Abstract
It is generally accepted that hydrogenosomes (hydrogen-producing organelles) evolved from a mitochondrial ancestor. However, until recently, only indirect evidence for this hypothesis was available. Here, we present the almost complete genome of the hydrogen-producing mitochondrion of the anaerobic ciliate Nyctotherus ovalis and show that, except for the notable absence of genes encoding electron transport chain components of Complexes III, IV, and V, it has a gene content similar to the mitochondrial genomes of aerobic ciliates. Analysis of the genome of the hydrogen-producing mitochondrion, in combination with that of more than 9,000 genomic DNA and cDNA sequences, allows a preliminary reconstruction of the organellar metabolism. The sequence data indicate that N. ovalis possesses hydrogen-producing mitochondria that have a truncated, two step (Complex I and II) electron transport chain that uses fumarate as electron acceptor. In addition, components of an extensive protein network for the metabolism of amino acids, defense against oxidative stress, mitochondrial protein synthesis, mitochondrial protein import and processing, and transport of metabolites across the mitochondrial membrane were identified. Genes for MPV17 and ACN9, two hypothetical proteins linked to mitochondrial disease in humans, were also found. The inferred metabolism is remarkably similar to the organellar metabolism of the phylogenetically distant anaerobic Stramenopile Blastocystis. Notably, the Blastocystis organelle and that of the related flagellate Proteromonas lacertae also lack genes encoding components of Complexes III, IV, and V. Thus, our data show that the hydrogenosomes of N. ovalis are highly specialized hydrogen-producing mitochondria.
Keywords: hydrogenosome, mitochondrion, horizontal gene transfer, evolution, adaptation, Nyctotherus
Introduction
Mitochondria are essential organelles in aerobic eukaryotes. They play a central role in ATP production as well as in many other cellular processes, such as apoptosis, cellular proliferation, heme synthesis, steroid synthesis, and Fe/S cluster assembly (Miller 1995; Lill and Kispal 2000; Scheffler 2001; Duchen 2004). Aerobic mitochondria perform oxidative phosphorylation, that is, they use oxygen as terminal electron acceptor, oxidizing NADH and FADH2 while producing ATP with the aid of an ATP synthase that exploits the proton gradient generated by the electron transport chain. Eukaryotes living under anaerobic circumstances have evolved a spectrum of organelles that are related to mitochondria and have names, such as anaerobic mitochondria (both temporarily and permanently anaerobic), hydrogenosomes, mitosomes, mitochondrial remnants, modified mitochondria, or mitochondria-like organelles, reflecting their metabolic peculiarities (Tielens et al. 2002; Barbera et al. 2007; Tachezy and Dolezal 2007; Tachezy and Smid 2007; Tielens and van Hellemond 2007; Hjort et al. 2010).
Mitochondria that produce ATP via oxidative phosphorylation have a mitochondrial genome, as the protein complexes of proton-pumping electron transport chains contain essential subunits whose genes were never translocated to the nuclear genome during evolution but always remained part of the mitochondrial genome. The canonical (textbook) mitochondrion uses oxygen as final electron acceptor, but there are also anaerobically functioning mitochondria that use compounds other than oxygen as final electron acceptor. Most organisms with anaerobic mitochondria use an endogenously produced electron acceptor, such as fumarate (Tielens et al. 2002). Some anaerobic functioning mitochondria possess a hydrogenase besides a proton-pumping electron transport chain, and therefore, they can use protons as final electron acceptors resulting in the production of hydrogen. These organelles are the so-called hydrogen-producing mitochondria and examples are found in Nyctotherus ovalis and Blastocystis sp. Hydrogen production in the latter organelles, however, has not been demonstrated so far (Lantsman et al. 2008; Stechmann et al. 2008). These organelles and those of Proteromonas, for which currently nothing is known about hydrogen production, are considered to be bona fide mitochondria as they use a proton-pumping electron transport chain and hence possess a mitochondrial genome encoding essential parts of it (Perez-Brocal et al. 2010).
Hydrogenosomes are mitochondrion-related organelles that, by definition, produce ATP and hydrogen using protons as electron acceptors. In contrast to hydrogen-producing mitochondria, hydrogenosomes have no mitochondrial genome, do not possess a proton-pumping electron transport chain, and produce ATP exclusively via substrate-level phosphorylation. Hydrogenosomes are found in various unrelated anaerobic eukaryotes, such as anaerobic flagellates, chytridiomycete fungi, and several anaerobic ciliates (Hackstein et al. 2001; Embley et al. 2003; Embley and Martin 2006; Hackstein, Baker, et al. 2008; Hackstein, de Graaf, et al. 2008; de Graaf, Duarte, et al. 2009).
Mitosomes produce neither hydrogen nor ATP, but in general have retained components of a Fe/S cluster synthesizing machinery, which is now believed to be the reason why mitochondria, hydrogenosomes, and mitosomes are essential to eukaryotic life (Henze and Martin 2003; Yarlett 2004; Tachezy and Dolezal 2007; Goldberg et al. 2008). Even for Entamoeba (Aguilera et al. 2008) that has been regarded as the potential exception to this pattern, evidence is now mounting that its mitosomes also assemble Fe/S clusters (Maralikova et al. 2010), although Mi-ichi et al. (2009) could not confirm this finding using a proteomics approach. All eukaryotic organisms studied so far in sufficient molecular and ultrastructural detail appear to possess either mitochondria, hydrogenosomes or mitosomes. Furthermore, no species has been found that contains both mitochondria and mitosomes or mitochondria and hydrogenosomes, supporting the hypothesis that these organelles have a common origin. Despite this common origin, mitochondria, hydrogenosomes, and mitosomes are very diverse: not only in the size of their proteomes but also by virtue of their protein composition and function (Gabaldón and Huynen 2004). However, classical aerobic mitochondria share two characteristics: They have retained a genome, allowing unambiguous documentation of their descent from an α-proteobacterium by phylogenetic analyses (Lang et al. 1999), and they contain an electron transport chain with Complexes I through IV as well as an ATPase (Complex V). In the evolution of mitochondria, hydrogenosomes and mitosomes from an α-proteobacterium, a large number of genes and proteins have been lost, gained from various sources, or retargeted to other organelles, thus shaping the huge diversity of current mitochondria and their homologs in various species (Gabaldón and Huynen 2004). Within the evolutionary “gap” between species with genome-containing mitochondria and species with mitosomes or hydrogenosomes (both lacking an organellar genome), the species N. ovalis provides an interesting link. Nyctotherus ovalis is an anaerobic ciliate that lives in the hindgut of various cockroach species. It has numerous hydrogen-producing organelles that are intimately associated with endosymbiotic methane–producing archaea that use the hydrogen produced by the organelles. Despite the hydrogenosomal metabolism of this organelle, we have found that it actually has a genome (Akhmanova, Voncken, van Alen, et al. 1998), that similar to mitochondrial genomes (Gray 2005), encodes elements of an electron transport chain (Boxma et al. 2005). Phylogenetic analyses of the organellar genome of N. ovalis showed that it has evolved from the genome of an aerobic mitochondrion of a ciliate ancestor (Boxma et al. 2005), providing direct evidence that hydrogenosomes can evolve from mitochondria. This blurs the distinction between mitochondria and hydrogenosomes. A comparable situation is found in the mitochondrion-like organelles of the phylogenetically only distantly related Stramenopile Blastocystis that is clearly distinct from the ciliates, which belong to the Alveolata. Blastocystis also retains a mitochondrial genome and parts of an electron transport chain (Perez-Brocal and Clark 2008; Stechmann et al. 2008; Wawrzyniak et al. 2008). Because this organelle also hosts a [FeFe] hydrogenase, it can be regarded as a hydrogenosome with genome, although thus far no hydrogenase activity has been demonstrated in this species (Lantsman et al. 2008; Stechmann et al. 2008).
Also for other (genome-less) types of hydrogen-producing organelles evidence is accumulating that they have evolved from mitochondria (Embley and Martin 2006). In the absence of an organellar genome, the evidence for homology is based on phylogenetic analyses of proteomes. Based on phylogenetic analyses of Complex I subunits, the hydrogenosomes of the parabasalid Trichomonas are inferred to share a common ancestor with mitochondria (Hrdý et al. 2004). Also, the hydrogenosomes of anaerobic chytrids seem to have evolved from the mitochondria of their aerobic ancestors (Akhmanova, Voncken, Harhangi, et al. 1998). It is therefore likely that hydrogenosomes arose repeatedly by evolutionary tinkering as an adaptation to the particular requirements of hosts, which thrive in rather different anaerobic environments.
