Abstract
Resolving the phylogenetic relationships among birds is a classical problem in systematics, and this is particularly so when it comes to understanding the relationships among Neoaves. Previous phylogenetic inference of birds has been limited to mitochondrial genomes or a few nuclear genes. Here, we apply deep brain transcriptome sequencing of nine bird species (several passerines, hummingbirds, dove, parrot, and emu), using next-generation sequencing technology to understand features of transcriptome evolution in birds and how this affects phylogenetic inference, and combine with data from two bird species using first generation technology. The phylogenomic data matrix comprises 1,995 genes and a total of 0.77 Mb of exonic sequence. First, we find an unexpected heterogeneity in the evolution of base composition among avian lineages. There is a pronounced increase in guanine + cytosine (GC) content in the third codon position in several independent lineages, with the strongest effect seen in passerines. Second, we evaluate the effect of GC content variation on phylogenetic reconstruction. We find important inconsistencies between the topologies obtained with or without taking GC variation into account, each supporting different conclusions of past studies and also influencing hypotheses on the evolution of the trait of vocal learning. Third, we demonstrate a link between GC content evolution and recombination rate and, focusing on the zebra finch lineage, find that recombination seems to drive GC content. Although we cannot reveal the causal relationships, this observation is consistent with the model of GC-biased gene conversion. Finally, we use this unparalleled amount of avian sequence data to study the rate of molecular evolution, calibrated by fossil evidence and augmented with data from alligator transcriptome sequencing. There is a 2- to 3-fold variation in substitution rate among lineages with passerines being the most rapidly evolving and ratites the slowest. This study illustrates the potential of next-generation sequencing for phylogenomic studies but also the pitfalls when using genome-wide data with heterogeneous base composition.
Keywords: birds, phylogenetics, base composition, recombination rate, substitution rate, molecular dating
Introduction
A major leap in molecular phylogenetics is expected with the introduction of deep sequencing using next-generation sequencing technology, turning phylogenetics into phylogenomics (Shendure and Ji 2008; Emerson et al. 2010). When targeted to the coding regions of the genome, that is, the transcriptome, it can allow obtaining sequence data from thousands of genes in a single sequence run. Transcriptome data sets could be particularly appropriate for resolving deep relationships because although protein-coding genes represent slowly evolving sequences, they are thought to be less prone to accumulate a large amount of nonphylogenetic (i.e., homoplastic) signal compared with fast evolving markers like mitochondrial DNA (mtDNA) (Springer et al. 2001). However, the assumed models of sequence evolution are critical to such large data sets as systematic biases in, for example, base composition could lead to strong support of erroneous phylogenies (e.g., Delsuc et al. 2005; Jeffroy et al. 2006).
Classical substitution models assume that guanine + cytosine (GC) content remains constant across the phylogeny. Violation of the so-called “stationary assumption” through convergent evolution toward GC-rich or GC-poor sequences is known to affect phylogenetic reconstruction. Specifically, there is a tendency to group species of similar base composition irrespective of their historical relationships (Foster and Hickey 1999; Phillips et al. 2004; Delsuc et al. 2005; Blanquart and Lartillot 2008; Sheffield et al. 2009). The evolution of GC content variation in vertebrates has been widely studied by focusing on the spatial heterogeneities of GC content within the genome. In birds, as in mammals, the genomic GC content alternate between GC-rich and GC-poor regions, at a Mb scale, often referred to as isochores (e.g., Bernardi 2000; Eyre-Walker and Hurst 2001; ICGSC 2004). In birds, the evolution of GC content in different lineages is yet to be characterized. An analysis of CR1 repeat elements in the chicken genome revealed that the isochore structure is reinforced, that is, that GC-rich regions evolve toward an even higher GC content (Webster et al. 2006).
Here, we study the inference of avian phylogeny and the influence and evolution of base composition in birds using brain transcriptome sequence data obtained through deep Roche 454 GS-FLX sequencing of nine bird species (Künstner et al. 2010) and whole-genome sequences of two bird species, chicken and zebra finch (ICGSC 2004; Warren et al. 2010). Our data set, consisting of a 0.77 Mb exonic sequence matrix, contains species, which belong to several Neoavian orders and includes passerines, a parrot, a dove, two hummingbirds, and an emu. We developed a phylogenomic approach that handles thousands of randomly sequenced orthologous genes from multiple species. We found striking variation in base composition evolution in different lineages, which profoundly influenced phylogenetic inference.
Materials and Methods
Sequence Data
Nine bird and one crocodile species were subject to brain transcriptome sequencing: emu (Dromaius novaehollandiae), ruby-throated hummingbird (Archilochus colubris), Anna's hummingbirds (Calypte anna), budgerigar (Melopsittacus undulatus), collared-dove (Streptopelia risoria), golden-collared manakin (Manacus vitellinus), American crow (Corvus brachyrhynchos), blue tit (Cyanistes (Parus) caeruleus), pied flycatcher (Ficedula hypoleuca) and American alligator (Alligator mississippiensis) (according to methods described in Künstner et al. [2010]; accession number SRX012365; http://www.ncbi.nlm.nih.gov/sra). In addition, we use the draft genome sequences of chicken (Gallus gallus) (ICGSC 2004) and zebra finch (Taeniopygia guttata) (Warren et al. 2010), downloaded from ENSEMBL (http://www.ensembl.org). The source of brain samples, methods for RNA isolation, cDNA preparation, large-scale sequencing, and sequence analysis are given in Künstner et al. (2010), which also includes accession numbers for the bird sequences analyzed in this study. Briefly, we isolated RNA from adult brain samples, prepared normalized cDNA, and ran this on a Roche 454 GS-FLX platform. The reads, which averaged 250 bp in length, were assembled into contigs using the NEWBLER software v2.0 distributed with the 454-instrument. We assumed as unknown each base pair with a quality score <20 (from the NEWBLER assembler).
Ortholog Selection and Supermatrix Building
We performed a reciprocal best-hit Blast analysis between contigs from the transcriptome data and zebra finch genes to select candidate orthologs. Amino acid sequence alignments were performed using the MAFFT software with high-accuracy parameters (Katoh et al. 2002). Contigs with premature in-frame stop codons or with pairwise amino acid divergence >30% relative to zebra finch sequence were excluded. Alignments were considered if they contained data from at least three newly sequenced species plus the zebra finch. Moreover, for each alignment, we removed sites with data available for less than four species, and we also removed genes with data for less than ten codons or with no parsimony informative sites (defined as polymorphic sites with a substitution shared by at least two species). This left data for 5,218 ENSEMBL v55-modeled genes. We then applied a four-step procedure to exclude misaligned and putative paralogous sequences using in-house C++ programs based on the BIO++ libraries (Dutheil et al. 2006).
The median pairwise divergence between all sequences of an alignment (D) was used to estimate the expected number of substitutions for each sequence (K) as K = D × sequence size (bp). We computed a P value for the excess of divergence in each sequence as Pr(X ≤ k), where X follows a Poisson distribution P(K), and k is the observed median number of substitution between that sequence and all the other sequences. We removed sequences with Pr(X ≤ k) < 0.01. This step was intended to exclude overdivergent sequences that are likely to represent paralogs, which was also confirmed after examining several examples of removed genes. We have observed that this empirical criterion also performs well for the removal of misaligned sequences.
We removed sequences from Neognathae species that were more divergent to zebra finch than the emu ortholog was. If emu was not included in the alignment, we used the chicken sequence as an “outgroup.” The position of Paleognathae (to which emu belongs) as a sister group to all other extant bird lineages (Neognathae) and of Galloanserae (chicken) as a sister group to Neoaves within Neognathae is considered uncontroversial.
We compared each sequence with the consensus sequence, that is, a sequence that showed the most common base at each site. Problematic regions of individual sequences were defined as regions with more than three contiguous variable sites compared with the consensus. Such sites were coded as “N,” as were any variable sites within 5-bp upstream or downstream of the problematic region. This step was included to remove small frameshifts (not leading to a stop codon) created by sequencing errors; the 454-technology is prone to errors in short stretches of homopolymer repeats (Harismendy et al. 2009).
In the last cleaning step, we estimated the phylogeny for each gene separately using a maximum likelihood (ML) method (RAxML software version 7.0.4 and a general time reversible [GTR] + Gamma4 model; Stamatakis 2006). We excluded from the analysis all genes that with >80% bootstrap support (100 times) indicated the nonmonophyly of oscines, Passeriformes, or Neoaves, taken to reflect obvious paralogous problem. Less than 30 genes were removed in this step, and it turned out that this criterion did not lead to a change in the topologies obtained (data not shown).
