Abstract
Molecular dynamics simulations and free energy calculation have been performed to study how the single-chain variable fragment (scFv) binds Methamphetamine (METH) and Amphetamine (AMP). The structures of the scFv:METH and the scFv:AMP complexes are analyzed by examining the time-dependence of their RMSDs, by analyzing the distance between some key atoms of the selected residues, and by comparing the averaged structures with their corresponding crystallographic structures. It is observed that binding an AMP to the scFv does not cause significant changes to the binding pocket of the scFv:ligand complex. The binding free energy of scFv:AMP without introducing an extra water into the binding pocket is much stronger than scFv:METH. This is against the first of the two scenarios postulated in the experimental work of Celikel et al. Protein Science 18, 2336 (2009). However, adding a water to the AMP (at the position of the methyl group of METH), the binding free energy of the scFv:AMP-H2O complex, is found to be significantly weaker than scFv:METH. This is consistent with the second of the two scenarios given by Celikel et al. Decomposition of the binding energy into ligand-residue pair interactions shows that two residues (Tyr175 and Tyr177) have nearly-zero interactions with AMP in the scFv:AMP-H2O complex, whereas their interactions with METH in the scFv:METH complex are as large as -0.8 and -0.74 kcal/mol. The insights gained from this study may be helpful in designing more potent antibodies in treating METH abuse.
Keywords: scFv, molecular dynamics simulation, binding free energy, MM-GBSA
Introduction
The abuse of methamphetamine - a potent and highly addictive psychostimulant - is a very serious problem in the United States and the world. The National Drug Intelligence Center reports that (t)-methamphetamine (METH) is the second major drug threat to the United States, only behind cocaine.[1] Current pharmacological therapies for the treatment of the adverse health effects of METH-like stimulants relieve some organ-based symptoms, but specific medications designed to treat direct medical complications of METH abuse are just developing. Active immunotherapy involves injection of a patient with a drug-like hapten conjugated to an antigenic carrier protein. This approach has produced promising results in early clinical trials for the treatment of nicotine and cocaine addiction.[2; 3] With medical treatment of METH as the ultimate goal, a novel single-chain variable fragment (scFv) against METH has been engineered from anti-METH monoclonal antibody mAb6H4 and found to have similar ligand affinity and specificity as mAb6H4.[4] Its crystallographic structure has been determined that sheds light on the binding mechanics of drug molecules. However, there are questions to be answered concerning the binding mechanisms of METH vs a similar drug, amphetamine (AMP).
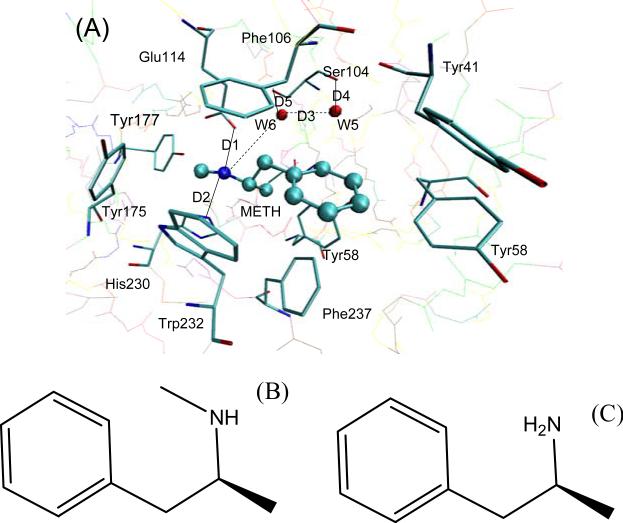
The scFv:METH complex is shown in Fig. 1 (A). The scFv consists of a variable light chain domain and a variable heavy chain domain, both possessing immunoglobulin fold. It has a deep pocket whose entrance is lined with seven aromatic residues, which encase 75% of the surface area of METH in a thermodynamically favorable arrangement. There are two water molecules near the bottom of the cavity.[5]
Fig. 1.
(A) The locations of the key residues two water molecules and the ligand in the scFv:METH complex. METH is displayed in a ball-and-stick representation, and the key residues are displayed in a stick representation. (B) Molecular structure of METH, and (C) Molecular structure of AMP.