Here, we describe the isolation and sequence analysis of the major part (41,666 bp) of the organellar genome of N. ovalis and compare it with the mitochondrial genomes of the ciliates Paramecium aurelia, Tetrahymena spp., and Euplotes minuta as well as with the mitochondrial/hydrogenosomal genome of the Stramenopiles Blastocystis sp. and Proteromonas lacertae. We show that the organellar genome of N. ovalis is a typical ciliate mitochondrial genome except for the obvious absence of genes encoding components of Complexes III, IV, and V. The lack of evidence for the presence of these genes in N. ovalis coincides with the definitive absence of these genes from the organellar genomes of Blastocystis sp. and P. lacertae (Perez-Brocal and Clark 2008; Stechmann et al. 2008; Wawrzyniak et al. 2008; Perez-Brocal et al. 2010). By comparing the hydrogenosomal proteins and metabolic pathways of N. ovalis with the various hydrogenosomes, mitosomes, mitochondria-like organelles, and mitochondria, we show that the N. ovalis organelle is as complex as some mitochondria.
Materials and Methods
Cell Isolation
Nyctotherus ovalis cells were isolated from the hindgut of the cockroach Blaberus sp. var. Amsterdam. Total DNA was isolated by dissolving living cells in 8 M guanidinium chloride and separation on a hydroxyapatite column using standard procedures (de Graaf, van Alen, et al. 2009).
The organellar DNA was isolated from the total DNA by pulsed field gel electrophoresis. This organellar DNA was used for a partial Sau 3A I digest and cloning in the Bam H1 restriction site from the vector pUC18c. From this library, 500 clones were sequenced in both directions and additional 500 in one direction. Less than 3% of these clones contained a mitochondrial sequence. Finally, larger pieces of the organellar genome were reconstructed, and the gaps were filled by long range polymerase chain reaction (PCR) with specific primers resulting in a contig of 41,666 bp representing the major part of the organellar genome (accession number: GU057832). Due to the very limited amount of DNA available, we did not attempt to sequence the chromosome ends.
Identification of N. ovalis sequences likely encoding hydrogenosomal proteins
Nyctotherus ovalis sequences translated in the six frames were compared with the mitochondrial proteomes of human, yeast, rat, and Tetrahymena. The mitochondrial proteomes were retrieved from the mitoproteome (http://www.mitoproteome.org), the SGD database (Cherry et al. 1998), the supplementary material from Sickmann et al. (2003), from Forner et al. (2006), and from Smith et al. (2007). The accession numbers of the N. ovalis sequences are displayed in supplementary table S1, Supplementary Material online. Nyctotherus ovalis sequences with a reciprocal best hit (BH) with one of the mitochondrial proteins (Smith Waterman, E < 0.01) were selected for further phylogenetic analysis to confirm orthology with a mitochondrial protein. Each nucleotide sequence selected was compared by the SWX algorithm with the 165 complete proteomes, and the first 100 hits were kept and used together with the N. ovalis sequence to produce a multiple alignment with Muscle v3.7 (default parameters; Edgar 2004). Positions of the alignments that did not contain gaps were automatically selected, and a tree was subsequently derived using PhyML (Guindon and Gascuel 2003) using the JTT model and an estimated number of invariable sites with four substitution rate categories. Hundred bootstraps were performed.
Phylogenetic Position
To determine the phylogenetic position of the N. ovalis hydrogen-producing mitochondrion, we composed a concatenated alignment phylogeny of the Complex I proteins that tend to be encoded on the mitochondrial genome (nad1, nad2, nad3, nad4, nad4L, nad5, and nad6), using the gamma-proteobacterium Escherichia coli and the α-proteobacterium Rickettsia prowazekii as outgroups. The sequences in each protein family were first aligned with Muscle (Edgar 2004). We then concatenated the alignment blocks, where gaps were inserted in the rare cases that a protein was missing from a certain species (e.g., nad6 from N. ovalis). Given the length of the alignment, we opted to compute a bootstrapped Maximum Likelihood (ML) phylogeny. For this, we first selected the best-fit model of amino acid replacement, using the Akaike information criterion (AIC) as goodness of fit measure, as implemented in ProtTest (version 2.2) (Abascal et al. 2005). Accordingly, a ML phylogeny, 100 times bootstrapped, was computed with PhyML (version 3.0.1) (Guindon and Gascuel 2003) using the previously selected LG model of amino acids substitution, with a discrete gamma distribution approximated by 4 rate-categories (+4G), estimated proportion of invariable sites (+I), and observed amino acid frequencies (+F).
Identification of Sequences Derived From HGT
In order to identify horizontal gene transfers (HGTs) present in the N. ovalis genome, we built phylogenies for all the proteins that had BHs with noneukaryotic proteins in a Blast search. We carefully selected the homologous data set from the 250 BHs in the nonredundant database, supplemented with ten phylogenetically well distributed, best Blast hits from eukaryotes only, aligned the sequences using ClustalW (version 2.0.10) (Thompson et al. 1994), and subsequently pruned the alignment for redundancy. Each alignment was then submitted to ProtTest (version 2.4) (Abascal et al. 2005) in order to statistically infer the best-fit model of amino acid substitution, according to the AIC. Finally, a ML phylogeny was computed with PhyML (version 3.0.1) (Guindon and Gascuel 2003) employing the previously chosen model and bootstrapped 100 times.
Subcellular Localization
Mitop2 (Andreoli et al. 2004) was used when the N-terminal part of the N. ovalis sequence was present to examine the presence of a putative Mitochondrial Import Signal.
N. ovalis cDNA and gDNA
Nyctotherus ovalis cDNA and genomic DNA (gDNA) libraries were constructed as described earlier (Ricard et al. 2008). The macronuclear gDNA clones were obtained after amplification of total ciliate DNA with telomere primers. In general, only gDNA clones with at least one telomere were used in order exclude contamination by bacterial sequences.
Results and Discussion
Gene Content and Structure of the Organellar Genome of N. ovalis
In 2005, Boxma et al. showed that a 14 Kb fragment of the organellar genome of the anaerobic hydrogen-producing ciliate N. ovalis that had been isolated from the cockroach Blaberus sp. var. Amsterdam was of mitochondrial origin (Boxma et al. 2005). Here, we describe an analysis of the nearly complete organellar genome. Pulsed field gel electrophoresis followed by Southern blot hybridization with two different 32P-labeled probes, nad7 and 12S (rns), indicated that the size of the organellar genome of N. ovalis exceeds 48 kb (supplementary fig. S1, Supplementary Material online).
Mitochondrial genes present in the organellar library were identified by Blast analysis (Altschul et al. 1997), and in the case of nonoverlap, subjected to long-range PCR to fill the gaps between the fragments. Using this approach, the major part of this genome (41,666 bp) has been sequenced and reconstructed as a single contig. This part of the organellar genome contains an almost complete set of genes found in the mitochondrial genomes of other ciliates (table 1, fig. 1). In addition, we found a stretch larger than 11 kb that possesses seven open reading frames (orfs) with no significant sequence similarity to any known genes.
Table 1.
Various Genes Encoded by the Organellar Genomes of Nyctotherus ovalis (Nov), Blastocystis sp. (Blas), Proteromonas lacertae (Pro), Euplotes minuta (Emi), and Plasmodium falciparum (Pfa).
| Gene | Nov | Blas | Pro | Emi | Pfa | Gene | Nov | Blas | Pro | Emi | Pfa |
| nad1 | ▪ | ▪ | ▪ | ▪ | □ | rps12 | ▪ | ▪ | ▪ | ▪ | □ |
| nad2 | ▪ | ▪ | ▪ | ▪ | □ | rps13 | □ | ▪ | □ | □ | □ |
| nad3 | ▪ | ▪ | ▪ | ▪ | □ | rps14 | ▪ | ▪ | ▪ | □ | □ |
| nad4 | ▪ | ▪ | ▪ | ▪ | □ | rps19 | □ | ▪ | ▪ | □ | □ |
| nad4L | ▪ | ▪ | ▪ | ▪ | □ | rpl2 | ▪ | ▪ | ▪ | ▪ | □ |
| nad5 | ▪ | ▪ | ▪ | ▪ | □ | rpl5 | □ | ▪ | ▪ | □ | □ |
| nad6 | □ | ▪ | ▪ | □ | □ | rpl6 | ▪ | ▪ | ▪ | ▪ | □ |
| nad7 | ▪ | ▪ | ▪ | ▪ | □ | rpl14 | ▪ | ▪ | ▪ | ▪ | □ |
| nad9 | ▪ | ▪ | ▪ | ▪ | □ | rpl16 | □ | ▪ | ▪ | ▪ | □ |
| nad10 | ▪ | □ | □ | ▪ | □ | trnA | □ | ▪ | □ | □ | □ |
| nad11 | □ | ▪ | ▪ | □ | □ | trnC | □ | ▪ | □ | □ | □ |
| rnl | ▪ | ▪ | ▪ | ▪ | ▪ | trnD | □ | ▪ | ▪ | □ | □ |
| rns | ▪ | ▪ | ▪ | ▪ | ▪ | trnE | □ | ▪ | ▪ | ▪ | □ |
| cob | □ | □ | □ | ▪ | ▪ | trnF | ▪ | ▪ | ▪ | ▪ | □ |
| cox1 | □ | □ | □ | ▪ | ▪ | trnH | □ | ▪ | □ | ▪ | □ |
| cox2 | □ | □ | □ | ▪ | □ | trnI | □ | ▪ | ▪ | □ | □ |
| cox3 | □ | □ | □ | □ | ▪ | trnK | □ | ▪ | ▪ | □ | □ |
| atp9 | □ | □ | □ | ▪ | □ | trnL | □ | ▪ | □ | □ | □ |
| ccmF/yejR | □ | □ | □ | ▪ | □ | trnM | □ | ▪ | ▪ | ▪ | □ |
| rps2 | □ | □ | ▪ | □ | □ | trnN | □ | ▪ | ▪ | □ | □ |
| rps3 | □ | ▪ | □ | ▪ | □ | trnP | □ | ▪ | ▪ | □ | □ |
| rps4 | □ | ▪ | ▪ | □ | □ | trnQ | □ | □ | ▪ | ▪ | □ |
| rps7 | □ | ▪ | □ | □ | □ | trnS | □ | □ | ▪ | □ | □ |
| rps8 | ▪ | ▪ | ▪ | □ | □ | trnV | □ | □ | ▪ | □ | □ |
| rps10 | □ | ▪ | ▪ | □ | □ | trnW | ▪ | ▪ | ▪ | ▪ | □ |
| rps11 | □ | ▪ | □ | □ | □ | trnY | ▪ | ▪ | ▪ | ▪ | □ |
NOTE.—▪: present in the organellar genome; □: absent in the organellar genome.