Using the list of zebra finch paralogous genes available in ENSEMBL, we identified the number of paralogs present in the set of genes removed by our cleaning process (2,328 of 3,223; 72.2%) and the number in the selected set of genes (1,264 of 1,995; 65.5%). The proportion of paralogs is significantly higher in the removed set of genes (chi-square = 8.227, degrees of freedom [df] = 1, P = 0.004), demonstrating that the empirical cleaning method is prone to more specifically remove paralogous sequences. However, it is important to note that some genuine orthologs may have unexpectedly high divergence and would therefore be removed by our cleaning process. Unfortunately, it is more or less impossible to tell apart overdivergent orthologs from paralogs with a partial set of genes generated by random transcriptome sequencing. In the end, one faces a trade-off between the risks of including paralogs and of excluding potentially informative orthologous sequences. Our approach could be considered conservative in this respect.
Phylogenetic Analyses
For analysis at the nucleotide level, we used ML with BPPML from the BPPSUITE package version 0.4.0 (Dutheil et al. 2006) and GARLI (version 0.951, Zwickl 2006). The model GTR + Gamma4 was selected as the best according to AIC using JMODELTEST, with a neighbor-joining tree as a fixed topology (Posada 2008). A model combining Gamma distribution and category of invariable site was not considered as it could generate a correlation between the proportion of invariable sites and the gamma shape parameter (see Yang 2007b, p. 113–114). We assessed the robustness of trees using bootstrap analysis. Analyses from both ML programs resulted in identical topologies and highly similar bootstrap support (data not shown).
For analyses at the protein level, we used PHYLOBAYES (Lartillot and Philippe 2004) and the site-heterogeneous mixture model CAT + Gamma4. Two independent Monte Carlo Markov Chains (MCMCs) were run in parallel for 10,000 cycles, saving one point every cycle and discarding the first 2,000 cycles as the burn-in cycles. Convergence was checked using the program BPCOMP of PHYLOBAYES that compares the frequency of the bipartitions obtained in the two independent chains (maxdiff < 0.05). Bayesian posterior probabilities were obtained from the 50% majority-rule consensus tree of the 8,000 MCMC sampled trees using the program READPB of PHYLOBAYES. We assessed the robustness of phylogenomic inference using both gene sampling and site sampling applying bootstrap processes. For each of the 100 bootstrap analyses, we estimated the 50% majority-rule consensus tree of the 1,000 MCMC sampled trees (with 500 points as a burn-in). The gene bootstrap support and the site bootstrap support were obtained using the 100 consensus trees obtained in each analysis.
Statistical analyses were preformed with R (R Development Core Team 2004) using the APE package (Paradis 2007) for plotting phylogenies.
Nonhomogeneous Model and GC Content Estimation
We extracted the 1st, 2nd, and 3rd codon positions of each alignment and calculated the GC content at each position separately (hereafter GC1, GC2, and GC3). We used the nonhomogeneous model of DNA sequence evolution proposed by Galtier and Gouy (1998) (T92 + Gamma5) and available in the BPPSUITE package (Dutheil and Boussau 2008) to infer both the ancestral GC3 at each node and the equilibrium GC3 (GC3*) for each branch. The GC3* corresponds to the model parameter θ and could be interpreted as GC3 under a branch length of infinite size, providing information on the on-going substitution process. The ancestral GC3 for the root is a model parameter (Galtier and Gouy 1998), whereas for the other nodes, it is estimated using an empirical Bayesian method implemented in the BIO++ library (Dutheil et al. 2006) and available in the BPPANCESTOR software of BPPSUITE. This approach takes the ML estimated from all the parameters (including topology and branch lengths) together with the sequence data to estimate a posterior probability of a given base at each site for each internal node (the highest probability corresponding to the estimated base). In order to assess the influence of missing data on GC3 and GC3* estimation, we performed the analysis on 100 bootstrapped matrices and present the 95% confidence interval (CI).
To test for a link between recombination rate and GC3 evolution, we performed two analyses.
First, we divided genes into two sets according to their position in the zebra finch genome: genes located in subtelomeric regions (<15 Mb from chromosome ends and including all microchromosomes of size <15 Mb) and subcentromeric regions (essentially central parts of macrochromosomes), respectively. Only autosomal genes with a known location were considered. These two gene sets can broadly be taken to represent high- and low-recombination landscapes, respectively (Groenen et al. 2009; Backström et al. 2010). When we instead considered the position in the chicken genome, only ∼10% of genes changed categories, confirming the extreme karyotypic conservation in most bird species (Griffin et al. 2007; Ellegren 2010). We estimated separately for the two gene sets both ancestral GC3 at each node and GC3* for each branch using the same method as above.
Second, we allocated genes in 5 Mb windows according to their position in the zebra finch genome. For each window, we removed species with less than 50 bp of sequence data. We also excluded windows that contained less than a total amount of 1.5 kb of exonic sequence, in both cases to reduce the variance in the parameter estimates. We estimated GC3* and the current GC3 in the lineage leading to the zebra finch, and following Meunier and Duret (2004), we correlated both the current GC3 and the GC3* with the mean recombination rate per window, using data from Backström et al. (2010).
Fossil Calibrations and Molecular Rate Estimation
We applied Bayesian methods that allow substitution rates to vary between branches, the so-called relaxed clock method. We used the nucleotide data set with MCMCTREE software (version4.4c available in the PAML package; Yang and Rannala 2006; Rannala and Yang 2007; Yang 2007a) and an HKY85 + Gamma4 model of sequence evolution. HKY85 + Gamma4 was chosen because it is the most complex model implemented in MCMCTREE (GTR + Gamma4 is not yet implemented). Moreover, there should be no need to use a more complex model since the branch lengths estimated with GTR and HKY85 models are extremely similar, implying that the substitution rates estimated with the two models must be very similar as well (data not shown). We used the autocorrelated (Brownian motion) model of rate evolution (corresponding option clock = 3 in MCMCTREE control file; Thorne et al. 1998). We ran four chains during 200,000 MCMC steps. The divergence dates were estimated using the last 100,000 steps of each chain (i.e., burn-in of 100,000 steps).
Brain transcriptome data from the American alligator (A. mississippiensis) was used as outgroup sequence. As a starting point, we considered the bird–crocodile calibration point provided by Benton and Donoghue (2007) at 235–254 My. These authors also proposed to constrain the Paleognathae–Neognathae split to between 66 and 86.5 My. This calibration is in agreement with the fossil record, which shows no trace of Neornithes but includes several non-Neornithes taxa in the lower Cretaceous (between 145 and 100 My; see Chiappe and Dyke 2002; Fountaine et al. 2005). However, this calibration is younger than that estimated based on molecular dating, which places the Paleognathae–Neognathae split in the lower Cretaceous or around the lower/upper Cretaceous boundary (∼100 My) (Ericson et al. 2006; Pereira and Baker 2006; Slack et al. 2006; Baker et al. 2007).
The oldest passerine fossil known is from the early Eocene (55 My; Boles 1995). The oldest suboscine songbird fossil is from the lower Oligocene in Europe (28–34 My; Mayr and Manegold 2006). A recent description of a Certhioidea (crown group of Passerida) fossil from the early Miocene (20 My; Manegold 2008) provides a useful minimum constraint for the Passerida split in the late Miocene (23 My).
Based on the available data described above, we choose to use a set of four of calibration points: 1) the bird–crocodile split between 235 and 254 My, 2) the Paleognathae–Neognathae split between 66 and 86.5 My, 3) the oscine–suboscine split younger than 65.5 My, and 4) the first split within the Passerida older than 23 My. We also estimate the substitution rate variation between lineages using the local molecular clock approach implemented in BASEML (Yoder and Yang 2000; Yang 2007a) using a GTR + Gamma4 model of sequence evolution. To study the intensity of natural selection in specific lineages, we applied the branch model implemented in CODEML (Yang 2007a) allowing for different ratios of the nonsynonymous and the synonymous substitution rates (w) in different lineages (model = 2 of CODEML).
Results
Impact of Base Composition on Phylogenetic Reconstruction
In reciprocal Blast analyses, we mapped the 454-derived transcriptome contig sequences from nine bird species onto the zebra finch genome, which allowed orthologous assessment of genes sequenced in different genomic regions. Due to the random nature of shotgun transcriptome sequencing, different overlapping sets of cDNA sequences were present in different avian samples. To handle this combinatorial gene mixture, we built a matrix for making phylogenomic inference. The matrix contained sequence data from 1,995 genes present in the zebra finch genome, representing more than 10% of the estimated total number of genes in avian genomes (ICGSC 2004; Warren et al. 2010). These genes have a total length after concatenation of 774,279 bp of exonic sequence. The amount of missing data was unevenly distributed across species and high for some, from 3.5% for the zebra finch to 87.9% for the ruby-throated hummingbird (table 1); the missing data for the zebra finch was the result of removing partial finch sequences during the supermatrix building process (see Materials and Methods). However, we still had mostly over 100 kb of exonic sequence per species, which is unprecedented in studies of avian molecular evolution in terms amount of sequence data.