The AMP molecule differs from METH only in the absence of the methyl group attached to the ammonium ion. The molecular structures of METH and AMP are shown in Fig. 1 (B) and (C). The aromatic ring of AMP and its ammonium ion could both participate in favorable interactions similar to that of the scFv:METH complex. Additionally, AMP is smaller in size than METH. It should not cause any steric hindrances when binding to scFv. However, there is a 4.55 kcal/mol difference in binding free energy between AMP and METH.[5] Celikel et al. postulated two scenarios for what may cause the weaker binding free energy of AMP: (1) The smaller molecule AMP has an overall shift in the ligand binding cavity: (2) A water molecule enters into the void space occupied by the methyl group of METH. [5]
Molecular dynamics (MD) simulations serve as a powerful tool for understanding mechanisms and dynamics of the protein-ligand complex. Several computational methods exist to estimate ligand binding affinities and selectivities, with various levels of computational expense and accuracy: [6] thermodynamic integration (TI), linear response (LR), free energy perturbation (FEP) [7; 8], fluctuation–dissipation theorem (FDT)[9; 10], and molecular mechanics generalized Born surface area (MM-GBSA). The MM-GBSA method is a fast and versatile tool for calculating the binding free energies of a given protein-ligand complex, which incorporates the effects of thermal averaging with a force field/continuum solvent models to post-process series of representative snapshots from MD trajectories. The MM-GBSA method has been successful in the recent studies of ligand binding interactions with multi-drug resistance. [11-17]
In this work, MD simulation and binding free energy calculations are performed to analyze the binding mechanics of the scFv:METH, scFv:AMP and scFv:AMP-H2O complexes. We are interested in the two scenarios postulated by Celikel et al. [5], and attempt to show which of the scenarios is more reasonable for the scFv:AMP binding mechanics.
Methods and experiment
MM/GBSA Calculations
In this work, the binding free energies are calculated using the MM-GBSA method supplied with the AMBER 10 package. We chose a total number of 300 snapshots evenly from the last 3 ns on the MD trajectory, at an interval of 10 ps. The MM-GBSA method can be conceptually summarized as:
| (1) |
| (2) |
| (3) |
| (4) |
| (5) |
where Gcomplex, Gprotein and Gligand are the free energies of the complex, the protein and the ligand, respectively. The binding free energy (ΔGbind) contains both an enthalpic (ΔH) and an entropic(-TΔS) contributions (Equation 2). The enthalpic(ΔH) component is composed of the gas-phase molecular mechanics energy (ΔGMM) and the solvation free energy (ΔGsolv). The gas-phase molecular mechanics energy (ΔGMM) is further divided into the noncovalent van der Waals component (ΔEvdW) and the electrostatic energies component (ΔEele) (Equation 3); and the solvation free energy (ΔGsolv) is further divided into a polar component (ΔGpol) and a nonpolar component (ΔGnonpol) (Equation 4). The polar component (ΔGpol) is calculated with a GB module of the AMBER suite. In our calculation, the dielectric constant is set to 1 inside the solute and 80 in solvent, respectively. The nonpolar component (ΔGnonpol) is determined by equation 5, where SASA is the solvent-accessible surface area that is determined with the MSMS program, using a probe radius of 1.4 Å. The values γ and β are empirical constants; In these calculations, the values of 0.00542 kcal/(mol·Å2) and 0.92 kcal/mol were used, respectively.
Finally, we approximated the conformational entropic contributions to the binding free energy by using the normal-mode analysis with the AMBER NMODE module. Because the entropic calculations for large systems are extremely time-consuming, 150 snapshots (every other snapshots of the 300 snapshots) for each system are used to estimate the contribution of the entropies to decrease the computational time. The complexes, proteins, and ligands are minimized with a distance-dependent dielectric constant, ε = 4Rij (Rij is the distance between two atom i and j).
Ligand-Residue Interaction Decomposition
The interactions between the ligand and each residue in scFv are analyzed using the MM-GBSA decomposition process applied in the MM-GBSA module of AMBER10.0. The binding interaction of each ligand-residue pair includes four terms: the van der Waals contribution (ΔGvdW), the electrostatic contribution (ΔGele), the polar solvation contribution (ΔGpol), and the nonpolar solvation contribution (ΔGninpol), respectively.
| (6) |
where ΔEvdW and ΔEele can be computed using the Sander program in AMBER10.0. The polar solvation contribution(ΔGpol )is calculated by using the generalized Born (GB) module, and the parameters for the GB calculation are developed by Onufriev et al. [18] ΔGnonpol is the non-polar contribution to solvation free energy. All energy components in Equation 6 are calculated using the same snapshots as used in the free energy calculation.
