Abstract
Centromere-drive is a process where centromeres compete for transmission through asymmetric "female" meiosis for inclusion into the oocyte. In symmetric "male" meiosis, all meiotic products form viable germ cells. Therefore, the primary incentive for centromere-drive, a potential transmission bias, is believed to be missing from male meiosis. In this article, we consider whether male meiosis also bears the primary cost of centromere-drive. Because different taxa carry out different combinations of meiotic programs (symmetric + asymmetric, symmetric only, asymmetric only), it is possible to consider the evolutionary consequences of centromere-drive in the context of these differing systems. Groups with both types of meiosis have large, rapidly evolving centromeric regions, and their centromeric histones (CenH3s) have been shown to evolve under positive selection, suggesting roles as suppressors of centromere-drive. In contrast, taxa with only symmetric male meiosis have shown no evidence of positive selection in their centromeric histones. In this article, we present the first evolutionary analysis of centromeric histones in ciliated protozoans, a group that only undergoes asymmetric "female" meiosis. We find no evidence of positive selection acting on CNA1, the CenH3 of Tetrahymena species. Cytological observations of a panel of Tetrahymena species are consistent with dynamic karyotype evolution in this lineage. Our findings suggest that defects in male meiosis, and not mitosis or female meiosis, are the primary selective force behind centromere-drive suppression. Our study raises the possibility that taxa like ciliates, with only female meiosis, may therefore undergo unsuppressed centromere drive.
Electronic supplementary material
The online version of this article (doi:10.1007/s00239-011-9449-0) contains supplementary material, which is available to authorized users.
Keywords: Tetrahymena thermophila, Positive selection, Phylogeny, Asymmetric female meiosis, Karyotype
Introduction
Centromeres serve as attachment points to mitotic and meiotic spindles and mediate the faithful segregation of all eukaryotic chromosomes. While the chromosome segregation apparatus is evolutionarily conserved, centromeric DNA and surrounding heterochromatin are rapidly evolving, and can range in size and complexity over several orders of magnitude. This rapid evolution, in spite of conserved function, has been termed the "centromere paradox" (Henikoff et al. 2001). We have suggested that "centromere-drive" can provide a general explanation for this apparent paradox. Under the "centromere-drive" model, centromeres compete via microtubule attachments for preferential transmission in female meiosis in animals and plants, since only one of four meiotic products becomes the egg. This competition confers a selfish advantage to chromosomes that make more microtubule attachments, and can result in runaway expansions of centromeric satellite, and rapid fixations of satellite DNA that are more competitive at recruiting centromeric proteins (Henikoff et al. 2001; Malik 2009).
While beneficial to the "driving" chromosome, centromere-drive, or female meiotic transmission distortion in general, can have deleterious effects on the fitness of an organism. For instance, in human populations, Robertsonian fusions (fusions of acrocentric chromosomes at centromeres) are preferentially transmitted through females (Pardo-Manuel de Villena and Sapienza 2001b), but male carriers of Robertsonian fusions are partially or completely sterile (Daniel 2002). Similarly, homozygosity for a driving centromeric locus in the plant Mimulus guttatus leads to a reduction in pollen (male gamete) counts (Fishman and Saunders 2008; Fishman and Willis 2005). Therefore, deleterious consequences of centromere-drive have only been shown so far in male meiosis or gametogenesis. This may be a consequence if there is a strict requirement for balanced meiosis and equitable recruitment of centromeric proteins to produce fertile male gametes. One way to counter such deleterious consequences of "centromere-drive" is the evolution of suppressors, acting to restore meiotic parity (Henikoff et al. 2001; Malik and Henikoff 2009; Talbert et al. 2008).
Previous studies of the genes encoding proteins essential for meiosis have revealed that many of them show patterns of positive selection. For instance, in three taxa containing meioses with both symmetric retention of meiotic products (hereafter referred to as male meiosis) and asymmetric retention of meiotic products (hereafter referred to as female meiosis), i.e., Drosophila, primates and plants, centromeric histone (CenH3) genes have been shown to evolve under positive selection (Cooper and Henikoff 2004; Malik and Henikoff 2001; Schueler et al. 2010; Talbert et al. 2002). These observations of independent episodes of positive selection on genes, which are essential for chromosome segregation in highly diverged lineages, support the hypothesis that they are involved in recurrent genetic conflict. In addition, other centromeric proteins (e.g. CENP-C) as well as heterochromatin proteins that may also bind satellite DNAs and potentially curb centromere-drive (Bayes and Malik 2009), have been found to evolve under positive selection (Axelsson et al. 2010; Talbert et al. 2008; Talbert et al. 2004). The "centromere-drive" model thus provides a simple rationale to explain the complexity of plant and animal centromeres and for the rapid evolution of their centromeric proteins.
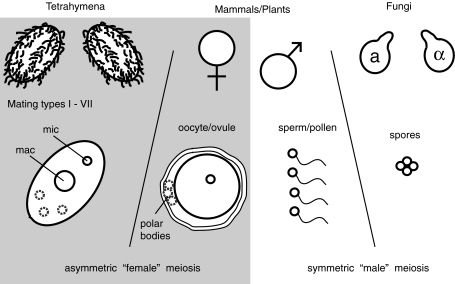
Examining taxa that carry out different complements of meiosis (female and/or male meioses) allow us to test the correlation between the meiotic programs and the evolutionary rates of centromeric DNA and proteins (Fig. 1). For instance, fungi lack female meiosis, such that every meiotic product is viable. Therefore, their centromeres lack the opportunity and impetus to compete against each other. In the absence of this asymmetry, there is no selective advantage, and perhaps a significant disadvantage, for centromeres to get larger because there is no transmission bias to exploit (Malik and Henikoff 2002). Thus, the model predicts that in the absence of female meiosis, centromeres should get considerably less complex and evolve toward an optimal evolutionary equilibrium with their cognate CenH3s (Malik and Henikoff 2002). Since there is no centromere-drive in this setting, and therefore, no deleterious effects to suppress, CenH3s are not predicted to evolve under positive selection. These predictions appear largely borne out by the patterns of evolution in the sensu stricto group of Saccharomyces budding yeast species (Talbert et al. 2004), although there remains a high degree of unexplained polymorphism and divergence in centromeric DNA sequences themselves (Bensasson et al. 2008).
Fig. 1.