Table 1.
Basic Sequence Statistics for Each Species.
| Species | Number of Genes | Exonic Sequence Length in bp (% of missing data) |
| Ruby-throated hummingbird | 271 | 93,594 (87.9) |
| Anna's hummingbird | 398 | 143,514 (81.5) |
| American crow | 555 | 189,681 (75.5) |
| Emu | 625 | 212,751 (72.5) |
| Pied flycatcher | 1,627 | 631,677 (18.4) |
| Chicken | 1,390 | 550,188 (28.9) |
| Golden-collared manakin | 794 | 285,240 (63.2) |
| Budgerigar | 528 | 154,815 (80.0) |
| Blue tit | 1,298 | 506,157 (34.6) |
| Collared dove | 375 | 133,455 (82.8) |
| Zebra finch | 1,912 | 746,739 (3.6) |
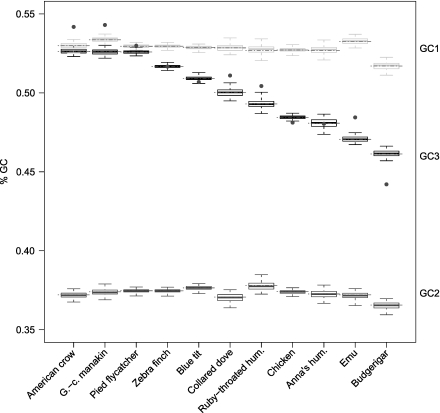
We found that the GC content in the third codon position (GC3) varied significantly across species (black boxes in fig. 1). Passerines had the most GC3-rich transcriptome (GC3 = 52.1%, bootstrap value range 50.9–52.6%). The budgerigar and the emu had the least GC3-rich (46.1% and 47.0%, respectively). In contrast, the GC1 and GC2 contents (1st and 2nd codon positions) were almost identical across species (gray and dark-gray boxes in fig. 1, respectively).
FIG. 1.
GC content at the 1st, 2nd, and 3rd codon position (GC1, GC2, and GC3) in light-gray, dark-gray, and black boxes, respectively. Dashed red lines show the observed value (number of Gs + Cs divided by the sequence length). Boxes give the quartiles of the distribution of the GC content obtained by bootstrapping of the original matrix (100 times); whiskers extend to the most extreme values obtained by bootstrapping. Grey dots are values corrected for missing data (see text).
There was also significant variation in GC3 among genes within a species. To evaluate this variation, we distributed genes in 5 Mb windows according to their position in the zebra finch genome. The between-window variation in GC3 was very large in all species, with an average standard deviation of 0.10 (fig. 2A), as compared with an average standard deviation of 0.002 of the bootstrap values (black boxes, fig. 1). This variation in GC3 among genes showed consistency across species since GC3 of particular windows varied from 33% to 77% when averaged across species. Finally, the between-species standard deviation of GC3 per window varied from 0.02 to 0.18 and was positively correlated to the mean GC3 per window (Spearman's Rho = 0.42, P = 1.3 × 10−9, n = 194, 5Mb windows: fig. 2B). These results demonstrate that the highest variation in GC3 among lineages was concentrated to the most GC3-rich regions of the genome. GC1 and GC2 showed much less variation between species than GC3 (standard deviation of 0.042 and 0.046, respectively).
FIG. 2.
(A) Distribution of GC3 in 5 Mb windows according to their position in the zebra finch genome. Boxes give the quartiles of the distribution; whiskers extend to the most extreme values obtained by bootstrapping. (B) Relationship between mean GC3 in 5 Mb windows and the corresponding standard deviation.
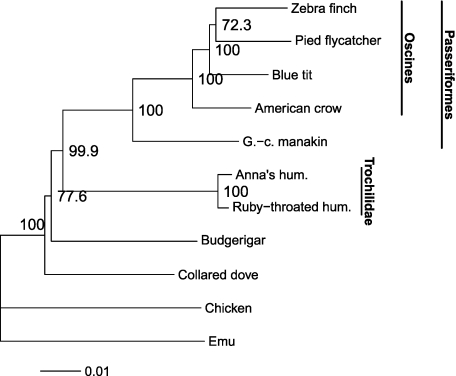
To make phylogenetic inference, we started with a classical ML analysis using nucleotide data (GTR + Gamma4 model) without any attempt to control for GC3 variation. This analysis confirmed without ambiguity the Neoavian (beyond chicken) and the passerine monophylies and the basal suboscine (manakin) and oscine songbird (other passerines) dichotomy of passerines (fig. 3). Within the passerines and in support of previous findings (Barker et al. 2004) we found strong support for the split between Corvoidea (i.e., crows) and Passerida (other oscines). A similar topology, except concerning the zebra finch/pied flycatcher relationship, was obtained using a three codon position partitioned GTR + Gamma4 model (data not shown).
FIG. 3.
ML tree obtained with a GTR + Gamma4 model and the complete nucleotide data set. Scale indicates substitution/site.
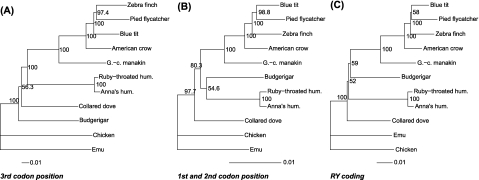
To investigate the influence of GC3 heterogeneity on phylogenetic inference, we split our data set according to codon position, separating the 3rd position from the 1st + 2nd positions. Interestingly, ML analyses of the two data sets differed with respect to the position of the budgerigar (a parrot) and on the zebra finch/blue tit/pied flycatcher relationship. In agreement with data from DNA hybridization (Sibley and Ahlquist 1990) and whole mitochondrial genome sequence analyses (Pratt et al. 2009), the 3rd codon position data set did not support budgerigar as a sister group to passerines (fig. 4A) (Ericson et al. 2006; Hackett et al. 2008). This topology was most similar to the one obtained with the full data set (fig. 3). In contrast, the 1st + 2nd codon position data set supported a closer relationship between budgerigar and passerines (fig. 4B), in partial agreement with recent results obtained with nuclear markers (Ericson et al. 2006; Hackett et al. 2008), but this topology grouped budgerigar and hummingbird but with moderate bootstrap support (55%; fig. 4B). The 3rd codon position favored the zebra finch/flycatcher clade reported in Barker et al. (2004), whereas the 1st + 2nd codon position data set supported a blue tit/pied flycatcher cade reported in Johansson et al. (2008), each with comparable strong bootstrap support (>97; fig. 4A and B). The blue tit/pied flycatcher /zebra finch relationship within the basal Passerida radiation is known to be notoriously difficult to resolve (for a review, see Johansson et al. 2008). The different topologies obtained with the two data sets were not due a difference in the number of informative sites; jackknifing the 3rd codon position data set to a similar number of sites as for the 1st + 2nd codon position data set strongly supported the same topology as obtained using the full 3rd codon position data set.
FIG. 4.
Phylogenetic trees based on transcriptome data. (A) ML tree obtained with a GTR + Gamma4 model and the third codon position data set. (B) ML tree obtained with a GTR + Gamma4 model and the 1st and 2nd codon position data set. (C) ML tree obtained with a GTR + Gamma4 model and the nucleotide data set with GC recorded as RY. Values are site bootstrap support (1,000 times). Scale indicates substitution/site; note the difference in the scale between trees (A) and (B).
The above analyses suggest that GC3 content in the full nucleotide data set is significantly influencing the overall topology of the avian tree with important contradictions to GC1 + 2. To this hypothesis, we estimated the phylogeny using the full nucleotide data set with GC3 content normalized. To do so, we used the RY sequence recoding method, with purines coded as R and pyrimidines as Y. RY recoding of the transcriptome placed the budgerigar as a sister group to passerines, more consistent with the result obtained with the 1st + 2nd codon position data set, again with moderate bootstrap support (59%; fig. 4C). This data set also supported the blue tit/flycatcher relationship (bootstrap support of 58%) as did the 1st + 2nd codon position data set. This finding indicates that the discrepancy between the phylogenetic results obtained with different codon positions is a result of the large heterogeneity in GC3.
To make an independent test of overall GC influence, we applied the nonhomogeneous model of nucleotide sequence evolution developed by Galtier and Gouy (1998). This model allows variation in GC content between branches (regardless of codon position) by relaxing the assumption of homogeneous base composition. We estimated the ML of several alternative topologies using a branch-specific nonhomogeneous model with the T92 + Gamma4 substitution model (supplementary fig. S1, Supplementary Material online). The highest likelihood was obtained for a topology (supplementary fig. S1C, Supplementary Material online) identical to the one obtained with the homogeneous model (fig. 3). This result indicates that relaxing the stationary assumption is not sufficient to disrupt the budgerigar position and that additional features such as, for example, long-branch attraction (Felsenstein 1978; Bergsten 2005), among-site rate variation (Yang 1996), and heterotachy (among branch/site rate variation) (Lopez et al. 2002) contribute to this result.
