in human cells. Curr Biol. 2002;12:439–445. doi: 10.1016/s0960-9822(02)00695-4. [DOI] [PubMed] [Google Scholar]
- 43.Zhang LF, et al. Perinucleolar targeting of the inactive X during S phase: evidence for a role in the maintenance of silencing. Cell. 2007;129:693–706. doi: 10.1016/j.cell.2007.03.036. [DOI] [PubMed] [Google Scholar]
- 44.Rego A, et al. The facultative heterochromatin of the inactive X chromosome has a distinctive condensed ultrastructure. J Cell Sci. 2008;121:1119–1127. doi: 10.1242/jcs.026104. [DOI] [PubMed] [Google Scholar]
- 45.Masui O, et al. Live-Cell Chromosome Dynamics and Outcome of X Chromosome Pairing Events during ES Cell Differentiation. Cell. 2011;145:447–458. doi: 10.1016/j.cell.2011.03.032. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Mohammad F, et al. Kcnq1ot1/Lit1 noncoding RNA mediates transcriptional silencing by targeting to the perinucleolar region. Mol Cell Biol. 2008;28:3713–3728. doi: 10.1128/MCB.02263-07. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Pandey RR, et al. Kcnq1ot1 antisense noncoding RNA mediates lineage-specific transcriptional silencing through chromatin-level regulation. Mol Cell. 2008;32:232–246. doi: 10.1016/j.molcel.2008.08.022. [DOI] [PubMed] [Google Scholar]
- 48.Wang L, et al. Silencing near tRNA genes requires nucleolar localization. J Biol Chem. 2005;280:8637–8639. doi: 10.1074/jbc.C500017200. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Huang S, et al. The dynamic organization of the perinucleolar compartment in the cell nucleus. J Cell Biol. 1997;137:965–974. doi: 10.1083/jcb.137.5.965. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Matera AG, et al. A perinucleolar compartment contains several RNA polymerase III transcripts as well as the polypyrimidine tract-binding protein, hnRNP I. J Cell Biol. 1995;129:1181–1193. doi: 10.1083/jcb.129.5.1181. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Chen T, et al. A role for the GSG domain in localizing Sam68 to novel nuclear structures in cancer cell lines. Mol Biol Cell. 1999;10:3015–3033. doi: 10.1091/mbc.10.9.3015. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Huang S. Review: perinucleolar structures. J Struct Biol. 2000;129:233–240. doi: 10.1006/jsbi.2000.4247. [DOI] [PubMed] [Google Scholar]
- 53.Kamath RV, et al. Perinucleolar compartment prevalence has an independent prognostic value for breast cancer. Cancer Res. 2005;65:246–253. [PubMed] [Google Scholar]
- 54.Matera AG, et al. Non-coding RNAs: lessons from the small nuclear and small nucleolar RNAs. Nat Rev Mol Cell Biol. 2007;8:209–220. doi: 10.1038/nrm2124. [DOI] [PubMed] [Google Scholar]
- 55.Collier S, et al. A distant coilin homologue is required for the formation of cajal bodies in Arabidopsis. Mol Biol Cell. 2006;17:2942–2951. doi: 10.1091/mbc.E05-12-1157. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56.Liu JL, et al. Coilin is essential for Cajal body organization in Drosophila melanogaster. Mol Biol Cell. 2009;20:1661–1670. doi: 10.1091/mbc.E08-05-0525. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57.Walker MP, et al. Reduced viability, fertility and fecundity in mice lacking the cajal body marker protein, coilin. PLoS One. 2009;4:e6171. doi: 10.1371/journal.pone.0006171. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58.Strzelecka M, et al. Coilin-dependent snRNP assembly is essential for zebrafish embryogenesis. Nat Struct Mol Biol. 2010;17:403–409. doi: 10.1038/nsmb.1783. [DOI] [PubMed] [Google Scholar]
- 59.Darzacq X, et al. Cajal body-specific small nuclear RNAs: a novel class of 2′-O-methylation and pseudouridylation guide RNAs. EMBO J. 2002;21:2746–2756. doi: 10.1093/emboj/21.11.2746. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 60.Theimer CA, et al. Structural and functional characterization of human telomerase RNA processing and cajal body localization signals. Mol Cell. 2007;27:869–881. doi: 10.1016/j.molcel.2007.07.017. [DOI] [PubMed] [Google Scholar]
