Abstract
Tarocystatin (CeCPI) from taro (Colocasia esculenta cv. Kaohsiung no. 1), a group-2 phytocystatin, shares a conserved N-terminal cystatin domain (NtD) with other phytocystatins but contains a C-terminal cystatin-like extension (CtE). The structure of the tarocystatin–papain complex and the domain interaction between NtD and CtE in tarocystatin have not been determined. We resolved the crystal structure of the phytocystatin–papain complex at resolution 2.03 Å. Surprisingly, the structure of the NtD–papain complex in a stoichiometry of 1:1 could be built, with no CtE observed. Only two remnant residues of CtE could be built in the structure of the CtE–papain complex. Therefore, CtE is easily digested by papain. To further characterize the interaction between NtD and CtE, three segments of tarocystatin, including the full-length (FL), NtD and CtE, were used to analyze the domain–domain interaction and the inhibition ability. The results from glutaraldehyde cross-linking and yeast two-hybrid assay indicated the existence of an intrinsic flexibility in the region linking NtD and CtE for most tarocystatin molecules. In the inhibition activity assay, the glutathione-S-transferase (GST)-fused FL showed the highest inhibition ability without residual peptidase activity, and GST-NtD and FL showed almost the same inhibition ability, which was higher than with NtD alone. On the basis of the structures, the linker flexibility and inhibition activity of tarocystatins, we propose that the overhangs from the cystatin domain may enhance the inhibition ability of the cystatin domain against papain.
Electronic supplementary material
The online version of this article (doi:10.1007/s00425-011-1398-8) contains supplementary material, which is available to authorized users.
Keywords: CeCPI, Cystatin, Cysteine protease inhibitor, Inhibition activity, Linker flexibility, Phytocystatin
Introduction
Tarocystatin (CeCPI), a cysteine protease inhibitor, was originally identified from the corm of taro (Colocasia esculenta cv. Kaohsiung no. 1), with its low pathogen susceptibility and high productivity (Yang and Yeh 2005). Tarocystatin is mainly expressed as one of the abundant proteins in the periderm of mature corm and the mature corm inside. Expression of protease inhibitors in the periderm may defend against underground nematode and fungus attack or serve as storage proteins in the corm (Yang and Yeh 2005; Wang et al. 2008).
Tarocystatin can inhibit the cysteine proteases and has been grouped in the cystatin superfamily. Recently, the inhibitory ability of cysteine protease inhibitors in plants was used to enhance the antipest and antifungus abilities of plants (Martinez et al. 2003, 2005; Aguiar et al. 2006; Christova et al. 2006; Goulet et al. 2008; Senthilkumar et al. 2010). Several lines of evidence support that cystatins in plants regulate the activity of cysteine protease for physiological and developmental processes in seed germination, organogenesis, and programed cell death (Kumar et al. 1999; Arai et al. 2002; Rivard et al. 2007; Valdes-Rodriguez et al. 2007; Martinez et al. 2009) and are involved in the complicated stress response to salt, drought, and oxidation (Diop et al. 2004; Zhang et al. 2008; Megdiche et al. 2009).
Cystatins tightly bind to and reversibly inhibit the activity of cysteine proteases such as the C1 papain family and C13 legumain family (Finn et al. 2008), with 1:1 stoichiometry. Most cystatins are composed of only 1 cystatin domain of about 100 residues in a molecular mass ranging from 12 to 16 kDa. Some cystatin proteins may contain several repetitive cystatin domains to form multicystatins (Rawlings and Barrett 1990). Each functional cystatin domain has three conserved motifs for interacting with target cysteine proteases: (1) the first major binding loop (L1) with QxVxG; (2) the second binding loop (L2) with a conserved aromatic residue, W or H; and (3) the N-terminal trunk with a conserved G. The three types of animal cystatin families include type-1 stefins composed of only the cystatin domain with nearly 100 residues that are neither disulfide bonds nor glycosylation sites; type-2 cystatins of secreted extracellular cystatin proteins with 120–130 residues that contain a signal peptide in the N terminus and 2 disulfide bonds in the C terminus; and type-3 kininogens of repetitive cystatin domain proteins ranging from 700 to 1,200 residues made of several glycosylated type-2 cystatins (Barrett 1986; Turk and Bode 1991; Turk et al. 2008; Kordis and Turk 2009).
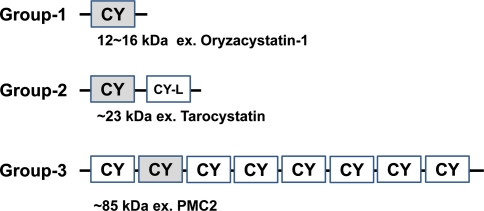
Cystatins in plants are distinctive with their specific conserved sequence [LVI]-[AGT]-[RKE]-[FY]-[AS]-[VI]-x-[EDQV]-[HYFQ]-N and are more closely related to type-2 cystatins than the other two animal cystatin classes. However, plant cystatins do not contain disulfide bonds and are similar to type-1 stefins. Because of the ambiguity, they should be grouped into a family of phytocystatins under the cystatin superfamily (Margis et al. 1998). From molecular evolutionary analysis, the phytocystatins can be further divided into three subgroups (Margis-Pinheiro et al. 2008): most phytocystatins belong to group-1 phytocystatins that contain only 1 cystatin domain with about 100 residues; group-2 phytocystatins (200–250 residues) share a highly conserved cystatin domain at the N terminus with an extended cystatin-like domain at the C terminus (the architecture of the dual domains of group-2 phytocystatins is distinctive in the cystatin superfamily); and group-3 phytocystatins are multicystatins with several repetitive cystatin domains (Fig. 1). In plants, the NMR structure of oryzacystatin-1 (OC-1) from rice (Oryza sativa) was the first available for group-1 phytocystatins (Nagata et al. 2000). Recently, a crystal structure of from potato (Solanum tuberosum) multicystatin 2 (PMC2) was resolved to provide new insights into the molecular functions of group-3 phytocystatins (Nissen et al. 2009). Until now, a few structures of human cystatins in complex with papain-like peptidases have been determined, such as the human stefin A–cathepsin H complex (Jenko et al. 2003) and human stefin B–papain complex (Stubbs et al. 1990). However, no structures of phytocystatins in complex with proteases have been resolved.
Fig. 1.
Three groups of phytocystatins. Phytocystatins can be divided into three groups on the basis of molecular mass: Group 1 is about 12–16 kDa and has one cystatin (CY) domain; group 2 is about 23 kDa and has one cystatin domain and an extended cystatin-like (CY-L) domain at the C terminus; group 3 is about 85 kDa and has several repetitive cystatin domains. The resolved cystatin domain structures are shaded in the gray box
Tarocystatin belongs to the group-2 phytocystatins and has 205 residues. The N-terminal cystatin domain (NtD; residues 1–98) has inhibitory ability against papain, and the roles of the C-terminal cystatin-like extension (CtE; residues 115–205) are controversial. Previous studies proposed that the CtE in group-2 phytocystatins might have possible roles including: (1) the CtE of tarocystatin showed weak papain activation properties and was proposed to possibly bind to papain (Wang et al. 2008); and (2) an SNSL motif in CtE of barley HvCPI-4 showed inhibitory activity against legumain (Martinez et al. 2007).
In this report, we describe the structures of the full-length (FL) and CtE of tarocystatin in complex with papain. Surprisingly, only the NtD was observed in the structure of the FL–papain complex and two residues of CtE remained in the structure of the CtE–papain complex. The structure of the tarocystatin–papain complex with an absent CtE in tarocystatin provides the structural insights to examine the binding mode of the cystatin domain against papain in plants. We also performed biochemical assays to identify the linker flexibility and inhibitory ability of tarocystatin against papain and found that an extra CtE of group-2 phytocystatins can increase the inhibitory ability of the cystatin domain in the N terminus.