Finally, we applied a CAT mixture model (Lartillot and Philippe 2004) using Bayesian analysis on proteome (amino acid) data, which has been shown to outperform empirical substitution matrices in the case of long-branch attraction problems (e.g., Lartillot et al. 2007; Delsuc et al. 2008; Philippe et al. 2009). Almost all relationships had high bootstrap support (fig. 5), except for the zebra finch/blue tit/pied flycatcher node. Notably, the topology inferred with the CAT mixture model placed budgerigar as the sister group to passerines (fig. 5). But unlike the nucleotide trees, it also brought doves closer to passerines.
FIG. 5.
Unrooted proteome tree. Majority-consensus tree of Bayesian phylogenetic inference conducted under the CAT + Gamma4 mixture model using the software PHYLOBAYES. Values behind the nodes are site bootstrap support (100 times). Values in front of the nodes in italics are genes bootstrap support (100 times). Branch lengths are ML estimate using WAG + Gamma8 model. Scale indicates substitution/site.
In summary, the position of budgerigar is strongly influenced by phylogenetic method and the data set used. Because the basal position is disrupted upon the use of 1) only the 1st + 2nd codon positions, 2) RY sequence recoding, and 3) a complex CAT mixture Bayesian model applied on proteome sequences, we suggest that the basal position of budgerigar indicated by the full data set was most likely an artifact of phylogenetic reconstruction at least in part caused by the similarity in base composition between the budgerigar and emu. Of course, we cannot exclude the possibility that there may have been other factors that contributed to this result.
Evolution of Base Composition in Birds
To further explore the variation in GC3 content among species and to understand the evolution of base composition in birds, we assessed the impact of missing data. We compared GC3 variation across species when estimated on the entire data set and when estimated on data sets based on sites only available in one species relative to all other species (replicated 11 times = the number of species). The rank order of all GC3 estimates using these reduced species-defined data sets correlated extremely well with the ranking obtained from the complete matrix (Spearman's Rho between 0.90 and 0.99, mean = 0.95; supplementary fig. S2, Supplementary Material online), showing that missing data does not explain the relative differences in GC3 among species. However, for some reduced data sets, GC3 of all species were systematically higher or lower than that obtained using the complete data set (supplementary fig. S2, Supplementary Material online). To quantify this deviation, we computed for each reduced data set the mean of the differences between GC3 of the complete data set and of the reduced data set. This value (deltaGC) provides an approximation of the GC3 deviation introduced by gene sampling in each species; a positive value, for example, indicates that the selected genes have, on average, a lower GC3 than the complete set of genes. The most extreme deviation was observed for the manakin and the budgerigar data sets with—1.7% and +1.9% deltaGC, respectively. Using these values, we calculated a corrected GC3 for each species by subtracting the deviation to the current GC3. After correcting for the missing data, there is still substantial variation among species in GC3 (fig. 1, grey dots). For example, the extreme GC3 content of the budgerigar transcriptome cannot be explained by biased sampling and even seems to have been underestimated in the original analyses (original estimate = 46.1%, corrected estimate = 44.4%).
The observed variation in GC3 among species must translate into differences in the substitution pattern of third codon position among lineages. To estimate the ancestral GC3 at each node and the equilibrium GC3 (GC3*) for each branch, we applied a nonhomogeneous model of DNA sequence evolution (Galtier and Gouy 1998) on third codon positions, using the proteome topology (fig. 5). This analysis showed an increase in GC3 in most branches of the Neognath phylogeny as manifested by a higher GC3* (fig. 6B) than ancestral GC3 (fig. 6A). The branch leading to the budgerigar has the smallest GC3* (52.4%) of all Neoaves branches, although this GC3* is still higher than the ancestral GC3 in Neoaves (43.0%). It suggests that the low-current GC3 estimated for the budgerigar is unlikely to come from a decrease of GC3 in the parrot lineage but rather represents a moderate increase of the low ancestral GC3. Interestingly, an increase in GC3 appears to have occurred independently in the Neoaves lineage and along the chicken branch, as GC3* at the ancestor node of all Neoaves (43.0 %, CI = 37.2–49.1; fig. 6B) was smaller than GC3 estimated at the root of the Neognathes (47.3%, CI = 47.0–47.6; fig. 6A). The ancestral Neoaves branch and the branch leading to ratites (emu) were the only lineages showing a GC3* below the ancestral root value (fig. 6A and B). These findings suggest that there has been convergence to a higher GC content in Neoaves and chicken lineages. The most marked increase in GC3* has taken place in the branch leading to passerines, with an extremely high GC3* (74.9%, CI = 72.2–77.3; fig. 6B). This implies a strong fixation bias toward Gs and Cs in the third codon position of passerines. The fact that GC3* is higher in all terminal passerine branches than current GC3 of passerines demonstrates an on-going process of GC enrichment in this order.
FIG. 6.
(A) Estimation of ancestral GC3 content at each node and current GC3 for each species and (B) Equilibrium GC3* estimated for each branch of the avian phylogeny. Values in brackets show the 95% bootstrap (100 times) CIs.
We sought a functional genomic explanation for the large-scale variation and convergence of GC3 in the avian transcriptome and considered recombination. Recombination may drive the evolution of GC content through GC-biased gene conversion (gBGC) (Galtier et al. 2001; Meunier and Duret 2004; Spencer et al. 2006). Of relevance here, recombination rates in avian genomes are highly heterogeneous and correlate negatively with both distance to chromosome ends (i.e., telomere) and chromosome size (ICGSC 2004; Groenen et al. 2009), with the telomere effect being particularly strong in zebra finch (Backström et al. 2010). To test if the increase in GC3 in passerines is linked to an increased recombination rate, we performed two additional analyses.
First, we analyzed two gene sets defined as subtelomeric and subcentromeric genes, respectively, according to their position in the zebra finch genome (see Materials and Methods). For these two gene sets, the ancestral GC3, current GC3, and GC3* were almost consistently higher in subtelomeric compared with subcentromeric regions (table 2). Moreover, we found that zebra finch genes located in subtelomeric regions were significantly more GC3-rich than orthologous chicken genes (55.4%, CI = 55.2–55.6 vs. 50.6%, CI = 50.3–50.8), a difference not found in subcentromeric regions (46.5% CI = 46.3–46.7 vs. 45.8% CI = 45.6–46.0). Moreover, the GC3* estimated for the ancestral passerine branch was strikingly higher for genes in subtelomeric compared with subcentromeric regions (82.6% CI = 80.0–85.0 vs. 57.7% CI = 54.0–60.1). These findings suggest that the strong increase in the GC3 in passerines is associated with a high-recombination rate environment.
Table 2.
Ancestral GC3, Current (observed) GC3, and Equilibrium GC3 (GC3*) in Subtelomeric (<15 Mb from chromosome ends) and Subcentromeric Regions (>15 Mb from chromosome ends) of Different Nodes and Branches of the Avian Phylogenetic Tree.
| Subtelomeric Regions, (n = 1,074) |
Subcentromeric Regions, (n = 756) |
|||||
| Node or Branch | Ancestral GC3 | Current GC3 | GC3* | Ancestral GC3 | Current GC3 | GC3* |
| Zebra finch | 55.4 (55.2–55.6) | 0.664 (0.644–0.670) | 46.5 (46.3–46.7) | 49.3 (48.3–50.7) | ||
| Chicken | 50.6 (50.3–50.8) | 52.5 (51.6–53.7) | 45.8 (45.6–46.0) | 50.8 (49.8–52.1) | ||
| Oscine/suboscine | 52.8 (52.5–53.1) | 82.3 (80.0–85.0) | 44.9 (44.7–45.1) | 57.7 (54.0–60.1) | ||
| Ancestral Neoaves | 49.3 (49.1–49.7) | 42.0 (35.5–49.8) | 44.1 (43.9–44.3) | 26.5 (20.4–31.9) | ||
Second, we took advantage of the regional recombination rates that have recently been estimated for the zebra finch genome (Backström et al. 2010), to seek a more direct relationship between evolution of GC3 and recombination rate. Using the 5 Mb windows defined above, we found a strong correlation between GC3 and the mean recombination rate per window (Spearman's Rho = 0.65, P < 2 × 10−16, n = 134, fig. 7A). Interestingly, GC3* was also strongly correlated with recombination rate (Spearman's Rho = 0.54, P = 11 × 10−11, n = 134, fig. 7B), suggesting that recombination might have a strong effect on the substitution pattern in the branch leading to the zebra finch. In agreement with a previous study of chicken, based on retrotransposon elements (Webster et al. 2006), we found a strong correlation between GC3 and GC3* (Spearman's Rho = 0.67, P < 2 × 10−16, n = 134) with GC3* almost always higher than current GC3 (in 102 of 134 windows). These finding indicates that the increase in GC3 is still an on-going process in zebra finch protein-coding genes.