Taxonomic diversity of meiosis in eukaryotes. Animals and plants undergo two types of meiosis: asymmetric (female) meiosis in which three of four meiotic products are eliminated (shaded), and male meiosis in which all four meiotic products have an equal likelihood of success. In contrast, some lineages (like Fungi) only undergo male meiosis, while other lineages like ciliated protozoans (including Tetrahymena) only undergo female meiosis. Each of these configurations is expected to differ in terms of their susceptibility to "centromere-drive" and its deleterious consequences (Henikoff et al. 2001; Malik 2005; Malik and Bayes 2006; Malik and Henikoff 2009)
An important second-facet of the centromere-drive model, evolutionary outcomes in the absence of male meiosis, has not been closely investigated. There are distinct possibilities for the evolution of centromeres and centromeric proteins in taxa with only female meiosis (Fig. 1). If centromere-drive is primarily, or exclusively, deleterious to the process of male meiosis, its absence could result in severely reduced selective pressure for suppressor alleles to arise, in spite of rampant centromere-drive occurring. Such lineages would evolve complex centromeres because of the competition in female meiosis. However, genes encoding their centromeric proteins would not be subject to positive selection. Alternately, deleterious effects of centromere-drive could have a large impact on additional processes, compelling the rapid evolution of suppressors. Mitosis might be sensitive to centromere-drive, if the driving centromeres could over-recruit a rate-limiting centromeric component away from homologous chromosomes, since centromere recognition and balanced distribution of chromosomes are vital to cell division. In addition, female meiosis itself could be hampered by the "selfish" evolution of centromeres for the same reasons. Understanding the evolutionary forces acting on centromeric proteins is a vital part of determining the impact of centromere-drive and how centromere-related factors regulate the fundamental cellular functions of mitosis and meiosis.
Ciliated protozoans like Tetrahymena thermophila provide an opportunity for studying selection on centromeric genes in the absence of male meiosis (Fig. 1). Ciliates possess a germline nucleus that undergoes canonical mitosis and meiosis. They also possess a somatic macronucleus that divides amitotically, and is derived from massive deletion and amplification of the micronuclear genome. Most pertinent is the meiotic program of ciliates, in which two mating cells pair and undergo female meiosis in which three of four meiotic products are eliminated, before the surviving meiotic product duplicates and is exchanged between the mating partners (Cervantes et al. 2006). All meiosis in ciliated protozoans is therefore "female". Such meiosis provides a substrate for centromere-drive and a system to ascertain the selective pressures shaping the evolution of genes encoding their centromeric proteins in the absence of male meiosis. We, and others have previously identified and characterized the Cna1p centromeric histone in Tetrahymena thermophila (Cervantes et al. 2006; Cui and Gorovsky 2006). CNA1 is essential for both mitotic and meiotic segregation, and its evolution provides a key point of comparison of the selective pressures and rates of evolution affecting CenH3 genes in different taxa.
In this article, we report evolutionary and cytological analyses of the centromeric histone CNA1 gene from a panel of related Tetrahymena species. In contrast to CenH3s from plants and mammals, we find no compelling evidence for positive selection in ciliates. Our findings are consistent with a model in which the absence of male meiosis removes the need to suppress centromere-drive, thus allowing centromeres to compete unchecked.
Materials and Methods
Strains
Tetrahymena species cultures were obtained from the American Type Culture Collection (ATCC) except for the following species: T. malaccensis, T. borealis, T. pigmentosa, T. hegewishii (gift from Peter Bruns lab) (Coyne and Yao 1996), T. vorax (gift from Dr. Howard Buhse), and T. mobilis (gift from Dr. Denis Lynn). DNA samples from 10 additional strains of T. thermophila were obtained (gift from Dr. Paul Doerder). All strains and their ATCC accession numbers are listed in Supplementary material Table S1.
Genomic DNA Isolation
Cultures were grown overnight in SPP media to saturation, lysates were prepared, and DNA phenol/chloroform extracted as previously described (Gaertig et al. 1993).
PCR
We obtained genomic DNA from these species, and PCR amplified and sequenced the histone H3-H4 spacer region, as previously described, to confirm the identity of the various species we had obtained (Brunk et al. 1990; Sadler and Brunk 1992). We used primers 5′-GCAAATGGCTAGGAAAGCTTATTAACC-3′ (CNA1 ATG forward) and 5′-TCATTTTTTAGTAGGGATAAATATACCTG-3′ (CNA1 Ter reverse) in a Touchdown PCR (annealing temperature from 55 to 40 degrees dropping 1 degree per cycle, then 55 degrees for 35 cycles) to obtain almost the entire coding sequence of the CNA1 gene from 10 T. thermophila (kind gift of Dr. Paul Doerder) and 4 T. elliotti strains (from ATCC) for McDonald–Kreitman analysis. This combination of CNA1 ATG forward and CNA1 Ter reverse also worked for the two most closely related species to T. thermophila (T. borealis, T. elliotti). However, additional primers had to be designed for a broader panel of Tetrahymena species (e.g., T. pyriformis, T. vorax) using sequence homology among the CNA1 sequences that had already been obtained. Thus, reverse primers, 5’-GGTAACTCTTTTTGCATGTAAAGC-3’ (NCE0078) and 5’-GGTAACTCTTTTAGCGTGTAAAGC-3’ (NCE0086) in combination with CNA1 ATG forward, and forward primer 5’-TATGAATCAGCAAGAGATAAAGTAATTAG-3’ (NCE0077) in combination with the CNA1 Ter reverse primer were used to obtain the CNA1 sequence in two amplicons. Despite this iterative strategy, we were unable to obtain sequences from Tetrahymena species that were very distantly related to T. thermophila (e.g., T. bergeri, T. corlissi, T. pigmentosa) likely due to the rapid divergence of the CNA1 nucleotide sequence in this clade. All sequences obtained have been deposited to Genbank under the accession numbers: HQ285826-HQ285837.
Phylogenetic and Evolutionary Analyses
To perform a McDonald–Kreitman test, 10 T. thermophila and 4 T. elliotti CNA1 sequences were aligned (using ClustalX), the number of nonsynonymous and synonymous changes were counted and tested for evidence of positive selection using Fisher’s exact test in the DnaSP program (Librado and Rozas 2009). Owing to low numbers of polymorphisms present in T. thermophila, a McDonald–Kreitman Poisson Random Field (MKPRF) test was performed (Bustamante et al. 2002) using an online server (http://cbsuapps.tc.cornell.edu/mkprf.aspx). MKPRF uses a Poisson Random Field to model the frequencies of polymorphism under negative, neutral, or positive selection. This method estimates the value of the selection parameter γ (=2Nes) (γ > 0 when there is an excess of nonsynonymous fixed differences and γ < 0 when there is an excess of nonsynonymous polymorphisms) and assesses significance of deviation from neutral expectations.