Materials and methods
Protein expression and purification of recombinant tarocystatin
The genes (GenBank: AF525880; kindly provided by Prof. Kai-Wun Yeh, National Taiwan University) encoding full-length tarocystatin (residues 1–205) from taro [Colocasia esculenta (L.) Schott., cv. Kaohsiung no. 1], N-terminal cystatin (NtD, residues 1–102) domain, and the C-terminal extension (CtE, residues 103–205) were amplified by PCR from previous construction (Yang and Yeh 2005; Wang et al. 2008) with four primers: F1 primer: 5′-CGGGATCCATGGCCTTGATGGGGGGC-3′, R1 primer: 5′-CGGAATTCCTAATCTGCTGGCGTAACCGAGGAT-3′, F2 primer: 5′-CGGGATCCCTCGGTGTAAAACGGGATGCG-3′, R2: 5′-CGGAATTCCTAGTTTCCAGAGTCTGAATGATCTTGC-3′. The genes, such as FL by F1 and R2 primers, NtD by F1 and R1 primers, and CtE by F2 and R2 primers, were further cloned into the BamHI–EcoRI sites of the vector pGEX4T1 (GE Healthcare). The three expression vectors were transformed into E. coli BL21 (DE3) cells, cultured by LB medium (100 μg/ml Ampicillin) for an A600 of ~0.6 and then induced with 1 mM isopropyl-β-d-thiogalactopyranoside (IPTG) at 30°C for 4 h. The crude cell lysate suspended in phosphate buffered saline was loaded onto a GSTtrap FF column (GE Healthcare). The column containing glutathione-S-transferase (GST) binding protein was washed with an 8× column volume of wash buffer (1× PBS, 5 mM ATP, 10 mM MgSO4). For removing the GST-fusion protein, the column was loaded with 30 U of thrombin at room temperature overnight. Finally, the FL, NtD and CtE without GST-fusion proteins were eluted by PBS buffer. The quantification of the recombinant proteins involved use of a BioRad protein assay kit with bovine serum albumin used as a standard.
Crystallization and X-ray data collection
Crystallization of the complex protein was performed by the hanging-drop vapor diffusion method at room temperature. Lyophilized papain was purchased as the commercial papaya latex enzyme (2× crystallized, Sigma-Aldrich, St Louis, MO, USA).
The protein solution containing the FL tarocystatin and papain was prepared by mixing tarocystatin at 8 mg/ml with papain at 8 mg/ml in a 1:1 molar ratio. The complex of tarocystatin–papain was crystallized from a drop containing 15% polyethylene glycol monomethyl ether (PEG MME) 2000, 0.05 M sodium acetate trihydrate, pH 4.6, 0.1 M ammonium sulfate against a reservoir of 30% PEG MME 2000, 0.1 M sodium acetate trihydrate, pH 4.6, and 0.2 M ammonium sulfate (screening kit from Hampton Research Corp.). The crystals of the complex appeared after approximately 1 month and disappeared after 6 weeks.
For the crystallization of the CtE–papain complex, a mixture of protein solution was prepared of CtE at 4 mg/ml with papain at 8 mg/ml in a 1:1 molar ratio. The complex of CtE–papain was crystallized from a drop containing 0.05 M Hepes, pH 7.5, and 35% (v/v) (±)-2-methyl-2,4-pentanediol (MPD) against a reservoir of 0.1 M Hepes, pH 7.5, and 70% (v/v) MPD (screening kit from Hampton Research Corp.). After 15 days, crystals could be observed in the drop.
A mixture of the reservoir solution with 100% glycerol in a 4:1 volume ratio was used as cryo-protectant for data collection. X-ray diffraction data were collected at 100 K and detected by a Quantum210 CCD detector at the BL13C1 beamline at NSRRC (Hsinchu, Taiwan). Diffraction data were processed with HKL2000 (Otwinowski and Minor 1997), and diffraction statistics are listed in Table 1.
Table 1.
Crystallography statistics of the tarocystatin–papain complex
| Crystals | Tarocystatin–papain complex | CtE–papain complex |
|---|---|---|
| Beamline | NSRRC BL13C1 | NSRRC BL13C1 |
| Wavelength (Å) | 0.97315 | 0.97315 |
| Data collection and processing | ||
| Space group | P212121 | P31 |
| Cell dimensions a, b, c (Å) | 36.07, 99.81, 165.57 | 48.89, 48.89, 200.87 |
| Resolution (Å)a | 30–2.03 (2.10–2.03) | 30–2.56 (2.76–2.56) |
| Completeness (%)a | 99.9 (95.7) | 99.8 (99.5) |
 a
a
|
35.54 (3.13) | 46.56 (3.08) |
| Unique reflections | 39,814 | 17,482 |
| Total reflections | 389,065 | 1,261,120 |
| R a,bmerge | 10.0 (41.3) | 7.6 (51.7) |
| Refinement | ||
| Resolution range (Å) | 30.0–2.03 | 30.0–2.6 |
| No. of reflections (working/test) | 33,886/3,804 | 13,417/1,476 |
| R-factor/R free (%) | 18.2/23.3 | 21.3/26.7 |
| Protein atoms | 4,656 | 3,350 |
| Solvent atoms | 576 | 171 |
| Model quality | ||
| RMS deviations in | ||
| Bond length (Å) | 0.012 | 0.007 |
| Bond angles (°) | 1.7 | 1.2 |
| Average B-factor (Å2) | ||
| Protein atoms | 23.6 | 59.5 |
| Solvent atoms | 33.6 | 52.8 |
| Ramachandran plot (%) | ||
| Most favored | 88.1 | 82.0 |
| Additionally allowed | 11.5 | 18.0 |
| Generously allowed | 0.2 | 0 |
| Disallowed | 0.2 | 0 |
aThe last shell values are indicated in parentheses
b
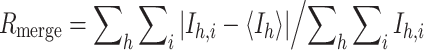 where
where 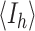 is the mean intensity of i observations for a given reflection h
is the mean intensity of i observations for a given reflection h
Structure determination and refinement
The FL–papain and CtE–papain complex structures were determined by the molecular replacement procedure in the program CNS (Brunger et al. 1998) with the structure of papain (PDB ID:1PPN) used as the search model. For the FL–papain complex structure, two obvious rotational and translational solutions for the papain structure were identified. The initial rigid body refinement of papain molecules gave an R factor of 41.1%. The clear continuous electron density for tarocystatin was given by Fourier maps, and the structure of tarocystatin was further built accordingly. For the CtE–papain complex structure, two rotational and two translational solutions for the papain structure showed distinct values from the other solutions. The rigid body refinement of papain in the CNS program gave a lower R factor, 29.6%.
Manual model rebuilding was subsequently performed by use of Coot (Emsley and Cowtan 2004), alternating with structure refinement by the CNS program, with 10% of the observed reflections randomly selected and set aside for calculation of the R free value. The final refined statistics are listed in Table 1. All molecular representations involved use of PyMOL (DeLano 2009). The structure of the FL–papain complex was submitted to the PISA server for analyzing the protein interface (Krissinel and Henrick 2007).