FIG. 7.
Relationship between recombination rate (in cM/Mb; sex-averaged; extracted from Backström et al. 2010) and (A) the current GC3 as well as (B) the equilibrium GC3 (GC3*). Sizes of the circles are proportional to the length of the alignments used in each 5 Mb window. Red lines indicate lowest fits of the data considering each window with the same weight.
Substitution Rate Variation among Lineages
We also used the transcriptome data set to address nucleotide substitution rate variation among avian lineages. A simple molecular clock model (i.e., one rate for all branches) was strongly rejected by a likelihood-ratio test (χ2 = 2508, df = 10, P < 2 × 10−16). To further investigate substitution rate variation among lineages, we applied a Bayesian relaxed molecular clock method using the MCMCTREE software (version 4.4c). This method estimates simultaneously substitution rates and divergence dates across the phylogeny, although we here focus on substitution rates only. We used the topology found in the proteome Bayesian analysis as the reference topology (fig. 5).
In order to be able to use the well-known bird/crocodile fossil calibration point at 235 and 254 My (Benton and Donoghue 2007), we incorporated brain transcriptome sequences of an American alligator. A reciprocal Blast analysis found 612 crocodile orthologs in the set of 1,995 avian genes used in the phylogenetic matrix. After removal of genes using the same selection process as for the bird data, we obtained 488 alligator genes spanning 161,251 bp in exonic length, with 79.1% of missing data for the alligator. In addition to the bird/crocodile split at 235–254 My, we use as calibration points several suggested divergence times within Aves (Paleognathae–Neognathae split, oscine–suboscine split, and first split within Passerida), as described in Materials and Methods.
Excluding the branch leading to the American alligator, the mean substitution rate per branch was 0.683 (±0.300) substitutions/site/billion years. Some general conclusions about substitution rate evolution across the avian phylogeny can be drawn (table 3; excluding short branches with large CIs). First, passerines evolve 45% more rapidly than other birds (0.768 vs. 0.530). This difference is even more pronounced when considering the flycatcher–zebra finch clade, having the highest rate of the whole tree with a mean of 0.895 (table 3). Second, the long ratite branch leading to emu had the lowest rate of substitution, with 0.335. There is thus 2.6-fold variation in substitution rate between the most extreme lineages. Excluding the bird/crocodile calibration point does not affect these results (data now shown). Moreover, we obtain similar substitution rate variation using the local molecular clock approach implemented in BASEML (clock = 2, model GTR + Gamma4; Yoder and Yang 2000). For example, using this approach we estimate that passerines evolve 48% faster than other birds.
Table 3.
Median Substitution Rate (substitution per site per billion of years) Estimated for Different Branches of the Avian Phylogeny.
| Clade | Branch | Substitution Rate |
| Passeriformes | Zebra finch | 0.87 (0.80, 1.25) |
| Pied flycatcher | 0.92 (0.85, 1.33) | |
| Blue tit | 0.58 (0.55, 0.76) | |
| American crow | 0.49 (0.46, 0.64) | |
| Golden-collared manakin | 0.55 (0.51, 0.71) | |
| Passerine root | 1.20 (1.03, 2.28) | |
| Nonpasserine Neoaves | Budgerigar | 0.58 (0.55, 0.70) |
| Collared dove | 0.51 (0.49, 0.62) | |
| Hummingbirdsa | 0.47 (0.40, 1.74) | |
| Neoaves root | 0.72 (0.62, 1.32) | |
| Galloanserae | Chicken | 0.56 (0.53, 0.66) |
| Palaeognathae | Emu | 0.34 (0.33, 0.40) |
NOTE.—95% CIs are provided in parentheses.
Ancestral + hummingbird branches.
To test if substitution rate estimation is related to gene sampling, we used the subset of sites only present in the emu (212,751 sites) and compared the results with that obtained using all data. We found that the rate estimates were extremely well correlated (Spearman's Rho = 0.96, P = 3.7 × 10−6, n = 22) and that the emu-defined subset of sites quantitatively reproduce the amount of variation estimated using the complete data set (for instance, for the emu-defined subset emu substitution rate = 0.343 and pied flycatcher = 0.840). This result shows that the low-substitution rate observed in the ratite branch was not the result of gene sampling bias. We also replicated rate estimates using only Anna's hummingbird- or Dove-specific sites and found highly comparable substitution rate estimation (data not shown).
Finally, because we estimated substitution rates using protein-coding genes, the observed rate variation among lineages could in theory be linked to mutation rate variation or differences in the regimes of natural selection acting on protein evolution (or both). In case of the latter, a selection model may imply that the overall rate of adaptive evolution or the overall rate of fixation of slightly deleterious mutations differ among avian lineages. Under both of these scenarios, the nonsynonymous substitution rate will vary among lineages, thereby contributing to the overall rate variation in protein-coding sequence. Lineage-specific variation in the role of natural selection has been suggested to occur due to long-term differences in the effective population size among lineages (Ellegren 2010).
We assessed possible variation in the intensity of natural selection across the bird phylogeny by estimating the nonsynonymous/synonymous substitution rate ratio (ω) for specific branches: all passerines, all nonpasserine Neoaves, the chicken, and the emu lineages. Passerines showed an intermediate ω of 0.0784, slightly lower than that for nonpasserine Neoaves (0.0847) but higher than that for emu (0.0586) and chicken (0.0626). This result indicates that the increase in passerine substitution rate is not linked to selective processes affecting nonsynonymous substitution only but more likely to an increase in mutation rate affecting both synonymous and nonsynonymous substitution rates.
Discussion
The Influence of Base Composition Variation on Neoavian Phylogeny
This is the first high-throughput next-generation sequencing application to avian phylogenomics that we are aware of. We discovered that even with many more genes than in all previous studies of avian phylogenetics based on standard DNA sequencing, the inferred phylogenies can substantially differ depending on sequence information content. Specifically, the GC3 content seemed to have a major affect on the placement of budgerigar in the phylogeny. Standard ML analysis using the full nucleotide data set did not reveal parrots (budgerigar) as a sister group to Passeriformes. In contrast, when excluding the third codon position or normalizing GC variation by RY coding, budgerigar was placed close to the passerines. In the nonnormalized analysis, the placement of budgerigar near the base of Neoaves is likely due to similarity of GC3 content in budgerigars and basal avian species.
However, the results obtained with a nonhomogeneous model indicated that the GC variation was not the only cause of the discrepancy between topologies. One possible additional factor may be that third codon position is fast evolving and therefore also more prone to long-branch attraction problems. The existence of multifactorial problems in phylogeny inference has been illustrated in other data sets (e.g., Sheffield et al. 2009). The statistical inconsistency of the homogeneous (stationary) model of nucleotide substitution is not surprising given that its underlying assumption is violated by heterogeneity in base composition. It has been shown that such inconsistency is exacerbated by low taxonomical sampling (Delsuc et al. 2005; Jeffroy et al. 2006). Taxon sampling is at present stage bound to be limited in phylogenomic analysis based on high-throughput sequence data due to the extensive costs associated with next-generation sequencing, particularly so when it comes to transcriptome sequencing using normalized cDNA.
The Evolution of Vocal Learning
The inferred phylogenies can lead to critically different conclusions on the evolution of convergent complex traits. For example, resolving the phylogenetic position of parrots has implications for understanding the evolution of vocal learning, a critical behavioral substrate for speech. A striking feature of this trait is that all three groups of birds that are vocal learners—oscine songbirds, parrots, and hummingbirds—have a set of seven similar forebrain nuclei distributed into two brain pathways that control song imitation (Jarvis and Mello 2000; Jarvis et al. 2000; Jarvis 2004). Vocal nonlearners (e.g., suboscine songbirds, dove, and chicken) do not have any of these brain nuclei (Brenowitz 1997; Feenders et al. 2008). Analogous vocal learning pathways are found in the human brain but not nonhuman primate (Jarvis 2004). Thus, vocal learning pathways are thought to represent a case of remarkable convergence for a complex trait that is a critical substrate for spoken human language (Doupe and Kuhl 1999; Wilbrecht and Nottebohm 2003; Jarvis 2004). The recent sister grouping of parrots and passerines was supported by the topology based on nuclear genes (Ericson et al. 2006; Hackett et al. 2008) but not by the analysis of complete mitochondrial genomic data (Gibb et al. 2007; Pratt et al. 2009) or past results based on DNA–DNA hybridizations (Sibley and Ahlquist 1990). Therefore, instead of three independent gains of vocal learning among birds as has been proposed for a long time based on past phylogenies (Nottebohm 1972; Jarvis 2004), topologies with parrots as sister to passerines (Ericson et al. 2007; Hackett et al. 2008; this study) raise the possibility that vocal learning may have evolved just twice in birds, once in hummingbirds and once in the common ancestor of parrots and passerines (and then lost in suboscine songbirds). However, the possibility of a “two-gains-and-one-loss” origin of vocal learning in Neoaves should be considered equally parsimonious as the “three-gains” hypothesis. Resolving the truth may not only require greater genome coverage and taxonomic sampling, but our findings indicate that it will also require comparing phylogenetic inference made with different models (varying in their assumption and complexity) and different subsets of the data (1st, 2nd, and 3rd codon positions, RY sequence recoding, or proteome sequence).