- 61.Jady BE, et al. Human telomerase RNA and box H/ACA scaRNAs share a common Cajal body-specific localization signal. J Cell Biol. 2004;164:647–652. doi: 10.1083/jcb.200310138. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 62.Zhu Y, et al. Telomerase RNA accumulates in Cajal bodies in human cancer cells. Mol Biol Cell. 2004;15:81–90. doi: 10.1091/mbc.E03-07-0525. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 63.Jady BE, et al. Cell cycle-dependent recruitment of telomerase RNA and Cajal bodies to human telomeres. Mol Biol Cell. 2006;17:944–954. doi: 10.1091/mbc.E05-09-0904. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 64.Tomlinson RL, et al. Telomerase reverse transcriptase is required for the localization of telomerase RNA to cajal bodies and telomeres in human cancer cells. Mol Biol Cell. 2008;19:3793–3800. doi: 10.1091/mbc.E08-02-0184. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 65.Tomlinson RL, et al. Cell cycle-regulated trafficking of human telomerase to telomeres. Mol Biol Cell. 2006;17:955–965. doi: 10.1091/mbc.E05-09-0903. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 66.Kiss T, et al. Biogenesis and intranuclear trafficking of human box C/D and H/ACA RNPs. Cold Spring Harb Symp Quant Biol. 2006;71:407–417. doi: 10.1101/sqb.2006.71.025. [DOI] [PubMed] [Google Scholar]
- 67.Spector DL, Lamond AI. Nuclear Speckles. Cold Spring Harb Perspect Biol. 2010;3:a000646. doi: 10.1101/cshperspect.a000646. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 68.Huang S, et al. In vivo analysis of the stability and transport of nuclear poly(A)+ RNA. J Cell Biol. 1994;126:877–899. doi: 10.1083/jcb.126.4.877. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 69.Carter KC, et al. A three-dimensional view of precursor messenger RNA metabolism within the mammalian nucleus. Science. 1993;259:1330–1335. doi: 10.1126/science.8446902. [DOI] [PubMed] [Google Scholar]
- 70.Visa N, et al. Intranuclear distribution of poly(A) RNA determined by electron microscope in situ hybridization. Exp Cell Res. 1993;208:19–34. doi: 10.1006/excr.1993.1218. [DOI] [PubMed] [Google Scholar]
- 71.Hutchinson JN, et al. A screen for nuclear transcripts identifies two linked noncoding RNAs associated with SC35 splicing domains. BMC Genomics. 2007;8:39. doi: 10.1186/1471-2164-8-39. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 72.Bernard D, et al. A long nuclear-retained non-coding RNA regulates synaptogenesis by modulating gene expression. EMBO J. 2010;29:3082–3093. doi: 10.1038/emboj.2010.199. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 73.Lin R, et al. Control of RNA processing by a large non-coding RNA over-expressed in carcinomas. FEBS Lett. 2011;585:671–676. doi: 10.1016/j.febslet.2011.01.030. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 74.Tano K, et al. MALAT-1 enhances cell motility of lung adenocarcinoma cells by influencing the expression of motility-related genes. FEBS Lett. 2010;584:4575–4580. doi: 10.1016/j.febslet.2010.10.008. [DOI] [PubMed] [Google Scholar]
- 75.Tripathi V, et al. The nuclear-retained noncoding RNA MALAT1 regulates alternative splicing by modulating SR splicing factor phosphorylation. Mol Cell. 2010;39:925–938. doi: 10.1016/j.molcel.2010.08.011. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 76.Shopland LS, et al. Clustering of multiple specific genes and gene-rich R-bands around SC-35 domains: evidence for local euchromatic neighborhoods. J Cell Biol. 2003;162:981–990. doi: 10.1083/jcb.200303131. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 77.Carter KC, et al. Discrete nuclear domains of poly(A) RNA and their relationship to the functional organization of the nucleus. J Cell Biol. 1991;115:1191–1202. doi: 10.1083/jcb.115.5.1191. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 78.Huang S, Spector DL. Nascent pre-mRNA transcripts are associated with nuclear regions enriched in splicing factors. Genes Dev. 1991;5:2288–2302. doi: 10.1101/gad.5.12a.2288. [DOI] [PubMed] [Google Scholar]
- 79.Moen PT, Jr, et al. Repositioning of muscle-specific genes relative to the periphery of SC-35 domains during skeletal myogenesis. Mol Biol Cell. 2004;15:197–206. doi: 10.1091/mbc.E03-06-0388. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 80.Brown JM, et al. Association between active genes occurs at nuclear speckles and is modulated by chromatin environment. J Cell Biol. 2008;182:1083–1097. doi: 10.1083/jcb.200803174. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 81.Brown JM, et al. Coregulated human globin genes are frequently in spatial proximity when active. J Cell Biol. 2006;172:177–187. doi: 10.1083/jcb.200507073. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 82.Hu Y, et al. Hsp70 gene association with nuclear speckles is Hsp70 promoter specific. J Cell Biol. 2010;191:711–719. doi: 10.1083/jcb.201004041. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 83.Cook PR. A model for all genomes: the role of transcription factories. J Mol Biol. 2010;395:1–10. doi: 10.1016/j.jmb.2009.10.031. [DOI] [PubMed] [Google Scholar]