FIG. 1.
Organellar gene maps of Nyctotherus ovalis and Euplotes minuta. Red: Complex I genes, blue: rRNA genes, green: ribosomal proteins, yellow: Complex III and IV genes, gray: unidentified open reading frames, pink: repeat region, dark gray: atp9 gene, and white: intergenic spacers. Capital letters indicate the various tRNA genes. Arrows: direction of transcription.
The organellar genome contains nine genes encoding elements of mitochondrial Complex I: nad1, nad2, nad3, nad4, nad4L, nad5, nad7, nad9, and nad10. This set of genes is also found in other ciliates (table 1). The nad1 gene is split in the mitochondrial genomes of all ciliates studied so far into a larger (nad1a) and a smaller (nad1b) part (de Graaf, van Alen, et al. 2009). In N. ovalis, we could identify the larger nad1a gene piece while the nad1b gene is likely encoded by orf224 (fig. 1). The other Complex I genes have a similar size as in the Tetrahymena species and P. aurelia. This is in contrast to the situation in E. minuta and E. crassus where many Complex I genes have extended 5′ ends (de Graaf, van Alen, et al. 2009). The nad4 gene of N. ovalis is a bit shorter than in other ciliates, and it is very likely that orf110 represents an additional part of the nad4 gene (fig. 1).
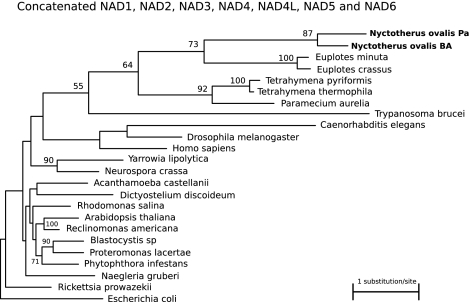
Phylogenetic analysis of seven concatenated Complex I genes reveals that the Complex I of N. ovalis is closely related to that of E. crassus and E. minuta and that it clusters together with the Complex I genes of Tetrahymena and Paramecium (fig. 2), whereas it is—as expected—only distantly related to Complex I of Blastocystis and Proteromonas.
FIG. 2.
ML phylogeny of the Nyctotherus ovalis hydrogen-producing mitochondrion, based on a concatenated alignment of seven mitochondrial Complex I encoded proteins (Nad1, Nad2, Nad3, Nad4, Nad4L, Nad5, and Nad6). Only bootstrap values of 50% and higher are indicated.
Organellar genes encoding components of Complexes III, IV, or V could not be identified. These genes are consistently present in the mitochondrial genomes of the aerobic ciliates P. aurelia, Tetrahymena spp., and E. minuta that have a size around 48 kb (table 1; de Graaf, van Alen, et al. 2009). Moreover, as a rule, all mitochondrial genomes sequenced to date host genes belonging to Complexes III, IV, and V (Burger et al. 2003). The loss of genes encoding Complexes III, IV, and V from the mitochondrial genome has been regarded as indicative of a loss from the species rather than a transfer to the nuclear genome in the mitochondrion-like organelles of Blastocystis (Perez-Brocal and Clark 2008; Stechmann et al. 2008; Wawrzyniak et al. 2008). The absence of Complex III and IV genes in the organellar genome of N. ovalis is consistent with the observation that activity of Complexes III and IV could not be observed in inhibitor studies (Boxma et al. 2005).
A total of 15 unique orfs were identified, seven of which form a cluster with a total length greater than 11 kb with no significant sequence similarity to any known genes. No open reading frames with sequence similarity to orfs of Tetrahymena spp. P. aurelia, and E. minuta were identified. We detected three transfer RNA genes in the organellar genome (trnF, trnY, and trnW), similar to the mitochondrial genome of P. aurelia in which four transfer RNA (tRNA) genes were found (Pritchard et al. 1990). It is possible that additional tRNA genes are present in the missing part of the organellar genome. A large complex repeat region is present in the organellar genome between orf479 and ssu/rns genes. This region contains 12 repeats of 34 nt followed by a (not perfectly) duplicated stretch of around 400 bp (supplementary fig. S2, Supplementary Material online). It is unlikely that this repeat region plays the same role in initiation of transcription as suggested for the mitochondrial genome of the E. minuta and E. crassus (de Graaf, van Alen, et al. 2009) because all genes found on the 41,666 bp part of the organellar genome of N. ovalis are oriented in the same direction, both before and after the repeat. In E. minuta and E. crassus, the repeat separates the open reading frames into two blocks, each having a single direction of transcription, from the repeat toward the ends of the chromosome.
We identified seven ribosomal proteins in the organellar genome of N. ovalis. This seems to be an average number for ciliates because nine ribosomal proteins were found in Tetrahymena spp., seven in P. aurelia, and six in E. minuta (de Graaf, van Alen, et al. 2009). Three of these ribosomal proteins were found in all four ciliate species with sequenced mitochondrial genomes: Rps12, Rpl2, and Rpl14. In contrast, the rps8 gene is unique for N. ovalis (table 1). Notably, as in E. minuta, all organellar ribosomal protein genes of N. ovalis have an N-terminal extension that is similar to a mitochondrial targeting signal (not shown, cf. Ueda et al. 2008; de Graaf, van Alen, et al. 2009). The overall A-T content of the organellar genome of N. ovalis (58.5%) is identical to the A-T content of the mitochondrial genome of P. aurelia. The organellar genes are not tightly packed, and intergenic spacers represent 11.7% of the genome. In contrast, the genomes of the other ciliates contain no more than about 4% of intergenic spacers. The A-T content of these spacers is almost identical to the overall A-T content: 57.8%. Spacer lengths vary from 0 to 2,159 bp with an average size of 320 bp. In six cases, genes have an overlap, ranging from 3–61 bp. The 11 kb fragment that harbors the cluster of seven orfs with no homology to other known genes contains large open reading frames with small intergenic spacers ranging from −3 to 9 bp. These seven genes do not appear to be the remains of genes encoding components of Complex III, Complex IV, and Complex V. Neither Blast searches against mitochondrial encoded proteins nor profile-based searches against domain databases (PFAM, SMART) produced hits, even at an insignificant E value cutoff of 10. Because none of the proteins has a pI > 10, it is also unlikely that they are ribosomal proteins.
Thus, the sequenced fragment of the organellar genome of N. ovalis exhibits all characteristics of a ciliate mitochondrial genome, except for the absence of genes encoding components of the mitochondrial Complexes III, IV, and V.
Nuclear Encoded Organellar Proteins
As in other mitochondria, most of the organellar proteins in N.ovalis are encoded by the nuclear genome, synthesized in the cytoplasm, and imported into the organelle. The signal to direct these proteins into the hydrogen-producing mitochondrion resembles mitochondrial targeting signals (Boxma et al. 2005) and can be an indication that a certain protein is targeted to the organelle. Furthermore, because mitochondrial proteins tend to preserve their cellular location in evolution (Calvo et al. 2006; Szklarczyk and Huynen 2009), the orthology of nuclear encoded mitochondrial proteins from species like Homo sapiens, Saccharomyces cerevisiae, and T. thermophila with N. ovalis proteins can be used to predict hydrogenosomal proteins. Here, we describe in more detail nuclear encoded proteins that are probably targeted to the hydrogen-producing mitochondrion of N. ovalis.