Protease activity assay of papain
The purified proteins from FL, NtD, and CtE of tarocystatin (250 pmol) were mixed with the nonactivated papain (250 pmol) in a final volume of 10 μl, respectively. All protein mixtures were incubated at room temperature for 24 h. Reactions were stopped by adding 1 μl of stop solution (62.5 mM Tris–HCl, pH 6.8, 5% SDS, 8.3% β-mercaptoethanol, 10% glycerol, 0.01% bromophenol blue) and heating to 100°C. Finally, 15% SDS-PAGE was used for electrophoretic analysis. Protein molecular weight markers were purchased from GE Healthcase.
Glutaraldehyde (GA) cross-linking and yeast two-hybrid assay
The purified proteins (1 μg) alone or paired were cross-linked with 0–0.01% (v/v) GA at room temperature, and the mixture was incubated in 5 μl of 50 mM PBS (pH 8.6) for 10 min at 37°C. The reaction was terminated by adding 1 μl of stop solution. Finally, the products were separated by SDS-PAGE (10% w/v polyacrylamide) and visualized by Coomassie brilliant blue staining.
The GAL4 yeast two-hybrid system (Clontech Co.) was used to analyze the protein–protein interaction. The paired genes were cloned into a pGBT9(+2) vector containing the GAL4 DNA binding domain or a pACT vector containing the GAL4 activation domain. The recombinant pGBT9 (+2) and pACT vectors were cotransformed into yeast host cells (AH109). The cotransformation of the above two vectors was checked by use of selection medium (SD/-Trp/-Leu) and examined by PCR. More restricted selection medium (SD/-Trp/-Leu/-His/+ 5 mM 3-AT) was used to incubate the yeast containing the recombinant vectors for 7 days at 30°C.
In-gel inhibitory activity assay
Inhibitory activity assay of tarocystatin involved SDS-PAGE (Michaud et al. 1996). A mixture of purified tarocystatin and 3 pmol activated papain (in a buffer with 5 mM l-cysteine) in a total volume of 10 μl was incubated at 37°C for 15 min and then mixed with the mild denaturing buffer (31.25 mM Tris–HCl, pH 6.8, 1% SDS, 1% sucrose, 0.005% bromophenol blue). The protein mixture was further subjected to 10% SDS-PAGE containing 0.25% gelatin. After electrophoresis, the gels were transferred to a solution with 2.5% v/v Triton X-100 for 30 min at room temperature for recovering the activity of papain and then incubated in an active buffer (100 mM sodium phosphate, pH 6.8, containing 8 mM EDTA, 10 mM l-cysteine and 0.2% Triton X-100) for 75 min at 37°C. The remained protease activity was observed as white bands with a blue background after Coomassie brilliant blue staining. The quantification of the bands involved use of the program Dolphin-1D (Wealtec Corp. USA).
PDB accession numbers
All coordinates and structural factors have been deposited in the Protein Data Bank with accession nos. 3IMA for the FL–papain complex and 3LFY for the CtE–papain complex.
Results
Overall structure of tarocystatin–papain complex
The protease-inhibitor mixture of the FL tarocystatin with papain was prepared for cocrystallization in a 1:1 molar ratio. The crystals of the FL–papain complex were grown after 1 month and formed an orthorhombic space group of P212121 with cell dimension a = 36.06 Å, b = 99.72 Å, c = 165.59 Å, α = β = γ = 90° (Table 1). The structure of the FL–papain complex was further resolved by the molecular replacement method, with the crystal structure of papain used as a search model (PDB ID: 1PPN). Two peak values were given in the cross rotation function and translation and gave a 41.1% R-factor by initial rigid body refinement. In the stage of refinement, a continuous electron density near the papain molecule was observed. Therefore, the structure of tarocystatin was built according to the continuous electron density. After model rebuilding and iterative refinement, the complex structure gave an 18.1% R-factor and 23.2% R free at resolution 2.03 Å (Table 1).
From the refined structure of the FL–papain complex, only the NtD of tarocystatin in complex with papain was built (Fig. 2a). The NtD–papain complex showed two structures in an asymmetric unit, including two papain molecules (chains A and C: residues 1–212), two NtDs of tarocystatin molecules (chain B: residues 2–9 and residues 16–91; chain D: residues 2–9 and residues 16–92), three acetic acid molecules and 576 water molecules. The cystatin folding of NtD was similar to that of other phytocystatins, such as OC-1 and PMC-2, with five antiparallel β-sheets wrapped up in a central α-helix. From the N terminus, one short β-sheet is from Ile7 to Val8, and the following α-helix is from residues Ala17 to Glu34. Four antiparallel longer β-sheets consisted of residues Gln39 to Val50, residues Ile54 to Glu64, residues Lys67 to Gln78, and residues Ser83 to Ser90. In addition, residues Ala79 to Asn82 in the loop 2 are shown to be 3/10 α-helix. The electron density of the residues from Val10 to Asn15 was broken, so models for these residues could not be defined.
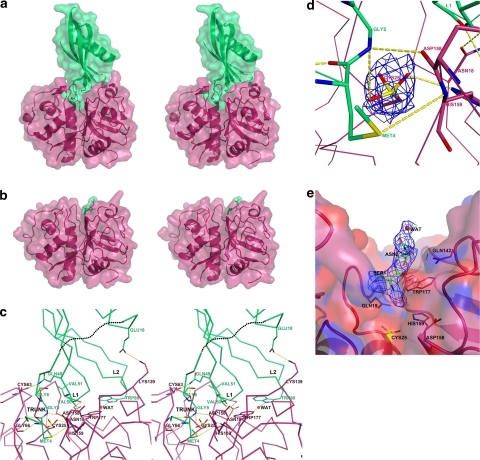
Fig. 2.
Crystal structures of tarocystatin–papain complex. a Stereo view of the structure of the tarocystatin–papain complex. The remaining NtD of tarocystatin is in lime green and papain is in warm pink. b The remnant residues of CtE are in lime green and papain is in warm pink. c Hydrogen bonding networks between tarocystatin and papain. Nine hydrogen bonds are highlighted by yellow dashed lines between NtD and papain (chain B to chain A): Met4 (N) to Gly66 (O), Met4 (O) to Gly66 (N), Met4 (SD) to His159 (N), Gly5 (N) to Asp158 (O), Gly5 (N) to OCS25 (OD2), Gln49 (NE2) to Cys63 (O), Val51(O) to Trp177 (NE1), Ser52 (N) to Gly20 (O), Ser52 (OG) to Asn18 (OD1). One water-mediated hydrogen bond is Trp80 (NE1) to Trp177 (O) and one salt bridge is Glu18 (OE2) to Lys139 (NZ). The residues from 10 to 15 of tarocystatin are not observed and are represented by black dots. d The 1σ of 2Fo-Fc electron density map for the oxidized sulfhydryl group of Cys25 of papain. e The dipeptide, Ser-Asn, of CtE is contoured by 1σ of the 2Fo-Fc electron density map
The structure of the chain A (papain) and chain B (NtD) complex is almost identical to that of chain C (papain) and chain D (NtD), with a root mean square deviation (RMSD) of 0.39 Å. The buried surface between NtD and papain is 2,080 Å2. Nine hydrogen bonds exist in the interface (chain B to chain A): Met4 (N) to Gly66 (O), Met4 (O) to Gly66 (N), Met4 (SD) to His159 (N), Gly5 (N) to Asp158 (O), Gly5 (N) to OCS25 (OD2), Gln49 (NE2) to Cys63 (O), Val51(O) to Trp177 (NE1), Ser52 (N) to Gly20 (O), Ser52 (OG) to Asn18 (OD1). One salt bridge is Glu18 (OE2) to Lys139 (NZ) and one water-mediated hydrogen bond is Trp80 (NE1) to Trp177 (O) (Fig. 2c). Cys25 in the papain structure is oxidized with three oxygen atoms to γS, which can be observed by clear electron density (Fig. 2d). The Matthews coefficient (Matthews 1968) of two NtD–papain complexes in per asymmetric unit is 2.21 Å3/dalton and the solvent content is 44.43% (Kantardjieff and Rupp 2003). On examining the molecular packing of the NtD–papain complex, no more space is left for CtE in the unit cell.