The Evolution of Heterogeneous Base Composition in Birds
Our study reveals an unexpected dynamic pattern of base composition evolution in protein-coding genes of birds. This pattern consists of an increase in GC3 in almost every branch of the avian phylogeny with marked exceptions in the ancestral branch leading to Neoaves and Neognathae and in the terminal Paleognathae (emu) lineage. Moreover, the increase is more pronounced in some lineages (like in passerines) than in others (like parrots). The pattern is compatible with the hypothesis that GC3 content in budgerigar and emu reflects the ancestral state, whereas other Neoaves lineages have evolved an increase in GC3.
The dynamic picture of GC3 content evolution we describe echos the revisited view of GC content evolution in mammals (Romiguier et al. 2010). Classically, the GC content of mammalian lineages has been thought to decline since the ancestor of placental mammals (Duret et al. 2002; Belle et al. 2004). However, an extensive analysis of 33 mammalian genomes has revealed a more dynamic picture where several lineages have undergone an independent increase in GC content (Romiguier et al. 2010).
Our results beg for an explanation to the variation in GC3 seen among avian lineages. One possible mechanism would be gBGC. The observation in several organisms of a correlation between GC content and recombination rate (see table 1 in Glémin 2010) has led some authors to propose a recombination-associated repair mechanism favoring GC over AT, in the form of gBGC. Since its original proposition, gBGC has been strengthened by direct experimental evidence such as analysis of meiotic recombination products in yeast (Mancera et al. 2008). Moreover, there are clear examples in mammals that show that an increased regional recombination rate has led to a drastic increase in regional GC content (Galtier 2004) and that GC* is directly correlated with recombination rate (Galtier 2004; Meunier and Duret 2004; Webster et al. 2005; Dreszer et al. 2007, Duret and Arndt 2008).
We found that the strong increase in GC3 content in passerines was concentrated to high-recombination rate (subtelomeric) regions. We also found a direct correlation between regional (5 Mb windows) GC3 content and recombination rate in zebra finch. Furthermore, the observed correlation between GC3* in the zebra finch lineage and zebra finch recombination rate supports the hypothesis that recombination rate is driving the increase in GC3. This correlation is in line with previous results obtained with chicken CR1 retrotransposon repeat elements, which showed that GC is maintained and even reinforced in the most GC-rich regions of the chicken genome (Webster et al. 2006). In conclusion, we cannot formally demonstrate that gBGC explains the evolution of base composition in birds. However, we can conclude that our observations are consistent with the expectations from the gBGC hypothesis.
The Rate of Molecular Evolution in Birds
There was a clear rate rank order of ratites–nonpasserines–passerines from slow to fast evolving. This variation in the substitution rate does not appear to be dependent of gene sampling or to an overall change in the intensity of natural selection acting on protein evolution. The accelerated evolution of the passerine transcriptome that we detect is consistent with previous observations made using exonic and intronic sequences (Hackett et al. 2008) and based on sequence motif evolution (Edwards et al. 2002). Nabholz et al. (2009) used mtDNA data and extensive taxonomic sampling to estimate that the neutral substitution rate of passerines is 2.3 times higher than that of other birds. This is similar to the figures estimated herein.
Conclusions
Here, we have presented an extensive phylogenomic analysis of avian transcriptome data obtained through next-generation sequencing technology. The analysis revealed several important results. 1) A confirmation that the stationary assumption of nucleotide evolution is critical to consider in phylogenetic reconstruction. 2) An unexpected dynamic pattern of GC3 evolution in the avian phylogeny with a marked increased in passerines and possible a link with recombination through gBGC. 3) Substantial variation in substitution rate among lineages where the passerines appear as the fastest evolving among the taxa studied here. More generally, our study demonstrates the usefulness of deep transcriptome sequencing for analyses of phylogenomics and molecular evolution. At the same time, it demonstrates limitations. Despite the very large amount of data collected, data matrices are likely to contain a lot of missing data in shotgun approaches directed toward the transcriptome (and of course even more so if genomic DNA is targeted). The use of normalized libraries (Barbazuk et al. 2007) would be one way to overcome this problem. Another issue is taxon sampling that in our case was not at par with what nowadays is common in phylogenetic studies based on individual genes or markers. The per-base costs for next-generation sequencing have shown a steady decrease over the few years the technology has been used, and there should be no reason to expect that they will not continue to do so. This, together with improved protocols for running many samples in parallel using tagged templates, should facilitate increased taxon sampling in phylogenomic work.
Supplementary Material
Supplementary figures S1–S2 are available at Molecular Biology and Evolution online (http://www.mbe.oxfordjournals.org/).
Acknowledgments
We thank Julien Dutheil for his advice on base composition analysis and the helpful comments made by two anonymous reviewers on a previous version of this manuscript. The work was funded by the Swedish Research Council, the European Research Council, and the Knut and Alice Wallenberg foundation (to H.E.) and by National Institute of Health (NIH) R01DC007218, an NIH Director's Pioneer Award, and the Howard Hughes Medical Institute (to E.D.J). The computations were performed on resources provided by the Swedish National Infrastructure for Computing at UPPMAX.