To further analyze the possibility of positive selection, we employed the PAML suite of programs (Yang 2007). CNA1 sequences from multiple Tetrahymena species were aligned using ClustalX (Thompson et al. 1997) with manual refinement of gap positions. A phylogenetic tree was created, and bootstrapped using either the Neighbor-joining method (Saitou and Nei 1987) in ClustalX, or using maximum-likelihood methods and the PhyML program (Guindon and Gascuel 2003). To identify diversifying selection, we required a significant difference in log likelihoods, by chi-squared testing, between nested PAML models M7 and M8. Model M7 allows codons to have dN/dS values according to a beta distribution (two parameters). Model M8 is the same as M7 except that it adds a discrete category of dN/dS with values >1. We tested for significance of diversifying selection by comparing twice the difference in log likelihoods (2lnλ) between M7 and M8 with two degrees of freedom. The genetic code was adjusted to the ciliate model where TAG and TAA both encode Glutamate (Q) instead of a termination codon (Horowitz and Gorovsky 1985) by setting the icode value to 5 in the PAML control file.
Sequences of centromeric histones from other lineages (Mammals—CENPA; Drosophila—Cid; and plants—HTR12) were obtained from Genbank (see Supplementary material Table S2). Lineage-based alignments were generated as described for CNA1, dS was calculated from the histone fold domain as a measure of divergence between sequences (Nei and Gojobori 1986), and PAML tests for positive selection was performed as described above (except the icode value was set to 0).
Western Blotting and Immunofluorescence
Tetrahymena species cultures were grown to the mid-log phase in SPP media, along with Paramecium tetraurelia (kind gift of Dr. Jim Forney), and processed for western blotting with polyclonal Cna1p affinity-purified antibody (1:1,000 dilution) as previously described (Cervantes et al. 2006). Immunofluorescence using Cna1p antibody (1:600 dilution) was performed with cells from Tetrahymena species as previously described for T. thermophila (Cervantes et al. 2006).
Results and Discussion
Molecular Evolution of CNA1
We sequenced the CNA1 gene from 10 T. thermophila and four T. elliotti strains. We found only one non-synonymous polymorphism in T. thermophila, consistent with recent estimates of a highly reduced effective population size at least among sampled North American T. thermophila (Katz et al. 2006), with high polymorphism levels being an exceptional finding in this species (Gerber et al. 2002). In four strains of T. elliotti, we found six synonymous and three non-synonymous polymorphisms (intriguingly, the same position is polymorphic in both T. elliotti and T. thermophila). Using the McDonald–Kreitman test (McDonald and Kreitman 1991), we found that the ratio of non-synonymous to synonymous pooled polymorphisms (3:6) was not significantly different from the non-synonymous to synonymous divergence (20:60, P-value >0.69, Table 1). Owing to the limited sample number, we also performed a MKPRF test (Bustamante et al. 2002) which was also not significant. This suggests that no accelerated fixation of non-synonymous changes indicative of positive selection had taken place since the divergence of T. thermophila and T. elliotti.
Table 1.
McDonald–Kreitman tests for positive selection on Tetrahymena CNA1
| Fixed (interspecies) | Polymorphic (intraspecies) | Ratios | P value* | |
|---|---|---|---|---|
| Replacement changes | 60 | 6 | 60:20::6:3 | 0.69 |
| Synonymous changes | 20 | 3 |
| MKPRF test** | ||||
|---|---|---|---|---|
| Posterior mean | Posterior S.D | Probability <0 | ||
| γ | −2.64 | 37.3 | 0.476 | |
* P-value calculated using Fisher’s exact two-tailed test
** As implemented in (Bustamante et al. 2002). See Materials and Methods
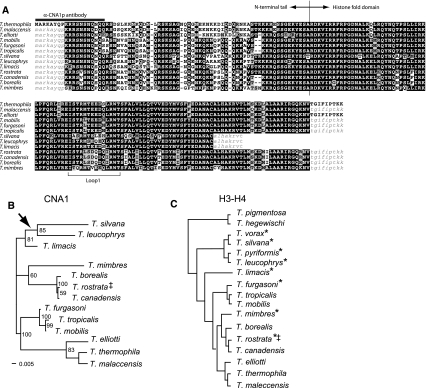
The low level of polymorphisms in T. thermophila and T. elliotti may have reduced the power of the McDonald–Kreitman test. To address whether CNA1 had been subject to positive selection, we sequenced CNA1 from a broad panel of Tetrahymena species (Supplementary material Table S1). We obtained genomic DNA from these species and PCR amplified and sequenced the histone H3-H4 spacer region, as previously described, to confirm the identity of the various species we had obtained (Brunk et al. 1990; Sadler and Brunk 1992). Next, we employed an iterative PCR approach (See Materials and Methods) to sequence the CNA1 locus from these Tetrahymena species. This strategy allowed us to obtain most of the CNA1 gene sequence from species as distantly related as—T. silvana, T. leucophrys, and T. mimbres. Despite multiple attempts with additional primers, we were unable to obtain CNA1 sequences from species more distant species. An amino acid alignment of the 13 CNA1 sequences is presented in Fig. 2a, and a maximum likelihood phylogeny based on CNA1 nucleotide sequences is presented in Fig. 2b. This phylogeny is largely congruent with the previous phylogenies of Tetrahymena species (Fig. 2c), which were based on either histone H3-H4 spacer regions (Brunk et al. 1990) (Sadler and Brunk 1992) or ribosomal RNA sequences (Sogin et al. 1986). The only notable discordance in topology between the CNA1 and H3-H4 trees is the relative placement of the T. furgasoni triad of species (also including T. tropicalis and T. mobilis) relative to the T. thermophila triad. However, this part of the topology was not well resolved in the previous analysis (Fig. 2c) (Brunk et al. 1990; Sadler and Brunk 1992); the CNA1 analysis finds an unambiguously closer relationship (100% bootstrap support) between the T. thermophila and T. furgasoni triads, than to T. borealis. In general, there was a higher degree of resolution between closely related species in the CNA1 phylogeny, albeit a lower depth of coverage of the various Tetrahymena species, compared with previous analyses.
Fig. 2.