Overall structure of the remnant residues of CtE–papain complex
The CtE from residues 103–205 was co-crystallized with papain for testing the binding between CtE and papain. The CtE and papain mixture was also prepared by a 1:1 molar ratio concentration. The crystals of the CtE–papain complex were grown after 15 days and formed in a trigonal space group of P31 with cell dimension a = 48.89 Å, b = 48.89 Å, c = 200.87 Å, α = β = 90° and γ = 120° (Table 1). The structure of CtE–papain was resolved by a molecular replacement method with the structure of papain used as the search template (PDB ID: 1PPN). In the initial stage of rigid body refinement, we obtained a low R-factor, 29.6%, which showed that only two papain structures with the remnant residues of CtE could be built (Fig. 2b). The structure of the CtE–papain complex gave a 21.3% R-factor and 26.7% R free at resolution 2.6 Å after model building and refinement (Table 1). The remnant residues of CtE are a dipeptide of Ser-Asn (Fig. 2e). The Matthews coefficient of the two remnant residues of the CtE–papain complex in per asymmetric unit is 2.96 Å3/dalton and the solvent content is 58.4%. Thus, the structure of the remnant residues of CtE reveals that CtE is easily digested by papain.
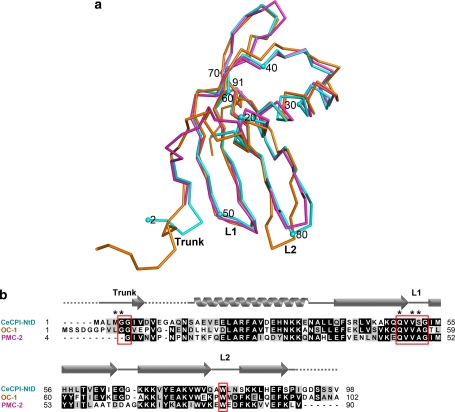
Structural comparison of NtD with OC-I and PMC-2
Until now, only two structures of cystatins from plants have been resolved. One is the OC-I (PDB ID: 1EQK) from rice, a member of group-1 phytocystatins, which was first resolved by NMR in 2000 (Nagata et al. 2000). The other structure is PMC-2 (PDB ID: 2W9Q), a domain from multicystatins in potato, which was resolved by X-ray crystallography at resolution 2.5 Å (Nissen et al. 2009). Comparing the difference in NtD with these two structures can provide the structure differences and the binding mode between phytocystatins and papain. The superimposition of the Cα atoms of NtD, OC-1 and PMC2 gave an average RMSD of 1.35 Å and 0.87 Å, respectively (Fig. 3a). The RMSD of the L1 loop QxVxG between NtD and OC-1 is 2.52 Å, and that of NtD and PMC2 is 0.68 Å. The RMSD of the L2 loop residue W between NtD and OC-1 is 4.17 Å, and that of NtD and PMC2 is 2.10 Å. The sequence alignment of NtD, OC-I, and PMC-2 revealed the similarity of the cystatin domain, except that the loop of residues 10–15 of NtD is highly flexible (Fig. 3b), which could explain why the electron density in the region is broken.
Fig. 3.
Structure comparison and sequence alignment of NtD of tarocystatin with OC-1 and PMC-2. a NtD of tarocystatin (CeCPI-NtD), OC-1, and PMC-2 are represented by cyan, orange, and magenta, respectively. The superimposition of the Cα atoms of NtD of tarocystatin, OC-1 and PMC2 gives an average RMS difference of 1.35 and 0.87 Å, respectively. The structure numbers indicate the residue number of NtD, and the conserved motifs are labeled by Trunk, L1 and L2. b The sequence alignment of CeCPI-NtD, OC-1, and PMC-2 reveals the similarity of the cystatin domain, except the loop of residues 10–15 of NtD is a highly flexible region. The residues of NtD interacting with papain by hydrogen bonding are labeled by asterisks. The secondary structure located above the sequence alignment is extracted from the structure of NtD, and the unresolved region is represented by dashed lines
The CtE of tarocystatin is easily digested by papain
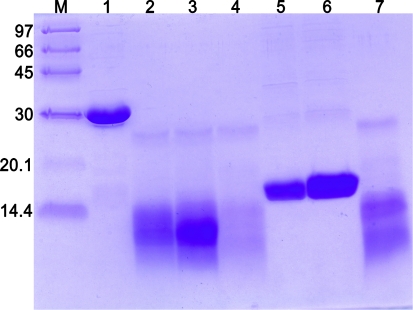
For confirming, the CtE of tarocystatin was digested by nonactivated papain to mimic crystallization condition. The FL, N-terminal fragment (NtD; residues 1–102) and the C-terminal extension (CtE; residues 103–205) were prepared. The proteins FL, NtD, and CtE were, respectively, mixed with nonactivated papain and incubated for 24 h. The results from SDS-PAGE showed that FL was digested by papain and remained as a cystatin domain about a 9 kDa band (Fig. 4, lane 2). A similar band could be observed by mixing NtD with papain (Fig. 4, lane 3). In the CtE mixed with papain, no proteins were left (Fig. 4, lane 4). The results further indicated that CtE is easily digested by papain.
Fig. 4.
Protease activity assay of papain. FL, NtD and CtE represent the full-length, N-terminal domain, and C-terminal extension of tarocystatin, respectively. The proteins from tarocystatin mixed with/without nonactivated papain were analyzed on 15% SDS-PAGE. Lane 1 FL only, lane 2 FL mixed with papain and remained about a band of 9 kDa, lane 3 NtD mixed with papain and remained a 9 kDa band, lane 4 CtE mixed with papain and no proteins left, lane 5 NtD only, lane 6 CtE only, lane 7 papain only, lane M standard protein marker
Linker flexibility in tarocystatin identified by interaction analysis with glutaraldehyde cross-linking and yeast two-hybrid assays
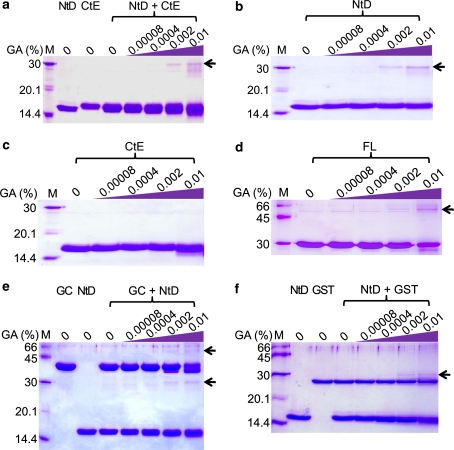
The results from biochemical analysis and structures of the NtD–papain and CtE–papain complexes suggested that the CtE is easily digested by papain. We proposed that the interaction between NtD and CtE might be weak. To examine the domain–domain interaction, we performed GA cross-linking and yeast two-hybrid assays. The CtE could be defined as containing a cystatin-like domain (residues 115–205) by a psiBLAST of the NCBI database (Wang et al. 2008).