References
- Backström N, Forstmeier W, Schielzeth H, et al. 11 co-authors. The recombination landscape of the zebra finch Taeniopygia guttata genome. Genome Res. 2010;20:485–495. doi: 10.1101/gr.101410.109. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Baker AJ, Pereira SL, Paton TA. Phylogenetic relationships and divergence times of Charadriiformes genera: multigene evidence for the Cretaceous origin of at least 14 clades of shorebirds. Biol Lett. 2007;3:205–209. doi: 10.1098/rsbl.2006.0606. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Barbazuk WB, Emrich SJ, Chen HD, Li L, Schnable PS. SNP discovery via 454 transcriptome sequencing. Plant J. 2007;51:910–918. doi: 10.1111/j.1365-313X.2007.03193.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Barker FK, Cibois A, Schikler P, Feinstein J, Cracraft J. Phylogeny and diversification of the largest avian radiation. Proc Natl Acad Sci U S A. 2004;101:11040–11045. doi: 10.1073/pnas.0401892101. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Belle E, Galtier N, Duret L, Eyre-Walker A. The decline of isochores in mammals: an assessment of the GC-content variation along the mammalian phylogeny. J Mol Evol. 2004;58:653–660. doi: 10.1007/s00239-004-2587-x. [DOI] [PubMed] [Google Scholar]
- Benton MJ, Donoghue PCJ. Paleontological evidence to date the tree of life. Mol Biol Evol. 2007;24:26–53. doi: 10.1093/molbev/msl150. [DOI] [PubMed] [Google Scholar]
- Bergsten J. A review of long-branch attraction. Cladistics. 2005;21:163–193. doi: 10.1111/j.1096-0031.2005.00059.x. [DOI] [PubMed] [Google Scholar]
- Bernardi G. Isochores and the evolutionary genomics of vertebrates. Gene. 2000;241:3–17. doi: 10.1016/s0378-1119(99)00485-0. [DOI] [PubMed] [Google Scholar]
- Blanquart S, Lartillot N. A site- and time-heterogeneous model of amino acid replacement. Mol Biol Evol. 2008;25:842–858. doi: 10.1093/molbev/msn018. [DOI] [PubMed] [Google Scholar]
- Boles WE. The World's oldest songbirds. Nature. 1995;374:21–22. [Google Scholar]
- Brenowitz EA. Comparative approaches to the avian song system. J Neurobiol. 1997;33:517–531. doi: 10.1002/(sici)1097-4695(19971105)33:5<517::aid-neu3>3.0.co;2-7. [DOI] [PubMed] [Google Scholar]
- Chiappe LM, Dyke GJ. The mesozoic radiation of birds. Annu Rev Ecol Syst. 2002;33:91–124. [Google Scholar]
- Delsuc F, Brinkmann H, Philippe H. Phylogenomics and the reconstruction of the tree of life. Nat Rev Genet. 2005;6:361–375. doi: 10.1038/nrg1603. [DOI] [PubMed] [Google Scholar]
- Delsuc F, Tsagkogeorga G, Lartillot N, Philippe H. Additional molecular support for the new chordate phylogeny. Genesis. 2008;46:592–604. doi: 10.1002/dvg.20450. [DOI] [PubMed] [Google Scholar]
- Doupe AJ, Kuhl PK. Birdsong and human speech: common themes and mechanisms. Annu Rev Neurosci. 1999;22:567–631. doi: 10.1146/annurev.neuro.22.1.567. [DOI] [PubMed] [Google Scholar]
- Dreszer TR, Wall GD, Haussler D, Pollard KS. Biased clustered substitutions in the human genome: the footprints of male-driven biased gene conversion. Genome Res. 2007;17:1420–1430. doi: 10.1101/gr.6395807. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Duret L, Arndt PF. The impact of recombination on nucleotide substitutions in the human genome. PLoS Genet. 2008;4:e1000071. doi: 10.1371/journal.pgen.1000071. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Duret L, Semon M, Piganeau G, Mouchiroud D, Galtier N. Vanishing GC-rich isochores in mammalian genomes. Genetics. 2002;162:1837–1847. doi: 10.1093/genetics/162.4.1837. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dutheil J, Boussau B. Non-homogeneous models of sequence evolution in the Bio++ suite of libraries and programs. BMC Evol Biol. 2008;8:255. doi: 10.1186/1471-2148-8-255. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dutheil J, Gaillard S, Bazin E, Glemin S, Ranwez V, Galtier N, Belkhir K. Bio++: a set of C++ libraries for sequence analysis, phylogenetics, molecular evolution and population genetics. BMC Bioinformatics. 2006;7:188. doi: 10.1186/1471-2105-7-188. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Edwards SV, Fertil B, Giron A, Deschavanne PJ. A genomic schism in birds revealed by phylogenetic analysis of DNA strings. Syst Biol. 2002;51:599–613. doi: 10.1080/10635150290102285. [DOI] [PubMed] [Google Scholar]
- Ellegren H Forthcoming. Evolutionary stasis: the stable chromosomes of birds. Trends Ecol Evol. 2010;25:283–291. doi: 10.1016/j.tree.2009.12.004. [DOI] [PubMed] [Google Scholar]
- Emerson KJ, Merz CR, Catchen JM, Hohenlohe PA, Cresko WA, Bradshaw WE, Holzapfel CM. Resolving postglacial phylogeography using high-throughput sequencing. PNAS. 2010;107:16196–16200. doi: 10.1073/pnas.1006538107. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ericson PGP, Anderson CL, Britton T, Elzanowski A, Johansson US, Kallersjo M, Ohlson JI, Parsons TJ, Zuccon D, Mayr G. Diversification of neoaves: integration of molecular sequence data and fossils. Biol Lett. 2006;2:543–547. doi: 10.1098/rsbl.2006.0523. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ericson PGP, Anderson CL, Mayr G. Hangin' on to our rocks 'n clocks: a reply to Brown et al. Biol Lett. 2007;3:260–261. [Google Scholar]
- Eyre-Walker A, Hurst LD. The evolution of isochores. Nat Rev Genet. 2001;2:549–555. doi: 10.1038/35080577. [DOI] [PubMed] [Google Scholar]
- Feenders G, Liedvogel M, Rivas M, Zapka M, Horita H, Hara E, Wada K, Mouritsen H, Jarvis ED. Molecular mapping of movement-associated areas in the avian brain: a motor theory for vocal learning origin. PLoS One. 2008;3:e1768. doi: 10.1371/journal.pone.0001768. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Felsenstein J. Cases in which parsimony or compatibility methods will be positively misleading. Syst Zool. 1978;27:401–410. [Google Scholar]
- Foster PG, Hickey DA. Compositional bias may affect both DNA-based and protein-based phylogenetic reconstructions. J Mol Evol. 1999;48:284–290. doi: 10.1007/pl00006471. [DOI] [PubMed] [Google Scholar]
- Fountaine TMR, Benton MJ, Dyke GJ, Nudds RL. The quality of the fossil record of mesozoic birds. Proc R Soc Lond Ser B Biol Sci. 2005;272:289–294. doi: 10.1098/rspb.2004.2923. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Galtier N. Recombination, GC-content and the human pseudoautosomal boundary paradox. Trends Genet. 2004;20:347–434. doi: 10.1016/j.tig.2004.06.001. [DOI] [PubMed] [Google Scholar]
- Galtier N, Gouy M. Inferring pattern and process: maximum-likelihood implementation of a nonhomogeneous model of DNA sequence evolution for phylogenetic analysis. Mol Biol Evol. 1998;15:871–879. doi: 10.1093/oxfordjournals.molbev.a025991. [DOI] [PubMed] [Google Scholar]
- Galtier N, Piganeau G, Mouchiroud D, Duret L. GC-content evolution in mammalian genomes: the biased gene conversion hypothesis. Genetics. 2001;159:907–911. doi: 10.1093/genetics/159.2.907. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gibb GC, Kardailsky O, Kimball RT, Braun EL, Penny D. Mitochondrial genomes and avian phylogeny: complex characters and resolvability without explosive radiations. Mol Biol Evol. 2007;24:269–280. doi: 10.1093/molbev/msl158. [DOI] [PubMed] [Google Scholar]
- Glémin S. Surprising fitness consequences of GC-biased gene conversion: I. Mutation load and inbreeding depression. Genetics. 2010;185:939–959. doi: 10.1534/genetics.110.116368. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Griffin DK, Robertson LB, Tempest HG, Skinner BM. The evolution of the avian genome as revealed by comparative molecular cytogenetics. Cytogenet Genome Res. 2007;117:64–77. doi: 10.1159/000103166. [DOI] [PubMed] [Google Scholar]
- Groenen MA, Wahlberg P, Foglio M, et al. 12 co-authors. A high-density SNP-based linkage map of the chicken genome reveals sequence features correlated with recombination rate. Genome Res. 2009;19:510–519. doi: 10.1101/gr.086538.108. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hackett SJ, Kimball RT, Reddy S, et al. 17 co-authors. A phylogenomic study of birds reveals their evolutionary history. Science. 2008;320:1763–1768. doi: 10.1126/science.1157704. [DOI] [PubMed] [Google Scholar]
- Harismendy O, Ng PC, Strausberg RL, et al. 11 co-authors. Evaluation of next generation sequencing platforms for population targeted sequencing studies. Genome Biol. 2009;10:R32. doi: 10.1186/gb-2009-10-3-r32. [DOI] [PMC free article] [PubMed] [Google Scholar]
- International Chicken Genome Sequencing Consortium (ICGSC) Sequence and comparative analysis of the chicken genome provide unique perspectives on vertebrate evolution. Nature. 2004;432:695–716. doi: 10.1038/nature03154. [DOI] [PubMed] [Google Scholar]