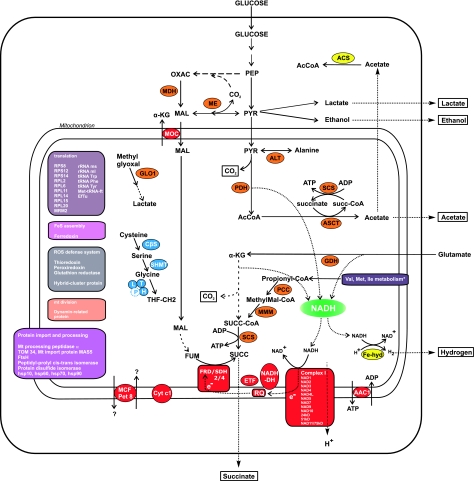
After our initial analysis (Boxma et al. 2005), we have obtained ∼4,500 additional N. ovalis gDNA and cDNA sequences (Ricard et al. 2008). To identify new proteins involved in the hydrogenosomal metabolism, we first performed a Bidirectional BH analysis. Thousand nine hundred and fourteen nonredundant cDNAs and 2,841 nonredundant gDNAs from N. ovalis were compared with the mitochondrial proteomes of human, yeast, and rat. This allowed us to identify orthologs of mitochondrial proteins for which 161 phylogenetic trees were constructed with PhyML and subsequently analyzed by hand to determine orthology relations (not shown). This phylogenetic analysis allowed us to predict a number of new hydrogenosomal proteins as well as to provide stronger evidence for some that had been predicted previously (Boxma et al. 2005). From the phylogenetic trees, we observed that most of the N. ovalis sequences with close relatives among the mitochondrial proteins also cluster with their counterparts from the aerobic ciliates T. thermophila and P. tetraurelia. This indicates their recent descent from proteins functioning in aerobic mitochondria (data not shown). Furthermore, to compare the metabolic complexity of the N. ovalis hydrogen-producing mitochondrion with that of mitochondria, genome-less hydrogenosomes, and mitosomes, we analyzed the distribution of N. ovalis proteins with sequence similarity to organellar proteins in other species. These were the mitochondria-possessing species Homo, Saccharomyces, Reclinomonas, Plasmodium, Tetrahymena, and Paramecium; the species possessing a mitosome Cryptosporidium, Encephalitozoon, Giardia, and Entamoeba; and the species with a hydrogenosome Piromyces, Trichomonas, Psalteriomonas, and Blastocystis (supplementary table S1, Supplementary Material online). Here, we describe some of the proteins and their roles. Figure 3 shows how they might function together, shaping the organellar metabolism:
FIG. 3.
Tentative reconstruction of the metabolism of the hydrogen-producing mitochondria of Nyctotherus ovalis. The metabolism is based on proteins that are orthologous to mitochondrial proteins (Methods) and proteins derived from HGT that are likely to have an organellar location based on their metabolic function. It includes the results of metabolic experiments described in Boxma et al. (2005). In red: metabolism linked to NADH (green) oxidation and electron transfer as well as solute carriers. In yellow are the proteins that seem to have been acquired by HGT. In blue: glycine metabolism. In orange: intermediate metabolism. Dotted arrows stand for metabolic steps we assume to be present based on biological experiments, broken arrows for the inferred metabolism for which we did not find the gene yet. AAC: ADP/ATP carrier; ACS: acetyl-CoA synthetase, AMP-forming (EC: 6.2.1.1); ALT: alanine amino transferase; ASCT: acetate:succinate CoA transferase; Cyt c1: cytochrome c1; CβS: cystathione β synthase; EfTu: elongation factor Tu; ETF: electron transfer flavoprotein; Fe-hyd: FeFe hydrogenase; FRD/SDH: fumarate reductase/succinate dehydrogenase; GDH: glutamate dehydrogenase; GLO1: glyoxalase I; MCF Pet 8: mitochondrial carrier family; MDH: malate dehydrogenase; ME: malic enzyme; Met tRNA ft: methionyl-tRNA formyltransferase; MMM: methyl malonyl CoA mutase; MOC: malate:oxoglutarate carrier; NADH-DH: NADH :quinone oxidoreductase; PCC: propionyl CoA carboxylase; PDH: pyruvate dehydrogenase; RQ: rhodoquinone; SCS: succinyl-CoA synthetase; SHMT: serine hydroxymethyl transferase; *includes several enzymes involved in branched-chain amino acid metabolism.
Pyruvate Dehydrogenase
The PDH complex converts pyruvate and CoA into acetyl-CoA while reducing NAD+ to NADH. In addition to the three subunits, which have already been described (E1α, E1β, and E2), we found a fourth subunit, a dihydrolipoyl dehydrogenase, also known as subunit E3, thereby completing the complex. The presence of a complete PDH complex contrasts strongly with the pyruvate catabolizing enzymes in other hydrogenosomes. Although the genome analysis of Blastocystis revealed a PDH and a pyruvate:ferredoxin oxidoreductase (PFO) (Stechmann et al. 2008), Lantsman et al. (2008) measured pyruvate:NADP+ oxidoreductase activity. Thus, the situation in Blastocystis is unclear. In Trichomonas pyruvate is decarboxylated by PFO (Hrdý et al. 2007). In contrast, in anaerobic chytridiomycete fungi (Piromyces sp. E2 and Neocallimastix sp. L2), pyruvate is catabolyzed by pyruvate-formate lyase (PFL) and not by PFO (Akhmanova et al. 1999). Furthermore, a large cDNA library of Piromyces constructed by the Department of Energy (DOE) Joint Genome Institute does not contain a single PFO but many PFL sequences (data not shown). The acetyl-CoA generated in N. ovalis by PDH is further metabolized by an acetate:succinate CoA-transferase (ASCT) belonging to the subfamily 1A with sequence similarity to the ASCT from Trypanosoma brucei (Tielens et al. 2010). Finally, ATP is generated by the action of succinyl-CoA synthetase (SCS), an enzyme that is also part of the tricarboxylic acid (TCA) cycle, but this cycle is not operative in N. ovalis (fig. 3). Also in Blastocystis and Trichomonas acetyl-CoA is metabolized to acetate by the cyclic action of ASCT and SCS. However, Blastocystis contains genes for ASCTs of the subfamilies 1B and 1C, and Trichomonas contains a gene for an ASCT of the subfamily 1C that does not exhibit sequence similarity to the subfamily 1A ASCT of N. ovalis (supplementary table S1, Supplementary Material online; Stechmann et al. 2008; Tielens et al. 2010).
TCA Cycle
Based on experiments with 14C labeled glucose (Boxma et al. 2005), the TCA cycle in N. ovalis is not complete, using only the malate–fumarate–succinate part in a reductive direction (fig. 3). Thus far, however, we have only found succinyl-CoA synthetase α and β subunits (the same enzyme as mentioned above) as well as the a and b subunit of succinate dehydrogenase (SDH)/fumarate reductase. These enzymes are also present in the mitochondrion-like organelle of Blastocystis in which succinyl-CoA synthetase, SDH, fumarate hydratase, and malate dehydrogenase have been identified (supplementary table S1, Supplementary Material online) (Stechmann et al. 2008; Wawrzyniak et al. 2008). These enzymes are universal in species with mitochondria. Note that among the gDNA sequences of N.ovalis, we did encounter a malate dehydrogenase that is orthologous to cytosolic malate dehydrogenases from other species and was therefore considered to be also cytoplasmic in N. ovalis as well.
Electron Transport Chain
Complex I (NADH-quinone oxidoreductase) consists of 14 subunits in eubacteria and of 33–45 subunits in mitochondria (Friedrich and Böttcher 2004; Gabaldon et al. 2005). We found 12 subunits of Complex I in N. ovalis (Nad1, Nad2, Nad3, Nad4, Nad4L, Nad5, Nad7, Nad9, Nad10, 24kDa*, 51kDa*, and 75kDa*), among them three that are encoded by the nuclear genome (marked by an asterisk); the others are encoded by the hydrogenosomal genome (fig. 1). All the N. ovalis Complex I proteins are part of the 14 proteins that compose the core bacterial Complex I. Also in Blastocystis, ten Complex I genes have been identified that are located on the organelle genome, and an additional six that are nuclear encoded (Perez-Brocal and Clark 2008; Stechmann et al. 2008; Wawrzyniak et al. 2008). In Proteromonas, ten Complex I proteins are encoded by the organellar genome (Perez-Brocal et al. 2010).
Of complex II, we detected SDHa and SDHb. In aerobic mitochondria, the four subunits of Complex II catalyze the oxidation of succinate to fumarate and the reduction of ubiquinone to ubiquinol. In N. ovalis, SDHa and SDHb have been proposed to function as fumarate reductase, reversing the reaction, and allowing the oxidation of quinols (rhodoquinol) (Boxma et al. 2005). In Blastocystis, all four components of Complex II have been identified (Stechmann et al. 2008).