System Setups
Atomic coordinates of the scFv:METH complex are obtained from the Protein Data Bank (PDB) (ID:3GKZ). This crystallographic structure is the starting structure of the MD simulation of the scFv:METH system. However, the crystallographic structure of scFv:AMP complex is not available. Two “crystallographic” structures will be fabricated instead. As shown in Fig. 1, the AMP molecule differs from METH by the absence of the methyl group attached to the ammonium ion of METH. So one of the starting structures of the scFv complex with AMP (scFv:AMP) is obtained by the directly replacing METH of the scFv:METH complex with AMP, and the other structure (scFv:AMP-H2O) is obtained similarly except one water molecular is placed at the location of the methyl group of METH. This method has been proved to be useful in studying the binding model of ligands with the same chemical scaffold.[19] The missing residues are simply ignored, because they are all located far from the active site in the crystallographic structures. All crystallographically resolved water molecules are retained in the starting model. Protons are added to the system by using the Leap module of AMBER10.
Addressing the lack of parameters needed for the ligand in the Cornell et al. force field,[20] the missing parameters are developed. Optimization of the ligands are first carried out at the HF/6-31G** level with the Gaussian 03 package [21]. The Restrained Electrostatic Potential (RESP) procedure [22], which is also part of the AMBER package, is used to calculate the partial atomic charges. Each ligand has one positive charge. GAFF [23] force field parameters and RESP partial charges are assigned using the ANTECHAMBER module in the AMBER 10 package. The standard AMBER force field for bio-organic systems (FF03) [24] is used to describe the protein parameters. To neutralize the charge of the systems, an appropriate number of chloride counterions are placed to the grids with the largest positive Coulombic potentials around the complexes. All solutes are surrounded by a truncated, octahedron periodic box of TIP3P [25] water molecules extended to a distance of 10 Å from the solute atoms.
Molecular Dynamics Simulations
Three simulations are carried out using the AMBER 10 [26] package with the Cornell et al. all-atom force field and the parameters are developed in this work. The particle mesh Ewald (PME) method is used to treat long-range electrostatic interactions, and the bond lengths involving bonds to hydrogen atoms are constrained using the SHAKE algorithm [27]. The time-step for the three MD simulations is 2 fs, with a direct-space, non-bonded cutoff of 12 Å.
The three systems are minimized with the SANDER module in a constant volume by 1000 cycles of steepest descent minimization followed by 1000 cycles of conjugated gradient minimization. These procedures ensure that the initial structures are maintained while the solvent is allowed to relax. After energy minimization, and applying the harmonic restraints with force constants of 2 kcal/(mol·Å2) to all solute atoms, canonical ensemble (NVT)-MD is then carried out for 70 ps, during which the systems are heated from 0 K to 300 K. The subsequent isothermal isobaric ensemble (NPT)-MD is used for 90 ps to adjust the solvent density. Finally, the 5 ns isothermal isobaric ensemble (NPT)-MD simulation is applied to both simulations without any restraints. The temperature is regulated at 300 K using the Langevin thermostat and the pressure is kept at 1.0 atm using isotropic positional scaling. Trajectories are analyzed at every 1 ps using the PTRAJ module.
Results and discussion
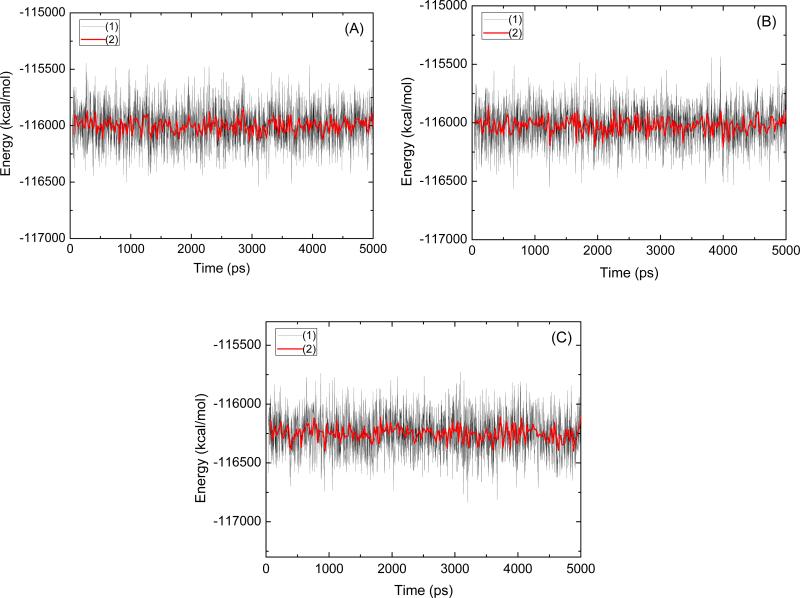
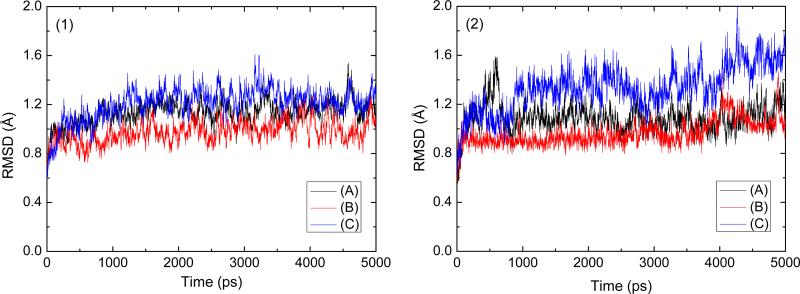