- Jarvis ED. Learned birdsong and the neurobiology of human language. Ann N Y Acad Sci. 2004;1016:749–777. doi: 10.1196/annals.1298.038. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jarvis ED, Mello CV. Molecular mapping of brain areas involved in parrot vocal communication. J Comp Neurol. 2000;419:1–31. doi: 10.1002/(sici)1096-9861(20000327)419:1<1::aid-cne1>3.0.co;2-m. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jarvis ED, Ribeiro S, Da Silva ML, Ventura D, Vielliard J, Mello CV. Behaviourally driven gene expression reveals song nuclei in hummingbird brain. Nature. 2000;406:628–632. doi: 10.1038/35020570. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jeffroy O, Brinkmann H, Delsuc F, Philippe H. Phylogenomics: the beginning of incongruence? Trends Genet. 2006;22:225–231. doi: 10.1016/j.tig.2006.02.003. [DOI] [PubMed] [Google Scholar]
- Johansson US, Fjeldsa J, Bowie RCK. Phylogenetic relationships within Passerida (Aves: Passeriformes): a review and a new molecular phylogeny based on three nuclear intron markers. Mol Phylogenet Evol. 2008;48:858–876. doi: 10.1016/j.ympev.2008.05.029. [DOI] [PubMed] [Google Scholar]
- Katoh K, Misawa K, Kuma K, Miyata T. MAFFT: a novel method for rapid multiple sequence alignment based on fast Fourier transform. Nucleic Acids Res. 2002;30:3059–3066. doi: 10.1093/nar/gkf436. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Künstner A, Wolf JBW, Backström N, et al. 13 co-authors. Comparative genomics based on massive parallel transcriptome sequencing reveals patterns of substitution and selection across 10 bird species. Mol Ecol. 2010;19(Suppl 1):266–276. doi: 10.1111/j.1365-294X.2009.04487.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lartillot N, Brinkmann H, Philippe H. Suppression of long-branch attraction artefacts in the animal phylogeny using a site-heterogeneous model. BMC Evol Biol. 2007;7(Suppl 1):S4. doi: 10.1186/1471-2148-7-S1-S4. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lartillot N, Philippe H. A Bayesian mixture model for across-site heterogeneities in the amino acid replacement process. Mol Biol Evol. 2004;21:1095–1109. doi: 10.1093/molbev/msh112. [DOI] [PubMed] [Google Scholar]
- Lopez P, Casane D, Philippe H. Heterotachy, an important process of protein evolution. Mol Biol Evol. 2002;19:1–7. doi: 10.1093/oxfordjournals.molbev.a003973. [DOI] [PubMed] [Google Scholar]
- Mancera E, Bourgon R, Brozzi A, Huber W, Steinmetz LM. High-resolution mapping of meiotic crossovers and non-crossovers in yeast. Nature. 2008;454:479–485. doi: 10.1038/nature07135. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Manegold A. Earliest fossil record of the Certhioidea (treecreepers and allies) from the early Miocene of Germany. J Ornithol. 2008;149:223–228. [Google Scholar]
- Mayr G, Manegold A. A small suboscine-like passeriform bird from the early oligocene of France. Condor. 2006;108:717–720. [Google Scholar]
- Meunier J, Duret L. Recombination drives the evolution of GC-content in the human genome. Mol Biol Evol. 2004;21:984–990. doi: 10.1093/molbev/msh070. [DOI] [PubMed] [Google Scholar]
- Nabholz B, Glémin S, Galtier N. The erratic mitochondrial clock: variations of mutation rate, not population size, affect mtDNA diversity across birds and mammals. BMC Evol Biol. 2009;9:54. doi: 10.1186/1471-2148-9-54. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nottebohm F. Origins of vocal learning. Am Nat. 1972;106:116–140. [Google Scholar]
- Paradis E. Analysis of phylogenetics and evolution with R. New York: Springer; 2007. [Google Scholar]
- Pereira SL, Baker AJ. A mitogenomic timescale for birds detects variable phylogenetic rates of molecular evolution and refutes the standard molecular clock. Mol Biol Evol. 2006;23:1731–1740. doi: 10.1093/molbev/msl038. [DOI] [PubMed] [Google Scholar]
- Philippe H, Derelle R, Lopez P, et al. (20 co-authors) Phylogenomics revives traditional views on deep animal relationships. Curr Biol. 2009;19:706–712. doi: 10.1016/j.cub.2009.02.052. [DOI] [PubMed] [Google Scholar]
- Phillips MJ, Delsuc F, Penny D. Genome-scale phylogeny and the detection of systematic biases. Mol Biol Evol. 2004;21:1455–1458. doi: 10.1093/molbev/msh137. [DOI] [PubMed] [Google Scholar]
- Posada D. jModelTest: phylogenetic model averaging. Mol Biol Evol. 2008;25:1253–1256. doi: 10.1093/molbev/msn083. [DOI] [PubMed] [Google Scholar]
- Pratt RC, Gibb GC, Morgan-Richards M, Phillips MJ, Hendy MD, Penny D. Toward resolving deep neoaves phylogeny: data, signal enhancement, and priors. Mol Biol Evol. 2009;26:313–326. doi: 10.1093/molbev/msn248. [DOI] [PubMed] [Google Scholar]
- Rannala B, Yang Z. Inferring speciation times under an episodic molecular clock. Syst Biol. 2007;56:453–466. doi: 10.1080/10635150701420643. [DOI] [PubMed] [Google Scholar]
- R Development Core Team. R: a language and environment for statistical computing. Vienna (Austria): R Foundation for Statistical Computing; 2004. [Google Scholar]
- Romiguier J, Ranwez V, Douzery EJ, Galtier N. Contrasting GC-content dynamics across 33 mammalian genomes: relationship with life-history traits and chromosome sizes. Genome Res. 2010;20:1001–1009. doi: 10.1101/gr.104372.109. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sheffield NC, Song H, Cameron SL, Whiting MF. Nonstationary evolution and compositional heterogeneity in beetle mitochondrial phylogenomics. Syst Biol. 2009;58:381–394. doi: 10.1093/sysbio/syp037. [DOI] [PubMed] [Google Scholar]
- Shendure J, Ji H. Next-generation DNA sequencing. Nat Biotechnol. 2008;26:1135–1145. doi: 10.1038/nbt1486. [DOI] [PubMed] [Google Scholar]
- Sibley CG, Ahlquist JE. Phylogeny and classification of birds: a study in molecular evolution. New Haven (CT): Yale University Press; 1990. [Google Scholar]
- Slack KE, Jones CM, Ando T, Harrison GLA, Fordyce RE, Arnason U, Penny D. Early penguin fossils, plus mitochondrial genomes, calibrate avian evolution. Mol Biol Evol. 2006;23:1144–1155. doi: 10.1093/molbev/msj124. [DOI] [PubMed] [Google Scholar]
- Spencer CC, Deloukas P, Hunt S, Mullikin J, Myers S, Silverman B, Donnelly P, Bentley D, Mcvean G. The influence of recombination on human genetic diversity. PLoS Genet. 2006;2:e148. doi: 10.1371/journal.pgen.0020148. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Springer M, Debry R, Douady C, Amrine H, Madsen O, de Jong W, Stanhope M. Mitochondrial versus nuclear gene sequences in deep-level mammalian phylogeny reconstruction. Mol Biol Evol. 2001;18:132–143. doi: 10.1093/oxfordjournals.molbev.a003787. [DOI] [PubMed] [Google Scholar]
- Stamatakis A. RAxML-VI-HPC: maximum likelihood-based phylogenetic analyses with thousands of taxa and mixed models. Bioinformatics. 2006;22:2688–2690. doi: 10.1093/bioinformatics/btl446. [DOI] [PubMed] [Google Scholar]
- Thorne J, Kishino H, Painter I. Estimating the rate of evolution of the rate of molecular evolution. Mol Biol Evol. 1998;15:1647–1657. doi: 10.1093/oxfordjournals.molbev.a025892. [DOI] [PubMed] [Google Scholar]
- Warren WC, Clayton DF, Ellegren H, et al. (74 co-authors) The genome of the zebra finch: special insights into vocal learning and communication. Nature. 2010;464:757–762. [Google Scholar]
- Webster MT, Axelsson E, Ellegren H. Strong regional biases in nucleotide substitution in the chicken genome. Mol Biol Evol. 2006;23:1203–1216. doi: 10.1093/molbev/msk008. [DOI] [PubMed] [Google Scholar]
- Webster MT, Smith NG, Hultin-Rosenberg L, Arndt PF, Ellegren H. Male-driven biased gene conversion governs the evolution of base composition in human Alu repeats. Mol Biol Evol. 2005;22:1468–1474. doi: 10.1093/molbev/msi136. [DOI] [PubMed] [Google Scholar]
- Wilbrecht L, Nottebohm F. Vocal learning in birds and humans. Ment Retard Dev Disabil Res Rev. 2003;9:135–148. doi: 10.1002/mrdd.10073. [DOI] [PubMed] [Google Scholar]
- Yang Z. Among-site rate variation and its impact on phylogenetic analyses. Trends Ecol Evol. 1996;11:367–372. doi: 10.1016/0169-5347(96)10041-0. [DOI] [PubMed] [Google Scholar]
- Yang Z. PAML 4: phylogenetic analysis by maximum likelihood. Mol Biol Evol. 2007a;23:7–9. doi: 10.1093/molbev/msm088. [DOI] [PubMed] [Google Scholar]
- Yang Z. Computational molecular evolution. Oxford: Oxford University Press; 2007b. [Google Scholar]
- Yang Z, Rannala B. Bayesian estimation of species divergence times under a molecular clock using multiple fossil calibrations with soft bounds. Mol Biol Evol. 2006;23:212–226. doi: 10.1093/molbev/msj024. [DOI] [PubMed] [Google Scholar]
- Yoder AD, Yang Z. Estimation of primate speciation dates using local molecular clocks. Mol Biol Evol. 2000;17:1081–1090. doi: 10.1093/oxfordjournals.molbev.a026389. [DOI] [PubMed] [Google Scholar]
- Zwickl D. Genetic algorithm approaches for the phylogenetic analysis of large biologiical sequence datasets under the maximum likelihood criterion [PhD thesis] [Austin (TX)]: University of Texas; 2006. [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.