Previous biochemical analyses have failed to detect the presence of Complex III/IV activity in the N. ovalis hydrogen-producing mitochondrion (Boxma et al. 2005), and accordingly, the analysis of its genome did not provide any evidence for the presence of Complex III/IV genes (fig. 1). Surprisingly, however, we did identify a nuclear-encoded homolog of cytochrome c1, an electron transporter related to Complexes III and IV. We sequenced the complete gene-sized-piece as well as the complete cDNA, confirming that the cytochrome c protein is transcribed. However, the cDNA appears to contain a stop codon (TGA), albeit one that is very rarely used. Therefore, with our current knowledge, we cannot exclude (Ricard et al. 2008) that the cDNA is translated notwithstanding the presence of a potential stop codon. Accordingly, an alignment of the cytochrome c sequence with homologs from various species reveals that the sequence is highly conserved but that the cysteines that normally hold the heme in cytochrome c1 are not present in the N. ovalis sequence, probably rendering the protein nonfunctional in electron transfer (supplementary fig. S3, Supplementary Material online). Therefore, if the gene is translated, it should exhibit an alternative function. This is the first report of a cytochrome c1 that has lost the two crucial cysteines as well as the histidine directly following the second cysteine. Only some Trypanosoma, Leishmania, and Euglena species miss the first cysteine, whereas the second cysteine and the histidine are universally conserved among species (Priest and Hajduk 1992).
We did not find genes encoding Complex V proteins in N. ovalis that could use the proton motive force generated by Complex I (Boxma et al. 2005). An organelle-encoded component of Complex V could not be identified in Blastocystis and Proteromonas either (Perez-Brocal and Clark 2008; Stechmann et al. 2008; Wawrzyniak et al. 2008; Perez-Brocal et al. 2010).
Another interesting protein involved in electron transfer is the α subunit of the electron transfer flavoprotein (ETF). ETF is an electron acceptor for various mitochondrial dehydrogenases, for example, dehydrogenases that oxidize amino acids. It transfers electrons to the electron transport chain via an ETF-ubiquinone oxidoreductase. The presence of ETF underscores the importance of the N. ovalis electron transport chain in its mitochondrial catabolism.
With the notable exception of Blastocystis and Proteromonas, complete functional Complexes I and II appear to be absent from hydrogenosomal species, such as Trichomonas, which has only retained two subunits of Complex I (the 24 and the 51 kD subunits) (Hrdý et al. 2004; Carlton et al. 2007; Stechmann et al. 2008). The electron transport chain is also absent from all mitosomal species. All these species lack a mitochondrial genome, whereas N. ovalis, Blastocystis, and Proteromonas possess elements of the electron transport chain as well as a mitochondrial genome. This supports the conclusion that all mitochondrial genomes carry genes encoding proteins of the electron transport chain (Burger et al. 2003), and likewise, that all organisms with an electron transport chain have a mitochondrial genome.
Propionate Metabolism
The predicted mitochondrial proteome contains a propionyl CoA carboxylase (EC: 6.4.1.3), that catalyzes the carboxylation reaction of propionyl CoA to D-methylmalonyl CoA, and a methylmalonyl CoA mutase (also found in Blastocystis, Stechmann et al. 2008) that converts methylmalonyl CoA to succinyl CoA. Both propionyl CoA carboxylase and methylmalonyl CoA mutase are absent from Paramecium and Tetrahymena (supplementary table S1, Supplementary Material online), indicating that the hydrogen-producing mitochondrion of N. ovalis does not just possess a subset of proteins present in aerobic mitochondria.
Fatty Acid Metabolism
In the fatty acid activation pathway, we detected a long-chain-fatty acid-CoA ligase, which catalyzes the reversible reaction:
ATP + a long-chain carboxylic acid + CoA <=> AMP + diphosphate + acyl-CoA.
This enzyme is present in all the 11 organisms in supplementary table S1, Supplementary Material online for which complete genomes are available, regardless of whether these organisms possess mitochondria, hydrogenosomes, or mitosomes. In contrast, the glycerol kinase is only present in the seven organisms that possess mitochondria or hydrogenosomes. Organisms with mitosomes lack glycerol kinase, with the exception of G. lamblia (supplementary table S1, Supplementary Material online).
Amino Acid Metabolism
Besides the glycine cleavage system (GCS, see below), we found several other enzymes involved in the metabolism of amino acids and specifically in the degradation of valine, methionine, and isoleucine. These were a branched-chain aminotransferase (EC: 2.6.1.42), the 2-oxoisovalerate dehydrogenase α and β (EC: 1.2.4.4) subunits, and a branched-chain α-keto acid dihydrolipoyl acyltransferase (EC: 2.3.1.-). The organelle of Blastocystis also contains enzymes involved in valine, leucine, and isoleucine metabolism (Stechmann et al. 2008). In addition, we identified a glutamate dehydrogenase and a cystathionine β-synthase. In general, amino acid metabolism appears much reduced in mitosomes and genome-less hydrogenosomes relative to mitochondria (supplementary table S1, Supplementary Material online).
GCS
The dihydrolipoyl dehydrogenase of PDH is also involved in the GCS that catalyzes the reversible reaction:
glycine + tetrahydrofolate + NAD+ ⇔ 5,10-methylene-tetrahydrofolate + NH3 + CO2 + NADH + H+
It provides the 5,10-methylene-tetrahydrofolate that is required for nucleotide synthesis. Of the GCS, we detected: the H-protein, a lipoic acid containing protein that carries the aminomethyl moiety of glycine that is bound to the sulfhydryl group by an S-C bond; the T-protein, which degrades reversibly the aminomethyl moiety attached to the H-protein to methylene-tetrahydrofolate and ammonia, using tetrahydrofolate in the process; and the L-protein, a dihydrolipoamide dehydrogenase that catalyzes the oxidation of the dihydrolipoyl group of the H-protein. We did not detect the substrate determining pyridoxal phosphate-containing protein (P-protein) of this complex, casting some doubt on whether the system actually uses Glycine. Nevertheless, we did find other components linked to this pathway: a cystathionine β-synthase, a serine hydroxymethyl transferase that converts serine to glycine, and a 5,10-methenyltetrahydrofolate synthetase. Interestingly, two proteins of the GCS have also been identified in the hydrogenosome of Trichomonas (L- and H-proteins) (Carlton et al. 2007), whereas the L, T, and H proteins also appear to be present in the genome of the minimal mitochondria-containing Plasmodium falciparum (supplementary table S1, Supplementary Material online). The mitochondrion-like organelle of Blastocystis contains all four components of the GCS, including the P-protein (Stechmann et al. 2008). The GCS is completely absent from mitosome-harboring species, such as Encephalitozoon cuniculi, E. histolytica, G. lamblia, and C. parvum.
Fe/S Cluster Synthesis
The only characteristic known so far that is shared by all mitochondria, mitosomes, and hydrogenosomes is the synthesis of Fe/S clusters (Lill and Mühlenhoff 2005; Tachezy and Dolezal 2007). Unfortunately, we found only a mitochondrial type I [2Fe2S] ferredoxin, which might provide reducing equivalents for Fe/S cluster synthesis as well as for other processes. Furthermore, we found the mitochondrial Hsp70, which has a role in Fe/S assembly and in other processes, such as protein import and folding. Although we have no reason to doubt their presence, we did not detect genes specific for Fe/S cluster assembly like Isu1/2, Nfs1, Isd11, Isa1/2, or frataxin and therefore have no specific evidence for canonical mitochondrial Fe/S cluster synthesis in N. ovalis.
Mitochondrial Protein Synthesis and Turnover
We found eight mitochondrial ribosomal proteins in N. ovalis: Rps 8, Rps 12, Rps 14; Rpl 2, Rpl 6, Rpl 14, Rpl 11, Rpl 15, and Rpl 20; the latter three are nuclear encoded. In agreement with Smits and coworkers (Smits et al. 2007), we did not find these mitochondrial ribosomal proteins in species lacking a mitochondrial genome, such as G. lamblia, E.cuniculi, Ent. histolytica, C. parvum, or T. vaginalis. Notably, 16 ribosomal proteins are encoded in the genome of the mitochondrion-like organelle of Blastocystis (Perez-Brocal and Clark 2008; Stechmann et al. 2008; Wawrzyniak et al. 2008). In N. ovalis, we found, besides the ribosomal proteins, FtsJ (MRM2), a well-conserved heat shock protein that is responsible for methylating 23S (rnl) rRNA.