Molecular evolution of CNA1 (a) Amino acid alignment of Cna1p from a panel of Tetrahymena species. Highlighted is the border between the N-terminal tail and histone fold domain, the sequence used as an epitope for antibody production, and the highly variable loop 1 region. Sequences corresponding to primer-binding sites are indicated in lower case. b Maximum likelihood phylogeny of CNA1 nucleic acid sequences. Bootstrap values above 50 are indicated at corresponding branches and the scale of substitutions/site is shown. The arrowhead indicates where the tree would root in the context of more diverse species as shown in c. c A phylogenetic tree of Tetrahymena species based on the histone H3-H4 spacer region (adapted from Brunk et al. 1990). The asterisks (*) denote species that lack a germline micronucleus. ‡The identification of this strain as T. rostrata has been challenged (Segade et al. 2009)
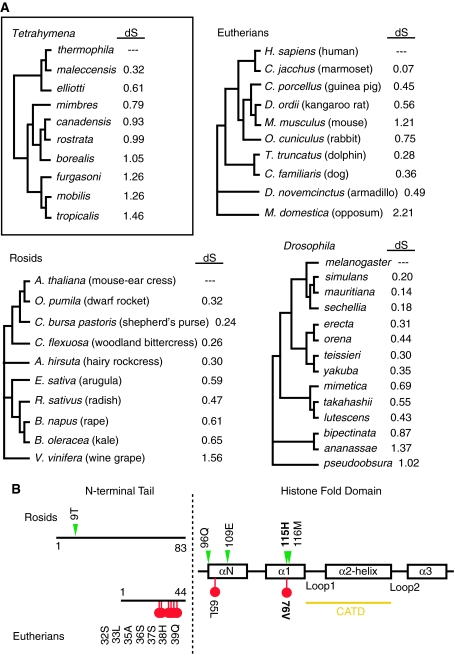
We were interested in determining whether there was any evidence of positive selection acting on our set of CNA1 sequences. In particular, our attention was drawn to regions of low conservation and apparently rapid evolution both in the N-terminal tail and the Loop1 regions (Fig. 2a), that have been previously been found to evolve under positive selection in CenH3s from taxa that undergo both symmetric and asymmetric meioses: primates, plants (Brassicaceae), and Drosophila (Cooper and Henikoff 2004; Malik and Henikoff 2001; Schueler et al. 2010; Talbert et al. 2002). However, these previous analyses were carried out in a dense taxonomic set with shallower evolutionary divergence compared with our Tetrahymena dataset. To address whether lower evolutionary divergence could affect signatures of positive selection acting on CenH3s, we assembled a deeper set of sequences from a clade of Mammals to expand the primate lineage, and a clade of Rosids (a subset of eudicot plants). A comparable analysis of Drosophila was not possible because of the presence of numerous N-terminal indels among CenH3 sequences of more diverged species than previously considered. Such divergence is consistent with adaptive evolution in this lineage, but hinders dN/dS analysis. The datasets of mammals and plants contained a sampling of species comparable to the divergence we observed in the Tetrahymena lineage, as judged by the rates of synonymous substitutions (dS) and overall tree length, and provided a better comparison of positive selection on CenH3 between clades (Fig. 3). For both Mammals and Rosids, we found clear evidence for positive selection using PAML analysis (Table 2), consistent with previous findings in more restricted phylogenetic samplings of primates and plants. However, our identification of individual sites evolving under positive selection differed slightly from previous analyses.
Fig. 3.
Evolution of centromeric histones in distinct taxonomic groups. a Cladograms of Tetrahymena, Mammals, Rosids, and Drosophila show rates of synonymous substitution (dS) as a gauge of divergence from T. thermophila, H. sapiens, A. thaliana, and D. melanogaster. b Amino acid positions under positive selection (Bayesian posterior probability >0.9) are shown for Rosids (triangles) and Mammals (circles). Amino acid site numbering is according to CenH3 from A. thaliana and H. sapiens
Table 2.
Maximum likelihood tests for positive selection on CenH3 proteins in three lineagesa
| Lineage | −2 (ln λ) | P value | % sites with dN/dS >1 | Average dN/dS for those sites with dN/dS >1 |
|---|---|---|---|---|
| Tetrahymena | ||||
| Full protein | 1.55 | 0.46 | 1.5 | 1.0 |
| HFD | 0.001 | 0.99 | 0.0 | 1.0 |
| Mammals | ||||
| Full protein | 42.48 | <0.0001 | 10.3 | 3.6 |
| HFD | 23.79 | <0.0001 | 7.9 | 2.6 |
| Rosids | ||||
| Full protein | 13.80 | 0.001 | 9.8 | 2.3 |
| HFD | 23.57 | <0.0001 | 13.8 | 2.1 |
aPAML models M7 vs. M8 with an f61 substitution table
We next carried out PAML analyses on the corresponding panel of 10 Tetrahymena species to assay whether positive selection had acted on CNA1 evolution. We found no evidence of positive selection on CNA1, because a "NS site" model that permitted codons to evolve under positive selection (M8) did not have a significantly different likelihood than a model disallowing positive selection (M7). Results from these analyses are shown in Table 2. The N-terminal tails of CenH3s differ in size as well as evolutionary constraint, which may result from divergent lineage-specific pressures. However, the histone fold domain (HFD) of CenH3 has the same binding partners in centromeric nucleosomes (Black and Bassett 2008; Palmer et al. 1991) and is presumed to have similar structural constraints across taxa. It is therefore especially striking to observe a complete lack of positive selection in the histone fold domain of CNA1 in contrast to previous observations of CenH3s from animals and plants (Cooper and Henikoff 2004; Malik and Henikoff 2001) and in our own analysis of Mammals and Rosids (Table 2). More conservative tests of positive selection in the HyPhy suite of programs, i.e., FEL, SLAC, and PARRIS also failed to return evidence of positive selection (Kosakovsky Pond and Frost 2005; Scheffler et al. 2006). We therefore conclude that, in contrast to lineages with both male and female meiosis, ciliated protozoans show no evidence for positive selection of their CenH3s.