Figure 5 shows the purity of the constructs (lanes without GA) and the results of the interaction of different segments by GA cross-linking assay. NtD and CtE could be cross-linked by >0.002% (v/v) GA (Fig. 5a), but most of the NtD and CtE belonged to a free form without interaction. Figure 5b shows the interaction of NtD and NtD cross-linked by >0.002% (v/v) GA, and Fig. 5c shows no interactions between CtE and CtE. To further confirm the interaction of NtD and NtD in the FL protein, the FL protein was treated with GA. A band shift of the interaction between NtD and NtD was observed with >0.002% (v/v) GA (Fig. 5d). However, the interaction of NtD and CtE in Fig. 5a could be the result of two possible interactions: (1) NtD and NtD or (2) NtD and CtE. A GST-fused CtE (GCtE) was prepared to interact with NtD. Two bands could be observed (Fig. 5e), the lower band representing the NtD–NtD interaction (about 30 kDa) and the upper band representing the GCtE–NtD interaction (about 60 kDa). Finally, GST and NtD were used as a negative control to exclude the interaction of GST and NtD. Only the NtD–NtD interaction could be observed (Fig. 5f).
Fig. 5.
Glutaraldehyde (GA) cross-linking analysis of NtD and CtE. The purified proteins FL, NtD and CtE were cross-linking by glutaraldehyde and resolved on 10% SDS-PAGE. The shifts of protein band are indicated by the arrow. Lane M standard protein marker. a NtD and CtE were cross-linked by >0.002% GA. b NtD and NtD were cross-linked by >0.002% GA. c CtE and CtE could not be cross-linked by GA. d FL and FL were cross-linked by >0.002% GA. e The interaction between GST-fused CtE (GCtE) and NtD was further confirmed. The lower band indicates the interaction of NtD–NtD, and the higher band indicates the interaction of GCtE–NtD. f The results confirm the interaction between NtD and NtD but not NtD and GST
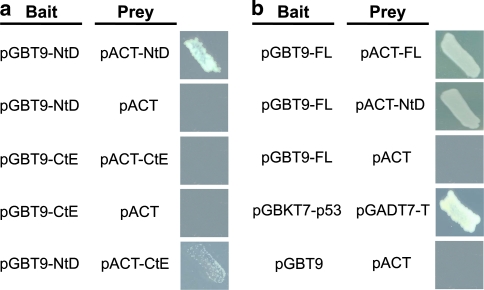
The domain–domain interaction of NtD, CtE and FL was further examined by yeast two-hybrid assay. The NtD–NtD interaction was identified with the constructions of the bait pGBT9-NtD and the prey pACT-NtD observed in the survival of yeast. A weak interaction of NtD–CtE was observed with a few yeast colonies. No interaction was found for the CtE–CtE interaction (pGBT9-CtE/pACT-CtE) and for the negative control constructions of pGBT9-NtD/pACT and pGBT9-CtE/pACT (Fig. 6a). An additional interaction of FL–FL and FL–NtD was shown with the interaction between pGBT9-FL/pACT-FL and pGBT9-FL/pACT-NtD. A positive control (pGBKT7-p53/pGADTT7-T) and two negative controls (pGBT9-FL/pACT, and pGBT9/pACT) were used to confirm the specific interaction between NtD and NtD, and NtD and CtE (Fig. 6b).
Fig. 6.
Domain–domain interaction by yeast two-hybrid assay. The bait represents the vector containing the GAL4 DNA binding domain, and the prey represents the vector containing the GAL4 activation domain. a The protein–protein interaction of NtD and NtD could be identified in the pGBT9-NtD and pACT-NtD constructs, whereas the constructs of pGBT9-NtD/pACT and pGBT9/pACT-NtD confirmed that the interaction of NtD and NtD was not caused by autoactivation. Lack of yeast grown with the construction of pGBT9-CtE and pACT-CtE showed that no interactions could be found between CtE and CtE. However, a few yeast colonies in pGBT9-NtD and pACT-CtE showed a weak interaction between NtD and CtE. b The constructs of NtD, CtE and FL were subcloned into pGBT9 and pACT vectors. An interaction could be observed in FL and FL, and NtD and FL. The constructs of pGBT9-FL/pACT and pGBT9/pACT-FL also confirmed that the interaction of FL was not caused by autoactivation. pGBKT7-p53/pGADT7-T and pGBT9/pACT served as the positive and negative control, respectively. Yeast was incubated in the SD/-Trp/-Leu/-His selection medium containing 5 mM 3-AT
Inhibition ability with different combinations of tarocystatin
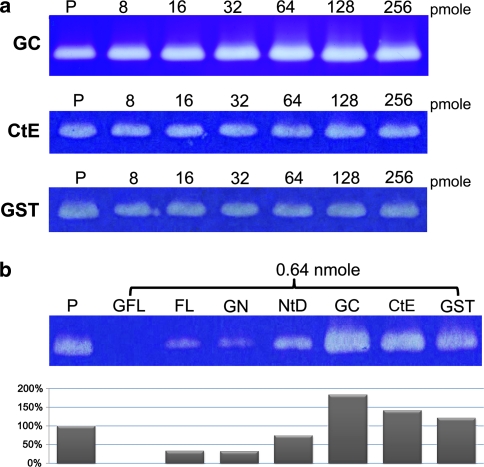
The inhibition ability of cystatin was characterized by an in-gel inhibition assay as described (Michaud et al. 1996). Wang et al. (2008) used an in-gel inhibition assay and demonstrated that CtE shows a weak activation property for papain but with GCtE. Here, we present a more detailed examination to observe the different inhibition abilities of tarocystatin against papain. All inhibition ability assays entailed 3 pmol papain activated by l-cysteine. We prepared several different combinations to monitor the inhibition ability: GST-fused FL (GFL; GST-NtD-CtE construction), FL (FL; NtD-CtE), GST-fused NtD (GNtD), NtD, GCtE, CtE, and GST. Figure 7a shows a serial concentration of 8–256 pmol of GCtE, CtE, and GST used to test the enhanced activity of papain. GCtE showed greater papain activity at >32 pmol, in contrast to CtE and GST with a little enhanced protease ability at >64 pmol.
Fig. 7.
In-gel inhibitory activity assay with different segments from tarocystatin. The brightness of the band indicates the protease activity of papain to digest the substrate gelatin and the brighter band represents the higher residual activity of protease. All inhibitory activity assays entailed 3 pmol papain. Lane P the positive control indicating the papain activity only. a With increased concentration, GCtE shows the more digestive ability of papain from 64 pmol. CtE and GST show only a little elevated protease activity. b The inhibition ability of different combinations of tarocystatin. The inhibition ability was GFL > FL > GNtD > NtD. GCtE shows the highest enhanced capacity, whereas CtE and GST show little enhanced capacity. Lane P represents the recovered activity of papain of 100%. Percentages indicate the recovered activity of papain relative to lane P
As compared with the inhibitory activity of these different segments, the inhibitory abilities of GFL, FL, GNtD, and NtD against papain were from high to low (Fig. 7b). GCtE showed a brighter band than those of CtE and GST. Therefore, the inhibition ability of the cystatin domain could be enhanced by an extra N-terminal domain (GNtD) or an extra C-terminal domain (FL by NtD-CtE) or both (GFL by GST-NtD-CtE) (Fig. 7b). From the quantification of the bands in the gel, the density of the band of lane P represented 100% recovered papain activity. The percentage of the other bands could be calculated in comparison with lane P; lane GFL indicates that the papain activity was totally inhibited. The remaining percentages are 33% for FL, 32% for GNtD and 75% for NtD, for an inhibitory ability of GFL > FL > GNtD > NtD. However, GCtE produced enhanced papain activity, with 183%, CtE with 141%, and GST with 121%, for enhanced ability of GCtE > CtE > GST. The Ki values of NtD and FL were further determined as 2.33 × 10−8 and 3.59 × 10−8 M, respectively. These values are similar to that ones previously reported (Wang et al. 2008).