Mitochondrial Carriers
We detected three mitochondrial carrier proteins: one orthologous to the 2-oxoglutarate/malate carrier that imports malate into the organelle and exports oxoglutarate out of the mitochondrial matrix, one orthologous to an ADP/ATP carrier, and one carrier that does not specifically cluster with any carrier of known specificity (PET8). The oxoglutarate/malate carrier (Coll et al. 2003) also transports other dicarboxylates and tricarboxylates (such as oxaloacetate, malate, malonate, succinate, citrate, isocitrate, cis-aconitate, and trans-aconitate). This carrier may also play an important role in several metabolic functions requiring organic acid flux to or from the mitochondria, such as nitrogen assimilation, transport of reducing equivalents into the mitochondria, and fatty acid elongation (Picault et al. 2002). Nyctotherus ovalis may use this carrier to import malate into the hydrogen-producing mitochondrion, which is transformed into fumarate and used by Complex II enzymes. In the sequenced genomes in supplementary table S1, Supplementary Material online, the oxoglutarate/malate carrier is present in several mitochondria-bearing species but absent from yeast, species with mitosomes, and species with hydrogenosomes without an organellar genome (supplementary table S1, Supplementary Material online). The oxoglutarate/malate carrier and a putative ADP/ATP carrier are also found in Blastocystis (Stechmann et al. 2008).
Additional mitochondrial proteins that are lacking in Trichomonas, and all species with mitosomes are MPV17 and ACN9, which have homologues in human and yeast mitochondria (not shown in fig. 3). They are, however, present in Blastocystis. Consistent with its presence in N. ovalis, MPV17 has been associated with mitochondrial genome stability (Spinazzola et al. 2006), while ACN9 has been associated with respiration (Steinmetz et al. 2002).
Protein Import and Processing
We detected the mitochondrial import machinery components TOM 34, MAS 5, MPP, FtsH, peptidyl-prolyl cis-trans isomerase, and protein disulfide isomerase. Hsps 10, 60, 70, and 90 were also identified.
Mitochondrial Division
One gene, a dynamin-related protein, has been found that might be engaged in mitochondrial division because it is an ortholog of the plant dynamin ADL2 that is known to be involved in mitochondrial division (Arimura et al. 2004).
ROS Defense
To our surprise, we found three systems used to cope with the detrimental effects of oxygen radicals. First of all, we found thioredoxins that act as antioxidants by facilitating the reduction of cysteines in other, oxidized proteins by thiol-disulfide exchange. Second, of the glutathione system, we found a glutathione reductase (EC 1.8.1.7) that reduces glutathione disulfide (GSSG) to the sulfhydryl form GSH. Finally, we found a peroxiredoxin.
The presence of enzymes supposed to react against oxygen radicals in an anaerobic environment might seem strange, but Reactive oxygen species (ROS) can be produced endogenously. Complex I is a major source of ROS, although in mitochondria Complex III seems to be the principal site of O2−• (superoxide) formation (Chen et al. 2003).
HGT
To detect HGT, we followed the same procedure as previously described (Ricard et al. 2006). After clustering the sequences to remove redundancy, we determined the BH of 1,914 cDNAs and 2,841 gDNAs against a set of 165 complete proteomes. We identified 31 cDNAs and 25 gDNAs with a bacterium as BH and 5 cDNAs and 6 gDNAs with an archaeon as BH. We analyzed the 67 N. ovalis sequences that have a bacterium or an archaeon as BH further, by reconstructing the evolution of the proteins in a phylogenetic tree generated with PhyML. When requiring that the N. ovalis sequence clustered with a high bootstrap value with only noneukaryotic sequences, four genes could be concluded to likely have been acquired through HGT (table 2; supplementary fig. S4a–d, Supplementary Material online). These genes are an acetyl-CoA synthase (AMP-forming), a β lactamase, a glycosylhydrolase, and an ornithine carbamoyltransferase. None of the proteins encoded by these genes has however an N-terminal targeting signal. In contrast, for the [FeFe] hydrogenase evidence for HGT and organellar location have been documented before (Boxma et al. 2007). This [FeFe] hydrogenase is remarkable because it is fused with the 24 and 51 kD subunits of a bacterial Complex I. This allows the use of NADH as electron donor. That we found only one gene obtained by HGT (the hydrogenase) that unequivocally encodes an enzyme located in the hydrogen-producing mitochondrion, suggests that the role of HGT in the transformation to hydrogen-producing mitochondrion is limited.
Table 2.
Nyctotherus ovalis Genes That Likely Have Been Acquired by HGT From Bacteria/Archaea.
| HGT (genus BH, id BH, sequence origin) | Accession Number | E.C. No | Hsa | Sce | Tth | Pte | Pfa | Cpa | Tva | Bla | Pla | Psp | Ehi | Gla | Ecu |
| Acetyl-CoA synthetase | AJ871315 | 6.2.1.1 | Y | Y | Y | Y | Y | Y | Y | Y | N | N | Y | ||
| (Archaeoglobus, YP_003399995, gDNA incl telomeres) | AM890088 | ||||||||||||||
| β lactamase | AM894317 | 3.5.2.6 | Y | N | Y | Y | N | Y | Y | N | N | N | N | ||
| (Pseudoalteromonas, YP_339054.1, gDNA incl telomere) | AM891292 | ||||||||||||||
| Glycosylhydrolase (glycosidase) | AM890556 | 3.2.1.- | N | N | N | N | N | N | N | N | N | N | N | ||
| (Geobacillus, YP_003244808.1, gDNA) | |||||||||||||||
| Ornithine carbamoyltransferase | AM896448 | 2.1.3.3 | Y | Y | N | N | Y | N | Y | Y | N | Y | N | ||
| (Aeromonas, YP_858515.1, cDNA incl. polyA) | |||||||||||||||
| [FeFe] hydrogenase | AY608627 | 1.12.7.2 | N | N | N | N | N | N | Y | Y | Y | Y | Y | Y | N |
| (Spirochaeta, YP_003873858, gDNA, telomeres) |
NOTE.—The proteins associated with the genes were selected based on having a BH with Bacteria or Archaea, followed by a phylogenetic analysis using the 250 BHs in the complete nonredundant database supplemented with the ten BHs in Eukaryotes and requiring a highly supported clustering of the N. ovalis gene with noneukaryotic genes, see Materials and Methods for details. With each HGT candidate are indicated the genus name of the species with the BH, the National Center for Biotechnology Information-identifier of the best hit, the source of the genetic material for the N. ovalis gene (cDNA or gDNA), and the extra evidence that the sequence is indeed derived from N. ovalis (the presence of telomeres specific for N. ovalis genes or of a poly A tail for a cDNA). Y indicates the presence of an ortholog of the mitochondrial protein in the corresponding species, N its absence. A blank cell is inserted if no answer is possible due to the incomplete genome. Species with mitochondria: (Hsa) Homo sapiens, (Sce) Saccharomyces cerevisiae, (Tth) Tetrahymena thermophila, (Pte) Paramecium tetraurelia, (Ram) Reclinomonas americana, and (Pfa) Plasmodium falciparum; species with hydrogenosomes: (Tva) Trichomonas vaginalis, (Bla) Blastocystis, (Pla) Psalteriomonas lanterna, and (Psp) Piromyces sp.; and species with a mitosomes: (Cpa) Cryptosporidium parvum, (Ehi) Entamoeba histolytica, (Gla) Giardia lamblia, and (Ecu) Encephalitozoon cuniculi.
Conclusions
With the bioinformatic analysis of the organellar genome and of the gDNAs and cDNAs, we are beginning to get a comprehensive view of the metabolism of the hydrogen-producing mitochondrion of N. ovalis. First of all, the organelle genome is a typical ciliate mitochondrial genome, with the exception of the notable lack of components of Complexes III, IV, and V of the electron transport chain. The latter feature is shared with the genome of the unrelated mitochondrion-like organelles of Blastocystis and Proteromonas. This phenomenon is a striking example of convergent evolution. Consequently, given the situation with Nyctotherus, Blastocystis, and Proteromonas, we can no longer use the production of ATP via oxidative phosphorylation as a defining feature of mitochondria, rather what sets them apart from mitosomes and hydrogenosomes is the proton-pumping electron transport chain of their mitochondrion-like organelles.
Moreover, the organelle of N. ovalis appears to host, besides an incomplete TCA cycle, other “classical” mitochondrial pathways, such as amino acid metabolism and fatty acid metabolism. A number of these proteins are typical for mitochondria and are not present in species with mitosomes or hydrogenosomes without a genome. These include, of course, proteins of the electron transport chain and of the mitochondrial ribosome but also enzymes, such as succinyl-CoA synthetase (however present in Trichomonas), pyruvate dehydrogenase, malate/α ketoglutarate antiporter, and 5,10-methenyltetrahydrofolate synthetase (see also Ginger et al. 2010). What emerges is a metabolism that is in some aspects (Complex III, Complex IV, and Complex V) reduced relative to the metabolism of aerobic ciliate mitochondria and that is strikingly similar to the metabolism of the mitochondrion-like organelle of Blastocystis. However, N. ovalis also possesses some extra proteins relative to aerobic ciliates. A few of these appear to have been acquired by HGT. The case of the [FeFe] hydrogenase is well documented (Boxma et al. 2007), but our analysis has also unearthed a few others, like an acetyl-CoA synthetase (AMP-forming). Nevertheless, their role in the adaptation to the anaerobic ecological niche of N. ovalis is not obvious. Not all N. ovalis proteins that we found and that are absent from the aerobic ciliates have been gained by HGT from bacteria. We also observe a pathway for propionate metabolism that contains a propionyl CoA carboxylase and a methylmalonyl CoA mutase. This pathway has not yet been observed in ciliates, and besides its presence in metazoa, has a patchy distribution among the eukaryotes, suggesting multiple loss events rather than HGT. Another example is a hybrid cluster protein (prismane) that is absent from the sequenced aerobic ciliates and that among the eukaryotes is mainly present in (faculatively) anaerobic species. However, the N. ovalis sequence does not specifically cluster with bacterial sequences (data not shown).