Comparative Analyses of CNA1 Expression and Localization
Our finding that centromeric histones do not evolve under positive selection in Tetrahymena suggested two disparate possibilities for chromosomal evolution. The first possibility is that Tetrahymena micronuclear chromosomes do not experience centromere-drive despite undergoing female meiosis. If this were the case, their centromeres would be expected to resemble those of the epigenetically maintained centromeres of fission yeast (Schizosaccharomyces pombe) and other fungal species that only undergo male meiosis, where the CenH3-bound portion is only a few kilobases in length (Clarke et al. 1986; Song et al. 2008; Wood et al. 2002). The lack of micronuclear genome assembly of Tetrahymena thermophila, coupled with the highly AT-rich and repetitive composition of micronuclear genomes have thus far precluded direct identification of their centromeres. Only a few regional centromeres have been characterized in detail in any genome to date (Schueler et al. 2001; Sun et al. 2003; Yan et al. 2008). However, several indirect observations suggest that micronuclear chromosomes in Tetrahymena are "regional" and "complex". First, cytological observations of Cna1p during meiosis and mitosis (Cervantes et al. 2006; Cui and Gorovsky 2006) are consistent with regional centromeres seen in human, Drosophila and Arabidopsis centromeres. Second, consistent with its presumed highly repetitive nature, centromeres are eliminated during the development of the somatic macronucleus (Cervantes et al. 2006; Cui and Gorovsky 2006). Given the analogy of centromere organization, it is inevitable that, like in animal and plant genomes, ciliate centromeres compete during female meiosis. Direct confirmation of the quality of Tetrahymena centromeres might be revealed by chromatin immuno-precipitation with Cna1p antibody.
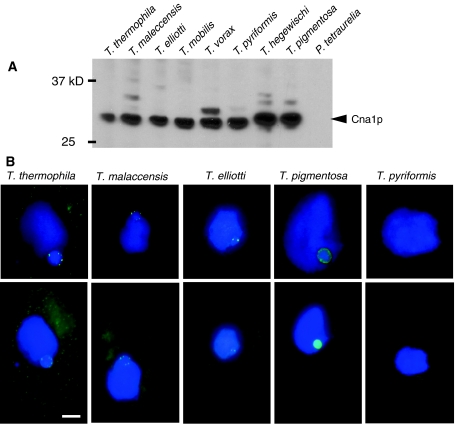
The current evidence for complex centromeres in Tetrahymena and lack of positive selection on CNA1 leads to the second possibility for chromosome evolution, where the absence of male meiosis could leave centromere-drive largely unsuppressed. Since it is not possible to carry out a comparison of centromeric sequences from different Tetrahymena species, we compared karyotypic evolution in Tetrahymena using a polyclonal antibody raised to the first 18 amino acids of Cna1p from T. thermophila (thick bar in Fig. 2a; (Cervantes et al. 2006). Noting the high degree of amino acid conservation in this region, we performed Western analysis of protein lysates from a panel of Tetrahymena species, including some species more divergent than ones from which we had obtained CNA1 sequence. We also included lysates from Paramecium tetraurelia, a more distantly related ciliated protozoan because analysis of its sequenced genome (Aury et al. 2006) suggested that its putative CenH3s have not preserved this amino acid stretch. Consistent with our expectations, we found that Western blots consistently identified bands at the expected size for Cna1p (Fig. 3a) in all tested Tetrahymena species, but not in P. tetaurelia. This finding demonstrates that all six amicronucleate species in our panel express Cna1p, despite a lack of meiosis. Moreover, evolutionary analysis of CNA1 from these amicronucleate species finds strong signs of purifying selection (Supplementary material Table S3; Fig. S1), supporting the assertion that the loss of micronuclei among these species were recent events and validates the use of these species for comparisons of evolutionary rates. Expression of Cna1p in the Tetrahymena panel also suggested the utility of our antibody for comparative cytological analyses in other Tetrahymena species, to address whether substantive karyotypic changes were evident in this species group.
We used the Cna1p antibody to stain cells from a panel of Tetrahymena species and found clear differences in Cna1p immunoreactivity. Germline micronuclei of T. thermophila contain Cna1p immuno-reactive dots consistent with its five metacentric chromosomes arranged peripherally in interphase cells (Fig. 4b) (Cervantes et al. 2006; Cui and Gorovsky 2006). For T. malaccensis, the species most closely related to T. thermophila (Fig. 2b, c), the Cna1p signal appeared identical to T. thermophila. However, for T. elliotti, the next most closely related species (Fig. 2b), we consistently observed only two Cna1p dots, suggesting a striking difference in karyotype (Fig. 4 middle panels). It is formally possible that this difference could reflect the loss of germline chromosomes for this strain maintained extensively in cell culture without sexual reproduction. This can occur because Tetrahymena do not need their germline micronucleus for survival, only for meiosis and post-meiosis development. Our attempts at detecting a meiotic complement of T. elliotti chromosomes, by mating different strains, were not successful, although fertility of T. elliotti lab strains has been reported (Nanney and Simon 1999). As expected, the amicronucleate species T. pyriformis showed no Cna1p signal above background reflecting an absence of micronuclear chromosomes with centromeres (Fig. 4b) despite expressing CNA1 (Fig. 4a). T. pigmentosa, a distant relative of T. thermophila, displayed an extensive peripheral micronuclear Cna1p signal (Fig. 4b). This pattern of immuofluorescence likely reflects an increase in chromosome number.
Fig. 4.
CNA1 expression and localization in Tetrahymena species. a Immunoblot analysis of Cna1p expression in a panel of Tetrahymena species and Paramecium tetraurelia. b Fixed cells from Tetrahymena species were labeled with DAPI to stain the larger macronuclei and smaller micronuclei, and anti-Cna1p antibody to visualize centromeres. Background fluorescence was reduced in close-up images of nuclei in top panels by deconvolution. Bar = 5 microns
Our cytological survey is more consistent with the rapid karyotypic evolution seen as a result of centromere drive, for instance in the mammalian lineage (Pardo-Manuel de Villena and Sapienza 2001a), than with the low rates of karyotypic evolution seen in lineages lacking female meiosis, including the sensu stricto species of Saccharomyces (Cardinali and Martini 1994). Considering the lack of positive selection on CNA1, we hypothesize that the exclusively female meiosis in ciliates has resulted in unsuppressed centromere-drive in this lineage.
Conclusions
Testing what biological forces drive key evolutionary aspects of chromosome organization can be a daunting challenge. This is especially true when the underlying DNA sequences are themselves hard to characterize and manipulate. Centromeres and pericentric heterochromatin exemplify this challenge; they are often composed of repetitive satellite arrays that vary in size, sequence, and length of repeats. We have proposed that the study of proteins that bind such sequences can often provide a facile, surrogate means to investigate the evolution of such repetitive DNA (Vermaak et al. 2009). Centromeric histones have proven to be highly informative for the study of centromeric DNA since they are the fundamental unit of centromeric chromatin (Allshire and Karpen 2008; Black and Bassett 2008). Indeed, the observation of positive selection acting on CenH3 genes in Drosophila and Arabidopsis allowed the proposal of the "centromere-drive" hypothesis in the first place (Henikoff and Malik 2002; Malik and Henikoff 2001; Talbert et al. 2002).