- 84.Sutherland H, Bickmore WA. Transcription factories: gene expression in unions? Nat Rev Genet. 2009;10:457–466. doi: 10.1038/nrg2592. [DOI] [PubMed] [Google Scholar]
- 85.Hu Q, et al. Enhancing nuclear receptor-induced transcription requires nuclear motor and LSD1-dependent gene networking in interchromatin granules. Proc Natl Acad Sci. 2008;105:19199–19204. doi: 10.1073/pnas.0810634105. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 86.Lin C, et al. Nuclear receptor-induced chromosomal proximity and DNA breaks underlie specific translocations in cancer. Cell. 2009;139:1069–1083. doi: 10.1016/j.cell.2009.11.030. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 87.Kumaran RI, et al. Chromatin dynamics and gene positioning. Cell. 2008;132:929–934. doi: 10.1016/j.cell.2008.03.004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 88.Pestic-Dragovich L, et al. A myosin I isoform in the nucleus. Science. 2000;290:337–341. doi: 10.1126/science.290.5490.337. [DOI] [PubMed] [Google Scholar]
- 89.Grummt I. Actin and myosin as transcription factors. Curr Opin Genet Dev. 2006;16:191–196. doi: 10.1016/j.gde.2006.02.001. [DOI] [PubMed] [Google Scholar]
- 90.Pederson T. As functional nuclear actin comes into view, is it globular, filamentous, or both? J Cell Biol. 2008;180:1061–1064. doi: 10.1083/jcb.200709082. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 91.Mao YS, Yin HL. Regulation of the actin cytoskeleton by phosphatidylinositol 4-phosphate 5 kinases. Pflugers Arch. 2007;455:5–18. doi: 10.1007/s00424-007-0286-3. [DOI] [PubMed] [Google Scholar]
- 92.Mani RS, et al. Induced chromosomal proximity and gene fusions in prostate cancer. Science. 2009;326:1230. doi: 10.1126/science.1178124. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 93.Kocanova S, et al. Activation of estrogen-responsive genes does not require their nuclear co-localization. PLoS Genet. 2010;6:e1000922. doi: 10.1371/journal.pgen.1000922. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 94.Belmont AS. Estrogen fueled, nuclear kiss: Did it move for you? Nucleus. 2010;1:440–443. doi: 10.4161/nucl.1.5.13051. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 95.Simon JA, Kingston RE. Mechanisms of polycomb gene silencing: knowns and unknowns. Nat Rev Mol Cell Biol. 2009;10:697–708. doi: 10.1038/nrm2763. [DOI] [PubMed] [Google Scholar]
- 96.Kerppola TK. Polycomb group complexes--many combinations, many functions. Trends Cell Biol. 2009;19:692–704. doi: 10.1016/j.tcb.2009.10.001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 97.Muller J, Kassis JA. Polycomb response elements and targeting of Polycomb group proteins in Drosophila. Curr Opin Genet Dev. 2006;16:476–484. doi: 10.1016/j.gde.2006.08.005. [DOI] [PubMed] [Google Scholar]
- 98.Grimaud C, et al. RNAi components are required for nuclear clustering of Polycomb group response elements. Cell. 2006;124:957–971. doi: 10.1016/j.cell.2006.01.036. [DOI] [PubMed] [Google Scholar]
- 99.Lanzuolo C, et al. Polycomb response elements mediate the formation of chromosome higher-order structures in the bithorax complex. Nat Cell Biol. 2007;9:1167–1174. doi: 10.1038/ncb1637. [DOI] [PubMed] [Google Scholar]
- 100.Bantignies F, et al. Polycomb-Dependent Regulatory Contacts between Distant Hox Loci in Drosophila. Cell. 2011;144:214–226. doi: 10.1016/j.cell.2010.12.026. [DOI] [PubMed] [Google Scholar]
- 101.Bantignies F, et al. Inheritance of Polycomb-dependent chromosomal interactions in Drosophila. Genes Dev. 2003;17:2406–2420. doi: 10.1101/gad.269503. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 102.Saurin AJ, et al. The human polycomb group complex associates with pericentromeric heterochromatin to form a novel nuclear domain. J Cell Biol. 1998;142:887–898. doi: 10.1083/jcb.142.4.887. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 103.Sing A, et al. A vertebrate Polycomb response element governs segmentation of the posterior hindbrain. Cell. 2009;138:885–897. doi: 10.1016/j.cell.2009.08.020. [DOI] [PubMed] [Google Scholar]