By definition, the organelle of N. ovalis is a hydrogenosome because it generates hydrogen with the aid of an [FeFe] hydrogenase (Lindmark and Müller 1973). We have shown here that this organelle is a mitochondrion that produces hydrogen, representing an intermediate state between mitochondria and genome-less hydrogenosomes. This allows us to outline a hypothetical scheme for the evolution of hydrogenosomes. A possible scenario for eukaryotes with a textbook mitochondrion genome and metabolism to evolve into an organism with hydrogenosomes could be as follows (where the order of events could be different):
Evolution of a reverse action of the TCA cycle (fumarate respiration) as shown in a number of anaerobic mitochondria (Tielens et al. 2002; Tielens and van Hellemond 2007).
Recruitment of a hydrogenase like that shown in Naegleria gruberi (Fritz-Laylin et al. 2010) and N. ovalis.
Loss of Complexes III, IV, and V of the electron transport chain as shown in N. ovalis, Blastocystis, and Proteromonas.
Loss of the remaining part of the organellar genome as shown, for example, in Trichomonas, Psalteriomonas, and other organisms with hydrogenosomes.
It is clear that hydrogenosomes, hydrogen-producing mitochondria, and mitochondrion-like organelles evolved several times from different aerobic progenitors. The eukaryotic cell is clearly characterized by the presence of a mitochondrion that has the capacity to adapt to anaerobic environments by reductive evolution to yield hydrogenosomes (and mitosomes).
Supplementary Material
Supplementary table S1 and figures S1–S4 are available at Molecular Biology and Evolution online (http://www.mbe.oxfordjournals.org/).
Acknowledgments
The authors would like to thank Klaas Schotanus for his help with the isolation of various N. ovalis sequences and Michel van de Pol for his help in drawing some of the figures. We also thank Prof. Alan Schwartz for revising the English phrasing. G.D.M.v.d.S. and G.R. were supported by the European Union fifth framework grant “CIMES” (QLK 3-2002-02151). I.S. was funded by the scholarship SFRH/BD/32959/2006 from the Portuguese Foundation for Science and Technology—FCT and by “Bolsas Rui Tavares 2010.” B.E.D. was funded by NWO HORIZON grant 050-71-058 and by NWO Veni grant 016.111.075. This work was supported by the Netherlands Genomics Initiative (Horizon Programme, project 050-71-555).
References
- Abascal F, Zardoya R, Posada D. ProtTest: selection of best-fit models of protein evolution. Bioinformatics. 2005;21:2104–2105. doi: 10.1093/bioinformatics/bti263. [DOI] [PubMed] [Google Scholar]
- Aguilera P, Barry T, Tovar J. Entamoeba histolytica mitosomes: organelles in search of a function. Exp Parasitol. 2008;118:10–16. doi: 10.1016/j.exppara.2007.08.004. [DOI] [PubMed] [Google Scholar]
- Akhmanova A, Voncken FG, Harhangi H, Hosea KM, Vogels GD, Hackstein JH. Cytosolic enzymes with a mitochondrial ancestry from the anaerobic chytrid Piromyces sp. E2. Mol Microbiol. 1998;30:1017–1027. doi: 10.1046/j.1365-2958.1998.01130.x. [DOI] [PubMed] [Google Scholar]
- Akhmanova A, Voncken FG, Hosea KM, Harhangi H, Keltjens JT, op den Camp HJ, Vogels GD, Hackstein JH. A hydrogenosome with pyruvate formate-lyase: anaerobic chytrid fungi use an alternative route for pyruvate catabolism. Mol Microbiol. 1999;32:1103–1114. doi: 10.1046/j.1365-2958.1999.01434.x. [DOI] [PubMed] [Google Scholar]
- Akhmanova A, Voncken F, van Alen T, van Hoek A, Boxma B, Vogels G, Veenhuis M, Hackstein JH. A hydrogenosome with a genome. Nature. 1998;396:527–528. doi: 10.1038/25023. [DOI] [PubMed] [Google Scholar]
- Altschul SF, Madden TL, Schaffer AA, Zhang J, Zhang Z, Miller W, Lipman DJ. Gapped BLAST and PSI-BLAST: a new generation of protein database search programs. Nucleic Acids Res. 1997;25:3389–3402. doi: 10.1093/nar/25.17.3389. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Andreoli C, Prokisch H, Hortnagel K, Mueller JC, Munsterkotter M, Scharfe C, Meitinger T. MitoP2, an integrated database on mitochondrial proteins in yeast and man. Nucleic Acids Res. 2004;32:D459–D462. doi: 10.1093/nar/gkh137. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Arimura S, Aida GP, Fujimoto M, Nakazono M, Tsutsumi N. Arabidopsis dynamin-like protein 2a (ADL2a), like ADL2b, is involved in plant mitochondrial division. Plant Cell Physiol. 2004;45:236–242. doi: 10.1093/pcp/pch024. [DOI] [PubMed] [Google Scholar]
- Barbera MJ, Ruiz-Trillo I, Leigh J, Hug LA, Roger AJ. The diversity of mitochondrion-related organelles amongst eukaryotic microbes. In: Martin WF, Müller M, editors. Originof mitochondria and hydrogenosomes. Berlin, Heidelberg (Germany): Springer-Verlag; 2007. pp. 239–275. [Google Scholar]
- Boxma B, de Graaf RM, van der Staay GW, et al. (15 co-authors) An anaerobic mitochondrion that produces hydrogen. Nature. 2005;434:74–79. doi: 10.1038/nature03343. [DOI] [PubMed] [Google Scholar]
- Boxma B, Ricard G, van Hoek AH, et al. (17 co-authors) The [FeFe] hydrogenase of Nyctotherus ovalis has a chimeric origin. BMC Evol Biol. 2007;7:230. doi: 10.1186/1471-2148-7-230. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Burger G, Gray MW, Lang BF. Mitochondrial genomes: anything goes. Trends Genet. 2003;19:709–716. doi: 10.1016/j.tig.2003.10.012. [DOI] [PubMed] [Google Scholar]
- Calvo S, Jain M, Xie X, Sheth SA, Chang B, Goldberger OA, Spinazzola A, Zeviani M, Carr SA, Mootha VK. Systematic identification of human mitochondrial disease genes through integrative genomics. Nat Genet. 2006;38:576–582. doi: 10.1038/ng1776. [DOI] [PubMed] [Google Scholar]
- Carlton JM, Hirt RP, Silva JC, et al. (65 co-authors) Draft genome sequence of the sexually transmitted pathogen Trichomonas vaginalis. Science. 2007;315:207–212. doi: 10.1126/science.1132894. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chen Q, Vazquez EJ, Moghaddas S, Hoppel CL, Lesnefsky EJ. Production of reactive oxygen species by mitochondria—central role of complex III. J Biol Chem. 2003;278:36027–36031. doi: 10.1074/jbc.M304854200. [DOI] [PubMed] [Google Scholar]
- Cherry JM, Adler C, Ball C, et al. (12 co-authors) SGD: Saccharomyces Genome Database. Nucleic Acids Res. 1998;26:73–79. doi: 10.1093/nar/26.1.73. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Coll O, Colell A, Garcia-Ruiz C, Kaplowitz N, Fernandez-Checa JC. Sensitivity of the 2-oxoglutarate carrier to alcohol intake contributes to mitochondrial glutathione depletion. Hepatology. 2003;38:692–702. doi: 10.1053/jhep.2003.50351. [DOI] [PubMed] [Google Scholar]
- de Graaf RM, Duarte I, Van Alen TA, Kuiper JW, Schotanus K, Rosenberg J, Huynen MA, Hackstein JHP. The hydrogenosomes of Psalteriomonas lanterna. BMC Evol Biol. 2009;9:287. doi: 10.1186/1471-2148-9-287. [DOI] [PMC free article] [PubMed] [Google Scholar]
- de Graaf RM, van Alen TA, Dutilh BE, Kuiper JW, van Zoggel HJ, Huynh MB, Görtz HD, Huynen MA, Hackstein JH. The mitochondrial genomes of the ciliates Euplotes minuta and Euplotes crassus. BMC Genomics. 2009;10:514. doi: 10.1186/1471-2164-10-514. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Duchen MR. Roles of mitochondria in health and disease. Diabetes. 2004;53(Suppl 1):S96–S102. doi: 10.2337/diabetes.53.2007.s96. [DOI] [PubMed] [Google Scholar]