To assess the quality of our MD simulations, energetic and structural properties are monitored along the entire 5 ns MD trajectory of each complex. Fig. 2 is the plot of the potential energies of these three systems as a function of time. The fluctuations of potential energies are less than 1000 kcal/mol for the whole course. Fig. 3 (1) shows the RMSDs of the protein backbone atoms for these three complexes. The RMSDs increase at the beginning of the MD, then level off in less a 1.5 ns. The figures of potential energy and the backbone RMSDs indicate that the solvated systems have reached equilibrium after 1.5 ns MD simulation. The last 3 ns averaged backbone RMSDs for the scFv:METH, scFv:AMP, and scFv:AMP-H2O complexes are 1.16 Å, 1.02 Å, and 1.35 Å, respectively, this is an indication that the generated MD trajectories of these complexes are quite stable. The averaged RMSD value of the scFv:AMP system is smaller than other two. This indicates that AMP's lack of the methyl group, when compare to METH, does not apparently change the backbone conformation of scFv. In order to show whether there is an overall shift in the ligand position, the heavy atoms’ RMSDs of the residues within 5 Å of the ligand is analyzed (Fig. 3 (2)). For the ligand position, the smaller RMSDs of the scFv:AMP complex, as compared with the RMSDs of the scFv:METH complex, indicate that there is not an overall shift in the ligand position. The averaged RMSD of the last 3ns MD simulation in the scFv:AMP-H2O complex is bigger than the one for the scFv:METH complex. This indicates that the water molecule in place of the methyl group of METH causes some structural change. The information is also demonstrated by the heavy atoms’ RMSDs of the residues within 5 Å of the ligand as shown in Fig. 3 (2).
Fig. 2.
The potential energies of the scFv:METH (A), scFv:AMP (B) and scFv:AMP-H2O (c) complexes observed in MD simulation as a function of time. The solid black line (1), the solid red line (2) represents a 20 ps running average.
Fig. 3.
Root-mean-square deviations (RMSD) observed in MD simulations as a function of time; (1) all the backbone atoms and (2) all the residue within 5 Å of the ligand atoms, (A) for scFv:METH, (B) for scFv:AMP, and (C) for scFv:AMP-H2O complexes.
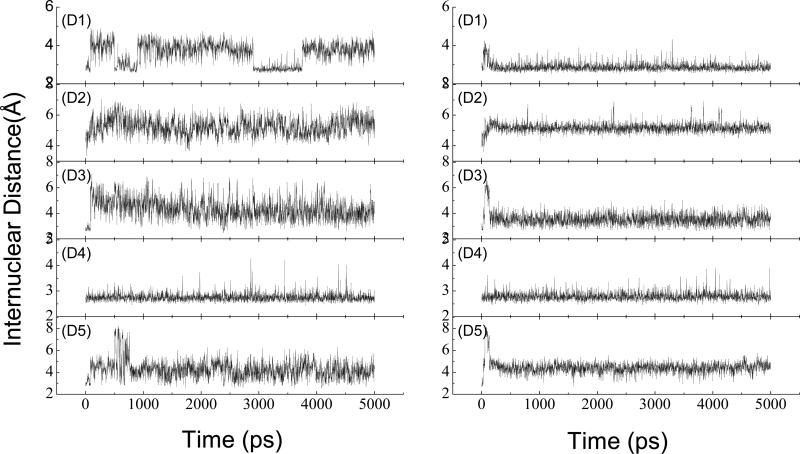
The crystallographic structure shows that there is a salt bridge between the ammonium ion of METH and the carboxyl oxygen of Glu114, and that there is a hydrogen bond between the ammonium ion of METH and the nitrogen atom of the His230 side chain. The salt bridge and the hydrogen bond are very important to anchoring METH in the pocket. The crystallographic structure also shows that there are two water molecules in the binding pocket, and the two water molecules form a hydrogen bond net with the nearby residues and METH. The selected internuclear distances versus the simulation time between atoms, as mentioned above, are illustrated in Fig. 4, and the atom name are defined as they are in the PDB files. D1 is the distance of the salt bridge. Note that the two oxygen atoms of Glu114 alternate in coming close to the N1 atom of METH in the scFv:METH complex. However, they are more fixed in the scFv:AMP complex as compared to the scFv:METH complex. D2 is the distance of the hydrogen bond formed between N1 of the ligand and the NE2 atom of His230; D3 is the distance of the oxygen atoms of the two water molecules; D4 is the distance between the oxygen atom of W5 and the oxygen of Ser104 main chain; and D5 is the distance between the oxygen atom of W6 and the side chain oxygen atom of Ser104. The fluctuations of D2, D3 and D5 in the scFv:METH complex are apparently bigger than in the scFv:AMP complex. For D4, there is no difference between the two complexes. The distances analysis suggests that there is not an overall structural shift in the ligand position in the scFv:AMP complex, and it seems that the interaction of AMP with scFv is stronger than the interaction of METH with scFv.