Discussion
Several structures of the cystatin superfamily in animals have been determined, such as chicken egg-white cystatin (Bode et al. 1988; Dieckmann et al. 1993), human stefin A (Martin et al. 1995; Tate et al. 1995), tetrameric structure of human stefin B (Jenko Kokalj et al. 2007) and domain swapping structure of human stefin C (Janowski et al. 2005). However, the structures of cystatin in complex with proteases such as papain are rare. The activated form of papain can digest most proteins, even protease inhibitors (Alphey and Hunter 2006). To date, only a few structures of cystatin in complexes with proteases have been resolved, such as stefin B with carboxymethylated papain in a deactivated form (Stubbs et al. 1990), and stefin A with cathepsin H (Jenko et al. 2003). Thus, the structures of inhibitor–protease complexes are difficult to obtain. We attempted to cocrystallize tarocystatin and papain and obtained crystals of the tarocystatin–papain complex. The crystals could be observed after 1-month treatment but disappeared after 6 weeks.
The lyophilized papain, which was dissolved in phosphate buffered saline (PBS) buffer without being activated by l-cysteine, was cocrystallized with tarocystatin. We assumed that the nonactivated papain might have no protease activity. To our surprise, only the cystatin domain (NtD) in complex with papain was observed, and the CtE was digested by the residual activity of papain. The digestion of CtE was further confirmed in Fig. 4. Thus, nonactivated papain still contained a little protease activity in the absence of l-cysteine activation. We further checked the activity of nonactivated papain and found that the residual activity was about 0.04% that of activation (Supplemental data Fig. S1). The residual protease activity is lower than that of carboxymethylated papain (~0.5% residual activity) (Stubbs et al. 1990). This is a clue to explain for why we could not resolve the structure of the full-length tarocystatin in complex with papain. In the other words, we might not be able to determine the complex structure by X-ray crystallography if the tarocystatin lack the CtE (Engh et al. 1993). In future study, the carboxymethylated papain, for deactivated residual protease activity, will be used to cocrystallize with tarocystatin to explore the overall structure of tarocystatin, because we could not crystallize tarocystatin alone for a long period.
Because few complex structures are available for comparison, we can only compare the structures of stefin B–papain and tarocystatin–papain. The RMSD between tarocystatin and stefin B is 1.9 Å, and the papains in these two complex structures are superimposed. Stefin B and tarocystatin have a similar binding mode to inhibit protease activity, although tarocystatin contains a CtE that was absent in the structure of the complex. From our results of the structures of tarocystatin–papain and CtE–papain, we propose that (1) the CtE of tarocystatin might be easily bound and digested by papain; (2) the interaction between NtD and CtE might be weak; or (3) the extended domain for the cystatin domain might increase its inhibition ability against papain.
The domain organization of tarocystatin has been predicted to be cystatin (NtD) and cystatin-like (CtE) domains. The architecture is similar to that of latexin, an endogenous protein inhibitor found in the rat brain (Pallares et al. 2005). The C-terminal subdomain of latexin has inhibition ability against carboxypeptidase A4. The N-terminal domain of latexin with folding of one α-helix and four β-sheets is similar to that of the C-terminal subdomain, which lacks inhibition ability. The overall structure of latexin can be stabilized by intramolecular interaction to form a funnel shape. Therefore, we performed GA cross-linking and yeast two-hybrid assays to examine the domain interaction in tarocystatin. The results of GA cross-linking revealed rare intramolecular interaction between NtD and CtE. The intermolecular interaction was weak from only a little amount of interaction between NtD and NtD. Therefore, most tarocystatin molecules might exist in a highly flexible form without domain–domain interaction, which explains why the crystals of full-length tarocystatin were difficult to obtain in our previous crystal screening conditions. The minor interaction between NtD and CtE and between NtD and NtD was further identified by yeast two-hybrid assay. The results were similar to the GA cross-linking assay, with the intermolecular interaction being higher between NtD and NtD than between NtD and CtE. The linker flexibility of tarocystatin differs from that of latexin, which exhibits intramolecular interaction. The flexibility of tarocystatin would increase the possibility of interaction against papain and might provide more efficient inhibition ability than the cystatin domain alone. However, the function of a small amount of intramolecular or intermolecular interaction is still unclear, especially, whether the domain interactions are involved in the regulation of inhibition ability of tarocystatin in vivo. More experiments are needed to confirm the possibilities.
CtE in the group-2 phytocystatins would provide higher inhibition ability (Wang et al. 2008). This result could explain the existence of a CtE in group-2 phytocystatins, which might be involved in the “arm race” between plants and pathogens (Christeller 2005) for enhancing the inhibition ability of cystatin. Here, we used several combinations of different segments to examine the inhibition ability of NtD which would be enhanced at the N terminus and at the C terminus by an extra domain and both. The inhibition ability of NtD, a cystatin domain resembling group-1 phytocystatins, represented the standard inhibitor. Adjoining a new domain at the N or C terminus of the cystatin domain, such as GNtD (GST-NtD) or FL (NtD-CtE), conferred almost the same inhibition ability, which was higher than that of NtD (cystatin domain only). GFL with the GST-NtD-CtE combination showed the highest inhibition ability (Fig. 7b). The GST protein was treated as a different domain from the cystatin domain in the inhibition ability assay for comparing with the CtE. The GST and CtE showed almost the same ability to enhance the protease activity of papain (Fig. 7a). In contrast to the inhibition ability, the combination of GCtE conferred a higher protease activity of papain than that with GST or CtE alone (Fig. 7a, b).
In the GA cross-linking assay, no domain interaction between GST and NtD or between GST and CtE could be observed (Fig. 5e, f). Therefore the domains GST, NtD and CtE remained flexible without domain interaction. Combined with the results from the biochemical assay and structures of the FL–papain and CtE–papain complexes, we propose that an extra domain from the cystatin domain, either at the N or C terminus, might serve as a bait to attract papain. When the extra domain is digested by papain, the cystatin domain would easily bind to and inhibit the activity of papain. The phenomenon might provide an explanation for the evolutionary tendency from a single cystatin domain (group-1 phytocystatins) and double cystatin domain (group-2 phytocystatins) toward a multicystatin domain (group-3 phytocystatin) for different inhibition ability. Results of in-gel inhibitory assay also provide a clue for designing new strategies for antipest or antifungus purposes.
In conclusion, group-2 phytocystatins are the different cystatins in plants containing a conserved cystatin domain in the N terminus and a cystatin-like domain in the extended C terminus. In this study, we resolved the structure of the phytocystatin–papain complex, which showed tarocystatin without the CtE. As well, the tarocystatin was digested by papain and remained as two residues in the structure of the CtE–papain complex. From the structures of the complexes, we provide structural information for group-2 phytocystatins against papain. The limited interaction between NtD and CtE was further confirmed by GA cross-linking and yeast two-hybrid assays and demonstrates its flexible nature. The linker flexibility between NtD and CtE might increase the inhibition role of the NtD against papain, which was confirmed by inhibition activity assay with combinations of various domains.
Electronic supplementary material
Below is the link to the electronic supplementary material.
Acknowledgments
The authors thank the supporting stuffs to help the X-ray data collection at the beamline BL13C1 of National Synchrotron Radiation Center (NSRRC), a national user facility supported by the National Science Council of Taiwan, ROC. The Synchrotron Radiation Protein Crystallography Facility is supported by the National Research Program for Genomic Medicine. Financial support of this work was from National Science Council, Taiwan to Y.S. Cheng (NSC. 96-2311-B-002-002).
Open Access
This article is distributed under the terms of the Creative Commons Attribution Noncommercial License which permits any noncommercial use, distribution, and reproduction in any medium, provided the original author(s) and source are credited.