Studying the evolutionary pressure on centromeric histones of Tetrahymena species allows us to consider the deleterious effects of "centromere-drive" and whether centromere complexity is always shaped by genetic conflict. Tetrahymena, like other ciliated protozoans, lack male meiosis, one of the processes that have been proposed to shoulder much of the cost of driving centromeres (Daniel 2002; Fishman and Saunders 2008; Henikoff et al. 2001; Malik 2009; Malik and Henikoff 2009). Our finding that CNA1 is not subject to positive selection, in contrast to taxa with both male and female meiosis, leads to the intriguing possibility that even though micronuclear centromeres are subject to centromere-drive in ciliates, this drive may be unsuppressed. Importantly, our findings also suggest that while mitosis and asymmetric (female) meiosis may be impacted by centromere-drive, defects in male meiosis appear to be the principal selective force favoring the evolution of suppressors.
It is possible that other factors besides CNA1 are selected to suppress centromeric drive in ciliates since additional centromeric proteins have been shown to evolve under positive selection in both animals and plants (Schueler et al. 2010; Talbert et al. 2004). Because Cna1p is the only centromeric protein known in ciliates, it is not currently possible to test whether positive selection of other centromeric proteins has "substituted" for positive selection on CNA1 in ciliates; the identification of additional centromeric proteins like CENP-C would provide a useful comparison (Talbert et al. 2004, 2008). However, CenH3 evolution provides an important litmus test for the suppression of centromere-drive because positive selection of CenH3s has been observed in every taxa considered so far with female and male meiosis. Thus, the absence of positive selection in CNA1 is especially noteworthy. Studying the selective pressures on CenH3 s in other taxa that also lack male meiosis will add further insight into centromere-drive and its consequences. As our present study demonstrates, naturally occurring taxonomic variation in meiosis provides one of the most powerful means available to decipher the central forces shaping centromeres and their binding partners in eukaryotic genomes.
Electronic supplementary material
Below is the link to the electronic supplementary material.
Acknowledgments
The authors thank the ATCC and our colleagues (Peter Bruns, Howard Buhse, Paul Doerder, Jim Forney, and Dennis Lynn) for sharing strains and the Cornell Computational Biology Service Unit for their computational support with MKPRF calculations. The authors thank Nitin Phadnis and Paul Talbert for their comments on the manuscript.
Funding
This study was supported by a Life Sciences Research Foundation postdoctoral fellowship from the Ellison Medical Foundation and K99 award from the National Institute of General Medical Sciences at the National Institutes of Health (grant number GM090042, to NCE), by a National Science Foundation pre-doctoral fellowship (to KCR), funds from Academia Sinica and National Research Council of Taiwan (to MCY), and by a R01 grant from the National Institute of General Medical Sciences at the National Institutes of Health (grant number GM74108, to HSM). HSM is an Early Career Scientist of the Howard Hughes Medical Institute.
Open Access
This article is distributed under the terms of the Creative Commons Attribution Noncommercial License which permits any noncommercial use, distribution, and reproduction in any medium, provided the original author(s) and source are credited.
Footnotes
Tetrahymena CNA1 sequences deposited in Genbank: HQ285826-HQ285837.
References
- Allshire RC, Karpen GH. Epigenetic regulation of centromeric chromatin: old dogs, new tricks? Nat Rev Genet. 2008;9:923–937. doi: 10.1038/nrg2466. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Aury JM, Jaillon O, Duret L, Noel B, Jubin C, Porcel BM, Segurens B, Daubin V, Anthouard V, Aiach N, Arnaiz O, Billaut A, Beisson J, Blanc I, Bouhouche K, Camara F, Duharcourt S, Guigo R, Gogendeau D, Katinka M, Keller AM, Kissmehl R, Klotz C, Koll F, Le Mouel A, Lepere G, Malinsky S, Nowacki M, Nowak JK, Plattner H, Poulain J, Ruiz F, Serrano V, Zagulski M, Dessen P, Betermier M, Weissenbach J, Scarpelli C, Schachter V, Sperling L, Meyer E, Cohen J, Wincker P. Global trends of whole-genome duplications revealed by the ciliate Paramecium tetraurelia. Nature. 2006;444:171–178. doi: 10.1038/nature05230. [DOI] [PubMed] [Google Scholar]
- Axelsson E, Albrechtsen A, van AP, Li L, Megens HJ, Vereijken AL, Crooijmans RP, Groenen MA, Ellegren H, Willerslev E, Nielsen R. Segregation distortion in chicken and the evolutionary consequences of female meiotic drive in birds. Heredity. 2010;105:290–298. doi: 10.1038/hdy.2009.193. [DOI] [PubMed] [Google Scholar]
- Bayes JJ, Malik HS. Altered heterochromatin binding by a hybrid sterility protein in Drosophila sibling species. Science. 2009;326:1538–1541. doi: 10.1126/science.1181756. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bensasson D, Zarowiecki M, Burt A, Koufopanou V. Rapid evolution of yeast centromeres in the absence of drive. Genetics. 2008;178:2161–2167. doi: 10.1534/genetics.107.083980. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Black BE, Bassett EA. The histone variant CENP-A and centromere specification. Curr Opin Cell Biol. 2008;20:91–100. doi: 10.1016/j.ceb.2007.11.007. [DOI] [PubMed] [Google Scholar]
- Brunk CF, Kahn RW, Sadler LA. Phylogenetic relationships among Tetrahymena species determined using the polymerase chain reaction. J Mol Evol. 1990;30:290–297. doi: 10.1007/BF02099999. [DOI] [PubMed] [Google Scholar]
- Bustamante CD, Nielsen R, Sawyer SA, Olsen KM, Purugganan MD, Hartl DL. The cost of inbreeding in Arabidopsis. Nature. 2002;416:531–534. doi: 10.1038/416531a. [DOI] [PubMed] [Google Scholar]