Fig. 4.
Selected distances as obtained from the MD simulation: the left for scFv:METH, the right for scFv:AMP. (D1) METH:N-Glu114:OE1, (D2) METH:N-His230:NE2, (D3)W5:O-W6:O, (D4) W5:O-Ser104:O, (D5) W6:O-Ser104:OG. The residue number corresponds to that in the X-ray structure.
The comparisons are carried out between the averaged structures of scFv:CETH as well as scFv:AMP complexes from the last 3 ns MD simulation and each starting sturctures. The averaged structures are very much in agreement with each starting structures; there is no significant change for the binding pocket residues and ligand. Note that the scFv:AMP complex shares the same starting scFv structure with the scFv:METH complex. This also indicates that the scFv:AMP complex is stable during the 5ns MD simulation, and that there are no large structural shifts for any of the pocket residues.
The MM-GBSA method has been used to calculate binding free energies. The calculated binding free energies (Table 1) are averaged from 300 snapshots, which are taken at even intervals from the last 3 ns of MD trajectories. The binding free energy of METH, AMP-H2O and AMP are -12.61, -8.30, and -16.94 kcal/mol, respectively. The absolute binding free energies have 1.63 and 1.87kcal/mol differences between the calculated and experimental values for the scFv:METH complex and scFv:AMP-H2O complex, respectively. The calculated relative binding free energy between scFv:MEHT and scFv:AMP-H2O is -4.31 kcal/mol which is in good agreement with the experimental result of -4.55 kcal/mol. However, for the scFv:AMP complex, the binding free energy is -16.94kcal/mol, which is much stronger than scFv:METH complex, in disagreement with the experimental results. We can conclude that the absence of the methyl group in AMP complex, relative to METH, causes a water molecule to enter into the cavity, thus decreasing the binding free energy. The electrostatic energy of METH is unfavorable by 35 kcal/mol than AMP because the polarity of METH is smaller than that of AMP. Permanent dipoles are 4.34 and 4.77 Debye for METH and AMP, respectively. The structure analysis shows that the water molecule is placed between the polar residue and the ligand in the scFv:AMP-H2O complex. The electrostatic screen caused by the additional water molecule is the reason for the smaller electrostatic energy of the scFv:AMP-H2O complex compare to the scFv:AMP complex.
Table 1.
Binding free energies computed by the MM-GBSA methoda
| Componentb | scFv:METH | scFv:AMP-H2O | scFv:AMP |
|---|---|---|---|
| Δ Eele | -107.14 ± 6.99 | -132.96 ± 6.43 | -142.15 ± 7.31 |
| Δ Evdw | -24.56 ± 2.23 | -20.39 ± 2.12 | -22.89 ± 2.31 |
| Δ Gnonpol | -3.67 ± 0.08 | -2.56 ± 0.07 | -3.23 ± 0.07 |
| Δ Gpol | 107.15 ± 5.95 | 132.21 ± 7.20 | 135.20 ± 6.05 |
| Δ Gele+pol | 0.01 | -0.75 | -6.95 |
| Δ Gvdw+nonpol | -28.23 | -22.95 | -26.12 |
| Δ H | -28.21 ± 2.13 | -23.70 ± 3.49 | -33.07 ± 2.89 |
| T Δ S | -15.60 ± 6.59 | -15.4 ± 4.88 | -16.13 ± 6.42 |
| Δ Gbind | -12.61 | -8.30 | -16.94 |
| Δ Gexpc | -10.98 | -6.43 | |
All values are given in kcal/mol, and the symbols are explained in the text.