Abbreviations
- NtD
N-terminal cystatin domain of tarocystatin
- CtE
C-terminal cystatin-like extension of tarocystatin
- FL
Full-length tarocystatin
- GST
Glutathione-S-transferase
References
- Aguiar JM, Franco OL, Rigden DJ, Bloch C, Jr, Monteiro AC, Flores VM, Jacinto T, Xavier-Filho J, Oliveira AE, Grossi-de-Sa MF, Fernandes KV. Molecular modeling and inhibitory activity of cowpea cystatin against bean bruchid pests. Proteins. 2006;63:662–670. doi: 10.1002/prot.20901. [DOI] [PubMed] [Google Scholar]
- Alphey MS, Hunter WN. High-resolution complex of papain with remnants of a cysteine protease inhibitor derived from Trypanosoma brucei. Acta Crystallogr Sect F Struct Biol Cryst Commun. 2006;62:504–508. doi: 10.1107/S1744309106014849. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Arai S, Matsumoto I, Emori Y, Abe K. Plant seed cystatins and their target enzymes of endogenous and exogenous origin. J Agric Food Chem. 2002;50:6612–6617. doi: 10.1021/jf0201935. [DOI] [PubMed] [Google Scholar]
- Barrett AJ. The cystatins: a diverse superfamily of cysteine peptidase inhibitors. Biomed Biochim Acta. 1986;45:1363–1374. [PubMed] [Google Scholar]
- Bode W, Engh R, Musil D, Thiele U, Huber R, Karshikov A, Brzin J, Kos J, Turk V. The 2.0 Å X-ray crystal structure of chicken egg white cystatin and its possible mode of interaction with cysteine proteinases. EMBO J. 1988;7:2593–2599. doi: 10.1002/j.1460-2075.1988.tb03109.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Brunger AT, Adams PD, Clore GM, DeLano WL, Gros P, Grosse-Kunstleve RW, Jiang JS, Kuszewski J, Nilges M, Pannu NS, Read RJ, Rice LM, Simonson T, Warren GL. Crystallography & NMR system: a new software suite for macromolecular structure determination. Acta Crystallogr D Biol Crystallogr. 1998;54:905–921. doi: 10.1107/S0907444998003254. [DOI] [PubMed] [Google Scholar]
- Christeller JT. Evolutionary mechanisms acting on proteinase inhibitor variability. FEBS J. 2005;272:5710–5722. doi: 10.1111/j.1742-4658.2005.04975.x. [DOI] [PubMed] [Google Scholar]
- Christova PK, Christov NK, Imai R. A cold inducible multidomain cystatin from winter wheat inhibits growth of the snow mold fungus, Microdochium nivale. Planta. 2006;223:1207–1218. doi: 10.1007/s00425-005-0169-9. [DOI] [PubMed] [Google Scholar]
- DeLano WL (2009) The PyMOL molecular graphics system. DeLano Scientific LLC, Palo Alto. http://www.pymol.org
- Dieckmann T, Mitschang L, Hofmann M, Kos J, Turk V, Auerswald EA, Jaenicke R, Oschkinat H. The structures of native phosphorylated chicken cystatin and of a recombinant unphosphorylated variant in solution. J Mol Biol. 1993;234:1048–1059. doi: 10.1006/jmbi.1993.1658. [DOI] [PubMed] [Google Scholar]
- Diop NN, Kidric M, Repellin A, Gareil M, d’Arcy-Lameta A, Pham Thi AT, Zuily-Fodil Y. A multicystatin is induced by drought-stress in cowpea (Vigna unguiculata (L.) Walp.) leaves. FEBS Lett. 2004;577:545–550. doi: 10.1016/j.febslet.2004.10.014. [DOI] [PubMed] [Google Scholar]
- Emsley P, Cowtan K. Coot: model-building tools for molecular graphics. Acta Crystallogr D Biol Crystallogr. 2004;60:2126–2132. doi: 10.1107/S0907444904019158. [DOI] [PubMed] [Google Scholar]
- Engh RA, Dieckmann T, Bode W, Auerswald EA, Turk V, Huber R, Oschkinat H. Conformational variability of chicken cystatin. Comparison of structures determined by X-ray diffraction and NMR spectroscopy. J Mol Biol. 1993;234:1060–1069. doi: 10.1006/jmbi.1993.1659. [DOI] [PubMed] [Google Scholar]
- Finn RD, Tate J, Mistry J, Coggill PC, Sammut SJ, Hotz HR, Ceric G, Forslund K, Eddy SR, Sonnhammer EL, Bateman A. The Pfam protein families database. Nucleic Acids Res. 2008;36:D281–D288. doi: 10.1093/nar/gkm960. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Goulet MC, Dallaire C, Vaillancourt LP, Khalf M, Badri AM, Preradov A, Duceppe MO, Goulet C, Cloutier C, Michaud D. Tailoring the specificity of a plant cystatin toward herbivorous insect digestive cysteine proteases by single mutations at positively selected amino acid sites. Plant Physiol. 2008;146:1010–1019. doi: 10.1104/pp.108.115741. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Janowski R, Kozak M, Abrahamson M, Grubb A, Jaskolski M. 3D domain-swapped human cystatin C with amyloidlike intermolecular beta-sheets. Proteins. 2005;61:570–578. doi: 10.1002/prot.20633. [DOI] [PubMed] [Google Scholar]
- Jenko Kokalj S, Guncar G, Stern I, Morgan G, Rabzelj S, Kenig M, Staniforth RA, Waltho JP, Zerovnik E, Turk D. Essential role of proline isomerization in stefin B tetramer formation. J Mol Biol. 2007;366:1569–1579. doi: 10.1016/j.jmb.2006.12.025. [DOI] [PubMed] [Google Scholar]
- Jenko S, Dolenc I, Guncar G, Dobersek A, Podobnik M, Turk D. Crystal structure of Stefin A in complex with cathepsin H: N-terminal residues of inhibitors can adapt to the active sites of endo- and exopeptidases. J Mol Biol. 2003;326:875–885. doi: 10.1016/S0022-2836(02)01432-8. [DOI] [PubMed] [Google Scholar]
- Kantardjieff KA, Rupp B. Matthews coefficient probabilities: improved estimates for unit cell contents of proteins, DNA, and protein-nucleic acid complex crystals. Protein Sci. 2003;12:1865–1871. doi: 10.1110/ps.0350503. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kordis D, Turk V. Phylogenomic analysis of the cystatin superfamily in eukaryotes and prokaryotes. BMC Evol Biol. 2009;9:266. doi: 10.1186/1471-2148-9-266. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Krissinel E, Henrick K. Inference of macromolecular assemblies from crystalline state. J Mol Biol. 2007;372:774–797. doi: 10.1016/j.jmb.2007.05.022. [DOI] [PubMed] [Google Scholar]
- Kumar GN, Houtz RL, Knowles NR. Age-induced protein modifications and increased proteolysis in potato seed-tubers. Plant Physiol. 1999;119:89–100. doi: 10.1104/pp.119.1.89. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Margis R, Reis EM, Villeret V. Structural and phylogenetic relationships among plant and animal cystatins. Arch Biochem Biophys. 1998;359:24–30. doi: 10.1006/abbi.1998.0875. [DOI] [PubMed] [Google Scholar]
- Margis-Pinheiro M, Zolet AC, Loss G, Pasquali G, Margis R. Molecular evolution and diversification of plant cysteine proteinase inhibitors: new insights after the poplar genome. Mol Phylogenet Evol. 2008;49:349–355. doi: 10.1016/j.ympev.2008.04.025. [DOI] [PubMed] [Google Scholar]