- Cardinali G, Martini A. Electrophoretic karyotypes of authentic strains of the sensu stricto group of the genus Saccharomyces. Int J Syst Bacteriol. 1994;44:791–797. doi: 10.1099/00207713-44-4-791. [DOI] [PubMed] [Google Scholar]
- Cervantes MD, Xi X, Vermaak D, Yao MC, Malik HS. The CNA1 histone of the ciliate tetrahymena thermophila is essential for chromosome segregation in the germline micronucleus. Mol Biol Cell. 2006;17:485–497. doi: 10.1091/mbc.E05-07-0698. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Clarke L, Amstutz H, Fishel B, Carbon J. Analysis of centromeric DNA in the fission yeast Schizosaccharomyces pombe. Proc Natl Acad Sci USA. 1986;83:8253–8257. doi: 10.1073/pnas.83.21.8253. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cooper JL, Henikoff S. Adaptive evolution of the histone fold domain in centromeric histones. Mol Biol Evol. 2004;21:1712–1718. doi: 10.1093/molbev/msh179. [DOI] [PubMed] [Google Scholar]
- Coyne RS, Yao MC. Evolutionary conservation of sequences directing chromosome breakage and rDNA palindrome formation in tetrahymenine ciliates. Genetics. 1996;144:1479–1487. doi: 10.1093/genetics/144.4.1479. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cui B, Gorovsky MA. Centromeric histone H3 is essential for vegetative cell division and for DNA elimination during conjugation in Tetrahymena thermophila. Mol Cell Biol. 2006;26:4499–4510. doi: 10.1128/MCB.00079-06. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Daniel A. Distortion of female meiotic segregation and reduced male fertility in human Robertsonian translocations: consistent with the centromere model of co-evolving centromere DNA/centromeric histone (CENP-A) Am J Med Genet. 2002;111:450–452. doi: 10.1002/ajmg.10618. [DOI] [PubMed] [Google Scholar]
- Fishman L, Saunders A. Centromere-associated female meiotic drive entails male fitness costs in monkeyflowers. Science. 2008;322:1559–1562. doi: 10.1126/science.1161406. [DOI] [PubMed] [Google Scholar]
- Fishman L, Willis JH. A novel meiotic drive locus almost completely distorts segregation in mimulus (monkeyflower) hybrids. Genetics. 2005;169:347–353. doi: 10.1534/genetics.104.032789. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gaertig J, Thatcher TH, McGrath KE, Callahan RC, Gorovsky MA. Perspectives on tubulin isotype function and evolution based on the observation that Tetrahymena thermophila microtubules contain a single alpha- and beta-tubulin. Cell Motil Cytoskeleton. 1993;25:243–253. doi: 10.1002/cm.970250305. [DOI] [PubMed] [Google Scholar]
- Gerber CA, Lopez AB, Shook SJ, Doerder FP. Polymorphism and selection at the SerH immobilization antigen locus in natural populations of Tetrahymena thermophila. Genetics. 2002;160:1469–1479. doi: 10.1093/genetics/160.4.1469. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Guindon S, Gascuel O. A simple, fast, and accurate algorithm to estimate large phylogenies by maximum likelihood. Syst Biol. 2003;52:696–704. doi: 10.1080/10635150390235520. [DOI] [PubMed] [Google Scholar]
- Henikoff S, Malik HS. Centromeres: selfish drivers. Nature. 2002;417:227. doi: 10.1038/417227a. [DOI] [PubMed] [Google Scholar]
- Henikoff S, Ahmad K, Malik HS. The centromere paradox: stable inheritance with rapidly evolving DNA. Science. 2001;293:1098–1102. doi: 10.1126/science.1062939. [DOI] [PubMed] [Google Scholar]
- Horowitz S, Gorovsky MA. An unusual genetic code in nuclear genes of Tetrahymena. Proc Natl Acad Sci USA. 1985;82:2452–2455. doi: 10.1073/pnas.82.8.2452. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Katz LA, Snoeyenbos-West O, Doerder FP. Patterns of protein evolution in Tetrahymena thermophila: implications for estimates of effective population size. Mol Biol Evol. 2006;23:608–614. doi: 10.1093/molbev/msj067. [DOI] [PubMed] [Google Scholar]
- Kosakovsky Pond SL, Frost SD. Not so different after all: a comparison of methods for detecting amino acid sites under selection. Mol Biol Evol. 2005;22:1208–1222. doi: 10.1093/molbev/msi105. [DOI] [PubMed] [Google Scholar]
- Librado P, Rozas J. DnaSP v5: a software for comprehensive analysis of DNA polymorphism data. Bioinformatics. 2009;25:1451–1452. doi: 10.1093/bioinformatics/btp187. [DOI] [PubMed] [Google Scholar]
- Malik HS. Mimulus finds centromeres in the driver’s seat. Trends Ecol Evol. 2005;20:151–154. doi: 10.1016/j.tree.2005.01.014. [DOI] [PubMed] [Google Scholar]
- Malik HS. The centromere-drive hypothesis: a simple basis for centromere complexity. Prog Mol Subcell Biol. 2009;48:33–52. doi: 10.1007/978-3-642-00182-6_2. [DOI] [PubMed] [Google Scholar]
- Malik HS, Bayes JJ. Genetic conflicts during meiosis and the evolutionary origins of centromere complexity. Biochem Soc Trans. 2006;34:569–573. doi: 10.1042/BST0340569. [DOI] [PubMed] [Google Scholar]
- Malik HS, Henikoff S. Adaptive evolution of Cid, a centromere-specific histone in Drosophila. Genetics. 2001;157:1293–1298. doi: 10.1093/genetics/157.3.1293. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Malik HS, Henikoff S. Conflict begets complexity: the evolution of centromeres. Curr Opin Genet Dev. 2002;12:711–718. doi: 10.1016/S0959-437X(02)00351-9. [DOI] [PubMed] [Google Scholar]
- Malik HS, Henikoff S. Major evolutionary transitions in centromere complexity. Cell. 2009;138:1067–1082. doi: 10.1016/j.cell.2009.08.036. [DOI] [PubMed] [Google Scholar]
- McDonald JH, Kreitman M. Adaptive protein evolution at the Adh locus in Drosophila. Nature. 1991;351:652–654. doi: 10.1038/351652a0. [DOI] [PubMed] [Google Scholar]
- Nanney D, Simon E. Laboratory and Evolutionary History of Tetrahymena thermophila. In: Asai D, Forney J, editors. Methods in Cell Biology. San Diego: Academic Press; 1999. pp. 3–26. [DOI] [PubMed] [Google Scholar]
- Nei M, Gojobori T. Simple methods for estimating the numbers of synonymous and nonsynonymous nucleotide substitutions. Mol Biol Evol. 1986;3:418–426. doi: 10.1093/oxfordjournals.molbev.a040410. [DOI] [PubMed] [Google Scholar]