- 104.Woo CJ, et al. A region of the human HOXD cluster that confers polycomb-group responsiveness. Cell. 2010;140:99–110. doi: 10.1016/j.cell.2009.12.022. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 105.Kagey MH, et al. The polycomb protein Pc2 is a SUMO E3. Cell. 2003;113:127–137. doi: 10.1016/s0092-8674(03)00159-4. [DOI] [PubMed] [Google Scholar]
- 106.MacPherson MJ, et al. The CTCF insulator protein is posttranslationally modified by SUMO. Mol Cell Biol. 2009;29:714–725. doi: 10.1128/MCB.00825-08. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 107.Zhang H, et al. The C. elegans Polycomb gene SOP-2 encodes an RNA binding protein. Mol Cell. 2004;14:841–847. doi: 10.1016/j.molcel.2004.06.001. [DOI] [PubMed] [Google Scholar]
- 108.Zhang H, et al. SUMO modification is required for in vivo Hox gene regulation by the Caenorhabditis elegans Polycomb group protein SOP-2. Nat Genet. 2004;36:507–511. doi: 10.1038/ng1336. [DOI] [PubMed] [Google Scholar]
- 109.Nunez E, et al. Nuclear organization in the 3D space of the nucleus - cause or consequence? Curr Opin Genet Dev. 2009;19:424–436. doi: 10.1016/j.gde.2009.07.005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 110.Fedorova E, et al. The nuclear organization of Polycomb/Trithorax group response elements in larval tissues of Drosophila melanogaster. Chromosome Res. 2008;16:649–673. doi: 10.1007/s10577-008-1218-6. [DOI] [PubMed] [Google Scholar]
- 111.Misteli T. Higher-order genome organization in human disease. Cold Spring Harb Perspect Biol. 2010;2:a000794. doi: 10.1101/cshperspect.a000794. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 112.Bass BL. RNA editing by adenosine deaminases that act on RNA. Annu Rev Biochem. 2002;71:817–846. doi: 10.1146/annurev.biochem.71.110601.135501. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 113.Rueter SM, et al. Regulation of alternative splicing by RNA editing. Nature. 1999;399:75–80. doi: 10.1038/19992. [DOI] [PubMed] [Google Scholar]
- 114.Chen LL, Carmichael GG. Gene regulation by SINES and inosines: biological consequences of A-to-I editing of Alu element inverted repeats. Cell Cycle. 2008;7:3294–3301. doi: 10.4161/cc.7.21.6927. [DOI] [PubMed] [Google Scholar]
- 115.Prasanth KV, et al. Regulating gene expression through RNA nuclear retention. Cell. 2005;123:249–263. doi: 10.1016/j.cell.2005.08.033. [DOI] [PubMed] [Google Scholar]
- 116.Hundley HA, et al. C. elegans and H. sapiens mRNAs with edited 3′ UTRs are present on polysomes. RNA. 2008;14:2050–2060. doi: 10.1261/rna.1165008. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 117.Kapranov P, et al. Genome-wide transcription and the implications for genomic organization. Nat Rev Genet. 2007;8:413–423. doi: 10.1038/nrg2083. [DOI] [PubMed] [Google Scholar]
- 118.Ponting CP, et al. Evolution and functions of long noncoding RNAs. Cell. 2009;136:629–641. doi: 10.1016/j.cell.2009.02.006. [DOI] [PubMed] [Google Scholar]
- 119.Mercer TR, et al. Long non-coding RNAs: insights into functions. Nat Rev Genet. 2009;10:155–159. doi: 10.1038/nrg2521. [DOI] [PubMed] [Google Scholar]
- 120.Wang X, et al. The Long Arm of Long Noncoding RNAs: Roles as Sensors Regulating Gene Transcriptional Programs. Cold Spring Harb Perspect Biol. 2010;3:a003756. doi: 10.1101/cshperspect.a003756. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 121.Wilusz JE, et al. Long noncoding RNAs: functional surprises from the RNA world. Genes Dev. 2009;23:1494–1504. doi: 10.1101/gad.1800909. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 122.Nagano T, Fraser P. No-Nonsense Functions for Long Noncoding RNAs. Cell. 2011;145:178–181. doi: 10.1016/j.cell.2011.03.014. [DOI] [PubMed] [Google Scholar]
- 123.McStay B. Nucleolar dominance: a model for rRNA gene silencing. Genes Dev. 2006;20:1207–1214. doi: 10.1101/gad.1436906. [DOI] [PubMed] [Google Scholar]
- 124.Pikaard CS. The epigenetics of nucleolar dominance. Trends Genet. 2000;16:495–500. doi: 10.1016/s0168-9525(00)02113-2. [DOI] [PubMed] [Google Scholar]
- 125.Tucker S, et al. Nucleolar dominance and ribosomal RNA gene silencing. Curr Opin Cell Biol. 2010;22:351–356. doi: 10.1016/j.ceb.2010.03.009. [DOI] [PMC free article] [PubMed] [Google Scholar]