Component: ΔEele: electrostatic energy in the gas phase; ΔEvdW: van der Waals energy; ΔGnonpol: non-polar solvation energy; ΔGpol: polar solvation energy; ΔGele+pol = ΔGele +ΔGpol; ΔGvdw+nonpol = ΔEvdw +ΔGnonpol; ΔH = ΔEele +ΔEvdW +ΔGnonpol +ΔGpol; ΔTS: total entropy contribution.
The experimental binding free energies are calculated using the equation ΔGexp= - RTlnKi.
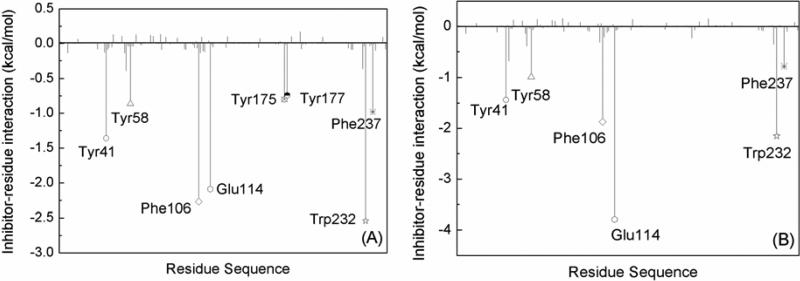
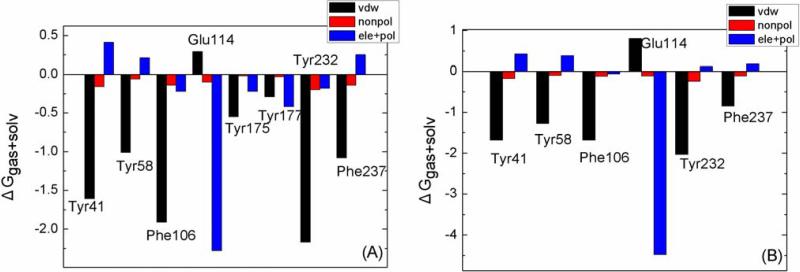
There is evidence that removing water mediated contacts, via introduction of function groups that replace the water, can increase binding in some cases [28; 29], while it can be unfavorable in others [30; 31]. Moreover, the environment surrounding the water molecule seems to play an important role. The binding free energies show that the water molecule inserted into the binding pocket is unfavorable for the binding between scFv and AMP. This result is consistent with the second scenario of Celikel et al. Further explaining the decline of binding free energy in greater detail, the binding free energy is decomposed into ligand-residue pairs for generating an ligand-residue interaction spectrum which is shown in Fig. 5. The decomposition approach is not only extremely useful to locate residues, which contribute to the protein-ligand interaction, but also helpful to elucidate the drug-resistant mechanism at the atomic level [16; 19; 32]. In this work, comparisons between the interaction spectrums of the scFv:METH complex and the scFv:AMP-H2O complex is performed. The labeled residues contribute larger energies. There is not a large difference for Tyr41, Tyr58, Phe106, Trp232 and Phe237 residues between the two complexes. Decomposing the free energy into different energy terms (Fig. 6) shows that the van de Waals interaction is the main driving force, and the nonpolar solvation interaction is favorable. For Glu114, the energy is apparently larger in the scFv:AMP-H2O complex than the scFv:METH complex (Fig. 5). The main energy term is electrostatic energy, and the increased energy mainly comes from the electrostatic energy (Fig. 6); the crystallographic structure shows that the ligand forms a hydrogen bond with the oxygen atom of side chain. The increase of the electrostatic energy indicates that the hydrogen bond is enhanced in the scFv:AMP-H2O complex relative to the scFv:METH complex. The hydrogen bond analysis also demonstrates that a hydrogen bond formed between the ammonium ion of the ligand and the oxygen atom of the Phe106 side chain. These results are also in agreement with the polar analysis of the ligand. The contributions of Tyr175 and Tyr177 residues disappear in the scFv:AMP-H2O complex, compared to the larger contribution in the scFv:MEHT complex. This is caused by the absent methyl group for the scFv:AMP-H2O complex, as shown in Fig. 1 (C); the methyl group mainly interacts with the Tyr175 and Tyr177 residues.
Fig. 5.
Decomposition of ΔGligand-residue on a per-residue basis for the (A) scFv:METH and (B) scFv:AMP-H2O complexes. The key residues with interaction energies larger than 0.7 kcal/mol are labeled.
Fig. 6.
Decomposition of ΔG on a per-residue basis for the key residues: (A) scFv:METH and (B) scFv:AMP-H2O.