- Palmer DK, O’Day K, Trong HL, Charbonneau H, Margolis RL. Purification of the centromere-specific protein CENP-A and demonstration that it is a distinctive histone. Proc Natl Acad Sci U S A. 1991;88:3734–3738. doi: 10.1073/pnas.88.9.3734. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pardo-Manuel de Villena F, Sapienza C. Female meiosis drives karyotypic evolution in mammals. Genetics. 2001;159:1179–1189. doi: 10.1093/genetics/159.3.1179. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pardo-Manuel de Villena F, Sapienza C. Transmission ratio distortion in offspring of heterozygous female carriers of Robertsonian translocations. Hum Genet. 2001;108:31–36. doi: 10.1007/s004390000437. [DOI] [PubMed] [Google Scholar]
- Sadler LA, Brunk CF. Phylogenetic relationships and unusual diversity in histone H4 proteins within the Tetrahymena pyriformis complex. Mol Biol Evol. 1992;9:70–84. doi: 10.1093/oxfordjournals.molbev.a040709. [DOI] [PubMed] [Google Scholar]
- Saitou N, Nei M. The neighbor-joining method: a new method for reconstructing phylogenetic trees. Mol Biol Evol. 1987;4:406–425. doi: 10.1093/oxfordjournals.molbev.a040454. [DOI] [PubMed] [Google Scholar]
- Scheffler K, Martin DP, Seoighe C. Robust inference of positive selection from recombining coding sequences. Bioinformatics. 2006;22:2493–2499. doi: 10.1093/bioinformatics/btl427. [DOI] [PubMed] [Google Scholar]
- Schueler MG, Higgins AW, Rudd MK, Gustashaw K, Willard HF. Genomic and genetic definition of a functional human centromere. Science. 2001;294:109–115. doi: 10.1126/science.1065042. [DOI] [PubMed] [Google Scholar]
- Schueler MG, Swanson W, Thomas PJ, Green ED. Adaptive evolution of foundation kinetochore proteins in primates. Mol Biol Evol. 2010;27:1585–1597. doi: 10.1093/molbev/msq043. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Segade P, Kher CP, Lynn DH, Iglesias R. Morphological and molecular characterization of renal ciliates infecting farmed snails in Spain. Parasitology. 2009;136:771–782. doi: 10.1017/S0031182009006040. [DOI] [PubMed] [Google Scholar]
- Sogin ML, Ingold A, Karlok M, Nielsen H, Engberg J. Phylogenetic evidence for the acquisition of ribosomal RNA introns subsequent to the divergence of some of the major Tetrahymena groups. EMBO J. 1986;5:3625–3630. doi: 10.1002/j.1460-2075.1986.tb04691.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Song JS, Liu X, Liu XS, He X. A high-resolution map of nucleosome positioning on a fission yeast centromere. Genome Res. 2008;18:1064–1072. doi: 10.1101/gr.075374.107. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sun X, Le HD, Wahlstrom JM, Karpen GH. Sequence analysis of a functional Drosophila centromere. Genome Res. 2003;13:182–194. doi: 10.1101/gr.681703. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Talbert PB, Masuelli R, Tyagi AP, Comai L, Henikoff S. Centromeric localization and adaptive evolution of an Arabidopsis histone H3 variant. Plant Cell. 2002;14:1053–1066. doi: 10.1105/tpc.010425. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Talbert PB, Bryson TD, Henikoff S. Adaptive evolution of centromere proteins in plants and animals. J Biol. 2004;3:18. doi: 10.1186/jbiol11. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Talbert PB, Bayes JJ, Henikoff S. Evolution of centromeres and kinetochores: A two-part fugue. In: De Wulf P, Earnshaw WC, editors. The Kinetochore. Berlin: Springer; 2008. pp. 193–230. [Google Scholar]
- Thompson JD, Gibson TJ, Plewniak F, Jeanmougin F, Higgins DG. The CLUSTAL_X windows interface: flexible strategies for multiple sequence alignment aided by quality analysis tools. Nucleic Acids Res. 1997;25:4876–4882. doi: 10.1093/nar/25.24.4876. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Vermaak D, Bayes JJ, Malik HS. A surrogate approach to study the evolution of noncoding DNA elements that organize eukaryotic genomes. J Hered. 2009;100:624–636. doi: 10.1093/jhered/esp063. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wood V, Gwilliam R, Rajandream MA, Lyne M, Lyne R, Stewart A, Sgouros J, Peat N, Hayles J, Baker S, Basham D, Bowman S, Brooks K, Brown D, Brown S, Chillingworth T, Churcher C, Collins M, Connor R, Cronin A, Davis P, Feltwell T, Fraser A, Gentles S, Goble A, Hamlin N, Harris D, Hidalgo J, Hodgson G, Holroyd S, Hornsby T, Howarth S, Huckle EJ, Hunt S, Jagels K, James K, Jones L, Jones M, Leather S, McDonald S, McLean J, Mooney P, Moule S, Mungall K, Murphy L, Niblett D, Odell C, Oliver K, O’Neil S, Pearson D, Quail MA, Rabbinowitsch E, Rutherford K, Rutter S, Saunders D, Seeger K, Sharp S, Skelton J, Simmonds M, Squares R, Squares S, Stevens K, Taylor K, Taylor RG, Tivey A, Walsh S, Warren T, Whitehead S, Woodward J, Volckaert G, Aert R, Robben J, Grymonprez B, Weltjens I, Vanstreels E, Rieger M, Schafer M, Muller-Auer S, Gabel C, Fuchs M, Dusterhoft A, Fritzc C, Holzer E, Moestl D, Hilbert H, Borzym K, Langer I, Beck A, Lehrach H, Reinhardt R, Pohl TM, Eger P, Zimmermann W, Wedler H, Wambutt R, Purnelle B, Goffeau A, Cadieu E, Dreano S, Gloux S, Lelaure V, Mottier S, Galibert F, Aves SJ, Xiang Z, Hunt C, Moore K, Hurst SM, Lucas M, Rochet M, Gaillardin C, Tallada VA, Garzon A, Thode G, Daga RR, Cruzado L, Jimenez J, Sanchez M, del Rey F, Benito J, Dominguez A, Revuelta JL, Moreno S, Armstrong J, Forsburg SL, Cerutti L, Lowe T, McCombie WR, Paulsen I, Potashkin J, Shpakovski GV, Ussery D, Barrell BG, Nurse P. The genome sequence of Schizosaccharomyces pombe. Nature. 2002;415:871–880. doi: 10.1038/nature724. [DOI] [PubMed] [Google Scholar]
- Yan H, Talbert PB, Lee HR, Jett J, Henikoff S, Chen F, Jiang J. Intergenic locations of rice centromeric chromatin. PLoS Biol. 2008;6:e286. doi: 10.1371/journal.pbio.0060286. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yang Z. PAML 4: phylogenetic analysis by maximum likelihood. Mol Biol Evol. 2007;24:1586–1591. doi: 10.1093/molbev/msm088. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.