- Martin JR, Craven CJ, Jerala R, Kroon-Zitko L, Zerovnik E, Turk V, Waltho JP. The three-dimensional solution structure of human stefin A. J Mol Biol. 1995;246:331–343. doi: 10.1006/jmbi.1994.0088. [DOI] [PubMed] [Google Scholar]
- Martinez M, Lopez-Solanilla E, Rodriguez-Palenzuela P, Carbonero P, Diaz I. Inhibition of plant-pathogenic fungi by the barley cystatin Hv-CPI (gene Icy) is not associated with its cysteine-proteinase inhibitory properties. Mol Plant Microbe Interact. 2003;16:876–883. doi: 10.1094/MPMI.2003.16.10.876. [DOI] [PubMed] [Google Scholar]
- Martinez M, Abraham Z, Gambardella M, Echaide M, Carbonero P, Diaz I (2005) The strawberry gene Cyf1 encodes a phytocystatin with antifungal properties. J Exp Bot 56:1821–1829 [DOI] [PubMed]
- Martinez M, Diaz-Mendoza M, Carrillo L, Diaz I. Carboxy terminal extended phytocystatins are bifunctional inhibitors of papain and legumain cysteine proteinases. FEBS Lett. 2007;581:2914–2918. doi: 10.1016/j.febslet.2007.05.042. [DOI] [PubMed] [Google Scholar]
- Martinez M, Cambra I, Carrillo L, Diaz-Mendoza M, Diaz I. Characterization of the entire cystatin gene family in barley and their target cathepsin L-like cysteine-proteases, partners in the hordein mobilization during seed germination. Plant Physiol. 2009;151:1531–1545. doi: 10.1104/pp.109.146019. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Matthews BW. Solvent content of protein crystals. J Mol Biol. 1968;33:491–497. doi: 10.1016/0022-2836(68)90205-2. [DOI] [PubMed] [Google Scholar]
- Megdiche W, Passaquet C, Zourrig W, Zuily Fodil Y, Abdelly C. Molecular cloning and characterization of novel cystatin gene in leaves Cakile maritima halophyte. J Plant Physiol. 2009;166:739–749. doi: 10.1016/j.jplph.2008.09.012. [DOI] [PubMed] [Google Scholar]
- Michaud D, Cantin L, Raworth DA, Vrain TC. Assessing the stability of cystatin/cysteine proteinase complexes using mildly-denaturing gelatin-polyacrylamide gel electrophoresis. Electrophoresis. 1996;17:74–79. doi: 10.1002/elps.1150170113. [DOI] [PubMed] [Google Scholar]
- Nagata K, Kudo N, Abe K, Arai S, Tanokura M. Three-dimensional solution structure of oryzacystatin-I, a cysteine proteinase inhibitor of the rice, Oryza sativa L. japonica. Biochemistry. 2000;39:14753–14760. doi: 10.1021/bi0006971. [DOI] [PubMed] [Google Scholar]
- Nissen MS, Kumar GN, Youn B, Knowles DB, Lam KS, Ballinger WJ, Knowles NR, Kang C. Characterization of Solanum tuberosum multicystatin and its structural comparison with other cystatins. Plant Cell. 2009;21:861–875. doi: 10.1105/tpc.108.064717. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Otwinowski Z, Minor W. Processing of X-ray diffraction data collected in oscillation mode. Methods Enzymol. 1997;276:307–326. doi: 10.1016/S0076-6879(97)76066-X. [DOI] [PubMed] [Google Scholar]
- Pallares I, Bonet R, Garcia-Castellanos R, Ventura S, Aviles FX, Vendrell J, Gomis-Ruth FX. Structure of human carboxypeptidase A4 with its endogenous protein inhibitor, latexin. Proc Natl Acad Sci USA. 2005;102:3978–3983. doi: 10.1073/pnas.0500678102. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rawlings ND, Barrett AJ. Evolution of proteins of the cystatin superfamily. J Mol Evol. 1990;30:60–71. doi: 10.1007/BF02102453. [DOI] [PubMed] [Google Scholar]
- Rivard D, Girard C, Anguenot R, Vezina LP, Trepanier S, Michaud D. MsCYS1, a developmentally-regulated cystatin from alfalfa. Plant Physiol Biochem. 2007;45:508–514. doi: 10.1016/j.plaphy.2007.03.028. [DOI] [PubMed] [Google Scholar]
- Senthilkumar R, Cheng CP, Yeh KW. Genetically pyramiding protease-inhibitor genes for dual broad-spectrum resistance against insect and phytopathogens in transgenic tobacco. Plant Biotechnol J. 2010;8:65–75. doi: 10.1111/j.1467-7652.2009.00466.x. [DOI] [PubMed] [Google Scholar]
- Stubbs MT, Laber B, Bode W, Huber R, Jerala R, Lenarcic B, Turk V. The refined 2.4 Å X-ray crystal structure of recombinant human stefin B in complex with the cysteine proteinase papain: a novel type of proteinase inhibitor interaction. EMBO J. 1990;9:1939–1947. doi: 10.1002/j.1460-2075.1990.tb08321.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tate S, Ushioda T, Utsunomiya-Tate N, Shibuya K, Ohyama Y, Nakano Y, Kaji H, Inagaki F, Samejima T, Kainosho M. Solution structure of a human cystatin A variant, cystatin A2-98 M65L, by NMR spectroscopy. A possible role of the interactions between the N- and C-termini to maintain the inhibitory active form of cystatin A. Biochemistry. 1995;34:14637–14648. doi: 10.1021/bi00045a004. [DOI] [PubMed] [Google Scholar]
- Turk V, Bode W. The cystatins: protein inhibitors of cysteine proteinases. FEBS Lett. 1991;285:213–219. doi: 10.1016/0014-5793(91)80804-C. [DOI] [PubMed] [Google Scholar]
- Turk V, Stoka V, Turk D. Cystatins: biochemical and structural properties, and medical relevance. Front Biosci. 2008;13:5406–5420. doi: 10.2741/3089. [DOI] [PubMed] [Google Scholar]
- Valdes-Rodriguez S, Guerrero-Rangel A, Melgoza-Villagomez C, Chagolla-Lopez A, Delgado-Vargas F, Martinez-Gallardo N, Sanchez-Hernandez C, Delano-Frier J. Cloning of a cDNA encoding a cystatin from grain amaranth (Amaranthus hypochondriacus) showing a tissue-specific expression that is modified by germination and abiotic stress. Plant Physiol Biochem. 2007;45:790–798. doi: 10.1016/j.plaphy.2007.07.007. [DOI] [PubMed] [Google Scholar]
- Wang KM, Kumar S, Cheng YS, Venkatagiri S, Yang AH, Yeh KW. Characterization of inhibitory mechanism and antifungal activity between group-1 and group-2 phytocystatins from taro (Colocasia esculenta) FEBS J. 2008;275:4980–4989. doi: 10.1111/j.1742-4658.2008.06631.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yang AH, Yeh KW. Molecular cloning, recombinant gene expression, and antifungal activity of cystatin from taro (Colocasia esculenta cv. Kaosiung no. 1) Planta. 2005;221:493–501. doi: 10.1007/s00425-004-1462-8. [DOI] [PubMed] [Google Scholar]
- Zhang X, Liu S, Takano T. Two cysteine proteinase inhibitors from Arabidopsis thaliana, AtCYSa and AtCYSb, increasing the salt, drought, oxidation and cold tolerance. Plant Mol Biol. 2008;68:131–143. doi: 10.1007/s11103-008-9357-x. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.