Conclusion
In this work, 5 ns MD simulations have been carried out to investigate the dynamics and stability of scFv with METH and AMP, and to calculate the binding free energy using the MM-GBSA method. The RMSD, distances, and averaged structure analysis show that the absence of the methyl group in AMP, relative to METH, does not cause an overall shift of the ligand-binding cavity. The free energies between scFv and the ligands show that the scFv:AMP complex does not decrease the binding affinity. The structural and energetic analysis demonstrate that the scenario of scFv:AMP complex without an additional water is infeasible. However, the results of scFv:AMP-H2O complex are in agreement with the experimental data.
Decomposing the free energy into ligand-residue interaction pairs has also been carried out; the results indicate that there are eight and six residues with large contribution for the scFv:METH and the scFv:AMP-H2O complexes, respectively. The main force for Try41, Tyr58, Phe106, Tyr232, and Phe237 is van de Waals in both complexes, and for Glu114 it is the electrostatic interaction. The contributions of Tyr175 and Tyr177 are significant in scFv:METH but very small in scFv:AMP-H2O. The information obtained from this study can be helpful for designing potent antibodies to reduce possible antigenicity and to tailor more effective haptens for anti-METH vaccines.
Acknowledgement
The authors acknowledge support from a NIH SC3 grant (Grant No. GM084834), the UTSA Computational Biology Initiative, and the Texas Advanced Computing Center.
References
- 1.NDIC . National Drug Intelligence Center; Johnstown, PA: 2008. [Google Scholar]
- 2.Martell BA, Mitchell E, Poling J, Gonsai K, Kosten TR. Biol Psychiatry. 2005;58:158–164. doi: 10.1016/j.biopsych.2005.04.032. [DOI] [PubMed] [Google Scholar]
- 3.Cornuz J, Zwahlen S, Jungi WF, Osterwalder J, Klingler K, van Melle G, Bangala Y, Guessous I, Muller P, Willers J, Maurer P, Bachmann MF, Cerny T. PLoS ONE. 2008;3:e2547. doi: 10.1371/journal.pone.0002547. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Peterson Eric C., Laurenzana Elizabeth M., Atchley William T., HPH Owens SM. JPET. 2008;325:124–133. doi: 10.1124/jpet.107.134395. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Celikel R, Peterson EC, Owens SM, Varughese KI. Protein Sci. 2009;18:2336–2345. doi: 10.1002/pro.244. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Wang W, Donini O, Reyes CM, Kollman PA. Annu. ReV. Biophys Biomol. Struct. 2001;30:211–243. doi: 10.1146/annurev.biophys.30.1.211. [DOI] [PubMed] [Google Scholar]
- 7.Essex JW, Severance DL, Tirado-Rives J, Jorgensen WL. J. Phys. Chem. B. 1997;101:9663–9669. [Google Scholar]
- 8.Roux B, Nina M, Pomès R, Smith JC. Biophys J. 1996;71:670–681. doi: 10.1016/S0006-3495(96)79267-6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Chen LY. J. Chem. Phys. 2008;129:144113. doi: 10.1063/1.2992153. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Chen LY, Bastien DA, Espejel HE. Phys Chem Chem Phys. 2010;12:6579–6582. doi: 10.1039/b926889h. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Stoica I, Sadiq SK, Coveney PV. J. Am. Chem. Soc. 2008;130:2639–2648. doi: 10.1021/ja0779250. [DOI] [PubMed] [Google Scholar]
- 12.Hou TJ, Yu R. J. Med. Chem. 2007;50:1177. doi: 10.1021/jm0609162. [DOI] [PubMed] [Google Scholar]
- 13.Ode H, Matsuyama S, Hata M, Hoshino T, Kakizawa J, Sugiura W. J. Med. Chem. 2007. 2007;50:1768–1777. doi: 10.1021/jm061158i. [DOI] [PubMed] [Google Scholar]
- 14.Hu G-D, Zhu T, Zhang S-L, Wang D, Zhang Q-G. Eur J Med Chem. 2010;45:227–235. doi: 10.1016/j.ejmech.2009.09.048. [DOI] [PubMed] [Google Scholar]
- 15.Hu G, Wang D, Liu X, Zhang Q. J Comput Aided Mol Des. 2010;24:687–697. doi: 10.1007/s10822-010-9366-0. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Wu EL, Han KL, Zhang JZH. Chem Eur J. 2008;14:8704–8714. doi: 10.1002/chem.200800277. [DOI] [PubMed] [Google Scholar]
- 17.Chen J, Yang M, Hu G, Shi S, Yi C, Zhang Q. J Mol Model. 2009;15:1245–1252. doi: 10.1007/s00894-009-0452-y. [DOI] [PubMed] [Google Scholar]
- 18.Onufriev A, Bashford D, Case DA. J. Phys. Chem. B. 2000;104:3712–3720. [Google Scholar]
- 19.Rafi SB, Cui G, Song K, Cheng X, Tonge PJ, Simmerling C. J Med Chem. 2006;49:4574–4580. doi: 10.1021/jm060222t. [DOI] [PubMed] [Google Scholar]
- 20.Cornell WD, Cieplak P, Bayly CI, Gould IR, Merz KM, Ferguson DM, Spellmeyer DC, Fox T, Caldwell JW, Kollman PA. J. Am. Chem. Soc. 2002;117:5179–5197. [Google Scholar]
- 21.Frisch MJ, Trucks GW, Schlegel HB, Scuseria GE, Robb MA, Cheeseman JR, Montgomery JA, Jr., Vreven T, Kudin KN, Burant JC, Millam JM, Iyengar SS, Tomasi J, Barone V, Mennucci B, Cossi M, Scalmani G, Rega N, Petersson GA, Nakatsuji H, Hada M, Ehara M, Toyota K, Fukuda R, Hasegawa J, Ishida M, Nakajima T, Honda Y, Kitao O, Nakai H, Klene M, Li X, Knox JE, Hratchian HP, Cross JB, Bakken V, Adamo C, Jaramillo J, Gomperts R, Stratmann RE, Yazyev O, Austin AJ, Cammi R, Pomelli C, Ochterski JW, Ayala PY, Morokuma K, Voth GA, Salvador P, Dannenberg JJ, Zakrzewski VG, Dapprich S, Daniels AD, Strain MC, Farkas O, Malick DK, Rabuck AD, Raghavachari K, Foresman JB, Ortiz JV, Cui Q, Baboul AG, Clifford S, Cioslowski J, Stefanov BB, Liu G, Liashenko A, Piskorz P, Komaromi I, Martin RL, Fox DJ, Keith T, Al-Laham MA, Peng CY, Nanayakkara A, Challacombe M, Gill PMW, Johnson B, Chen W, Wong MW, Gonzalez C, Pople JA. Gaussian, Inc.; Wallingford CT: 2004. [Google Scholar]
- 22.Cieplak P, Cornell WD, Bayly C, Kollman PA. J. Comput. Chem. 1995;16:1357–1377. [Google Scholar]
- 23.Wang JM, Wolf RM, Caldwell JW, Kollman PA, Case DA. J. Comput. Chem. 2004;25:1157. doi: 10.1002/jcc.20035. [DOI] [PubMed] [Google Scholar]
- 24.Wang W, Kollman PA. J. Mol. Biol. 2000;303:567. doi: 10.1006/jmbi.2000.4057. [DOI] [PubMed] [Google Scholar]
- 25.Jorgensen WL, Chandrasekhar J, Madura JD, Impey RW, Klein ML. J. Chem. Phys. 1983;79:926. [Google Scholar]
- 26.Case DA, Darden TA, Cheatham TE, III, Simmerling CL, Wang J, Duke RE, Luo R, Merz KM, Pearlman DA, Crowley M, Walker RC, Zhang W, Wang B, Hayik S, Roitberg A, Seabra G, Wong KF, Paesani F, Wu X, Brozell S, Tsui V, Gohlke H, Yang L, Tan C, Mongan J, Hornak V, Cui G, Beroza P, Mathews DH, Schafmeister C, Ross WS, Kollman PA. University of California; San Francisco: 2006. [Google Scholar]
- 27.Ryckaert JP, Ciccotti G, Berendsen HJC. J. Comput. Phys. 1977;23:327. [Google Scholar]
- 28.Weber PC, Pantoliano MW, Simons DM, Salemme FR. J. Am. Chem. Soc. 1994;116:2717–2724. [Google Scholar]
- 29.Chen JM, Xu SL, Wawrzak Z, Basarab GS, Jordan DB. Biochemistry. 1998;37:17735–17744. doi: 10.1021/bi981848r. [DOI] [PubMed] [Google Scholar]
- 30.Clarke C, Woods RJ, Gluska J, Cooper A, Nutley MA, Boons G-J. J. Am. Chem. Soc. 2001;123:12238–12247. doi: 10.1021/ja004315q. [DOI] [PubMed] [Google Scholar]
- 31.Scott DS, Katherine AE, Michael AG, Milos VN, Martin JS. Protein Sci. 2005;14:249–256. [Google Scholar]
- 32.Hou TJ, Yu R. J Med Chem. 2007;50:1177–1188. doi: 10.1021/jm0609162. [DOI] [PubMed] [Google Scholar]