- Edgar RC. MUSCLE: multiple sequence alignment with high accuracy and high throughput. Nucleic Acids Res. 2004;32:1792–1797. doi: 10.1093/nar/gkh340. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Embley TM, Martin W. Eukaryotic evolution, changes and challenges. Nature. 2006;440:623–630. doi: 10.1038/nature04546. [DOI] [PubMed] [Google Scholar]
- Embley TM, van der Giezen M, Horner DS, Dyal PL, Bell S, Foster PG. Hydrogenosomes, mitochondria and early eukaryotic evolution. IUBMB Life. 2003;55:387–395. doi: 10.1080/15216540310001592834. [DOI] [PubMed] [Google Scholar]
- Forner F, Foster LJ, Campanaro S, Valle G, Mann M. Quantitative proteomic comparison of rat mitochondria from muscle, heart, and liver. Mol Cell Proteomics. 2006;5:608–619. doi: 10.1074/mcp.M500298-MCP200. [DOI] [PubMed] [Google Scholar]
- Friedrich T, Böttcher B. The gross structure of the respiratory complex I: a Lego System. Biochim Biophys Acta. 2004;1608:1–9. doi: 10.1016/j.bbabio.2003.10.002. [DOI] [PubMed] [Google Scholar]
- Fritz-Laylin LK, Prochnik SE, Ginger ML, et al. (24 co-authors) The genome of Naegleria gruberi illuminates early eukaryotic versatility. Cell. 2010;140:631–642. doi: 10.1016/j.cell.2010.01.032. [DOI] [PubMed] [Google Scholar]
- Gabaldón T, Huynen MA. Shaping the mitochondrial proteome. Biochim Biophys Acta. 2004;1659:212–220. doi: 10.1016/j.bbabio.2004.07.011. [DOI] [PubMed] [Google Scholar]
- Gabaldon T, Rainey D, Huynen MA. Tracing the evolution of a large protein complex in the eukaryotes, NADH:ubiquinone oxidoreductase (Complex I) J Mol Biol. 2005;348:857–870. doi: 10.1016/j.jmb.2005.02.067. [DOI] [PubMed] [Google Scholar]
- Ginger ML, Fritz-Laylin LK, Fulton C, Cande WZ, Dawson SC. Intermediary metabolism in protists: a sequence-based view of facultative anaerobic metabolism in evolutionarily diverse eukaryotes. Protist. 2010;161:642–671. doi: 10.1016/j.protis.2010.09.001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Goldberg AV, Molik S, Tsaousis AD, Neumann K, Kuhnke G, Delbac F, Vivares CP, Hirt RP, Lill R, Embley TM. Localization and functionality of microsporidian iron-sulphur cluster assembly proteins. Nature. 2008;452:624–628. doi: 10.1038/nature06606. [DOI] [PubMed] [Google Scholar]
- Gray MW. Evolutionary biology: the hydrogenosome's murky past. Nature. 2005;434:29–31. doi: 10.1038/434029a. [DOI] [PubMed] [Google Scholar]
- Guindon S, Gascuel O. A simple, fast, and accurate algorithm to estimate large phylogenies by maximum likelihood. Syst Biol. 2003;52:696–704. doi: 10.1080/10635150390235520. [DOI] [PubMed] [Google Scholar]
- Hackstein JHP, Akhmanova A, Voncken FGJ, et al. (12 co-authors) Hydrogenosomes: convergent adaptations of mitochondria to anaerobic environments. Zoology. 2001;104:290–302. doi: 10.1078/0944-2006-00035. [DOI] [PubMed] [Google Scholar]
- Hackstein JHP, Baker SE, van Hellemond JJ, Tielens AG. Hydrogenosomes of anaerobic chytrids: an alternative way to adapt to anaerobic environments. In: Tachezy J, editor. Hydrogenosomes and mitosomes: mitochondria of anaerobic eukaryotes. Berlin, Heidelberg (Germany): Springer-Verlag; 2008. pp. 147–162. [Google Scholar]
- Hackstein JHP, de Graaf RM, van Hellemond JJ, Tielens AG. Hydrogenosomes of anaerobic ciliates. In: Tachezy J, editor. Hydrogenosomes and mitosomes: mitochondria of anaerobic eukaryotes. Berlin, Heidelberg (Germany): Springer-Verlag; 2008. pp. 97–112. [Google Scholar]
- Henze K, Martin W. Evolutionary biology: essence of mitochondria. Nature. 2003;426:172–176. doi: 10.1038/426127a. [DOI] [PubMed] [Google Scholar]
- Hjort K, Goldberg AV, Tsaousis AD, Hirt RP, Embley TM. Diversity and reductive evolution of mitochondria among microbial eukaryotes. Philos Trans R Soc Lond B Biol Sci. 2010;365:713–727. doi: 10.1098/rstb.2009.0224. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hrdý I, Hirt RP, Dolezal P, Bardonova L, Foster PG, Tachezy J, Embley TM. Trichomonas hydrogenosomes contain the NADH dehydrogenase module of mitochondrial complex I. Nature. 2004;432:618–622. doi: 10.1038/nature03149. [DOI] [PubMed] [Google Scholar]
- Hrdý I, Tachezy J, Müller M. Metabolism of trichomonad hydrogenosomes. In: Tachezy J, editor. Hydrogenosomes and mitosomes: mitochondria of anaerobic eukaryotes. Berlin, Heidelberg (Germany): Springer-Verlag; 2007. pp. 113–145. [Google Scholar]
- Lang BF, Gray MW, Burger G. Mitochondrial genome evolution and the origin of eukaryotes. Annu Rev Genet. 1999;33:351–397. doi: 10.1146/annurev.genet.33.1.351. [DOI] [PubMed] [Google Scholar]
- Lantsman Y, Tan KS, Morada M, Yarlett N. Biochemical characterization of a mitochondrial-like organelle from Blastocystis sp. subtype 7. Microbiology. 2008;154:2757–2766. doi: 10.1099/mic.0.2008/017897-0. [DOI] [PubMed] [Google Scholar]
- Lill R, Kispal G. Maturation of cellular Fe-S proteins: an essential function of mitochondria. Trends Biochem Sci. 2000;25:352–356. doi: 10.1016/s0968-0004(00)01589-9. [DOI] [PubMed] [Google Scholar]
- Lill R, Mühlenhoff U. Iron-sulfur-protein biogenesis in eukaryotes. Trends Biochem Sci. 2005;30:133–141. doi: 10.1016/j.tibs.2005.01.006. [DOI] [PubMed] [Google Scholar]
- Lindmark DG, Müller M. Hydrogenosome, a cytoplasmic organelle of the anaerobic flagellate Tritrichomonas foetus, and its role in pyruvate metabolism. J Biol Chem. 1973;248:7724–7728. [PubMed] [Google Scholar]
- Maralikova B, Ali V, Nakada-Tsukui K, Nozaki T, van der Giezen M, Henze K, Tovar J. Bacterial-type oxygen detoxification and iron-sulfur cluster assembly in amoebal relict mitochondria. Cell Microbiol. 2010;12:331–342. doi: 10.1111/j.1462-5822.2009.01397.x. [DOI] [PubMed] [Google Scholar]
- Mi-ichi F, Abu Yousuf M, Nakada-Tsukui K, Nozaki T. Mitosomes in Entamoeba histolytica contain a sulfate activation pathway. Proc Natl Acad Sci U S A. 2009;106:21731–21736. doi: 10.1073/pnas.0907106106. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Miller WL. Mitochondrial specificity of the early steps in steroidogenesis. J Steroid Biochem Mol Biol. 1995;55:607–616. doi: 10.1016/0960-0760(95)00212-x. [DOI] [PubMed] [Google Scholar]
- Perez-Brocal V, Clark CG. Analysis of two genomes from the mitochondrion-like organelle of the intestinal parasite Blastocystis: complete sequences, gene content, and genome organization. Mol Biol Evol. 2008;25:2475–2482. doi: 10.1093/molbev/msn193. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Perez-Brocal V, Shahar-Golan R, Clark CG. A linear molecule with two large inverted repeats: the mitochondrial genome of the stramenopile Proteromonas lacertae. Genome Biol Evol. 2010;2:257–266. doi: 10.1093/gbe/evq015. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Picault N, Palmieri L, Pisano I, Hodges M, Palmieri F. Identification of a novel transporter for dicarboxylates and tricarboxylates in plant mitochondria. Bacterial expression, reconstitution, functional characterization, and tissue distribution. J Biol Chem. 2002;277:24204–24211. doi: 10.1074/jbc.M202702200. [DOI] [PubMed] [Google Scholar]
- Priest JW, Hajduk SL. Cytochrome c reductase purified from Crithidia fasciculata contains an atypical cytochrome c1. J Biol Chem. 1992;267:20188–20195. [PubMed] [Google Scholar]
- Pritchard AE, Seilhamer JJ, Mahalingam R, Sable CL, Venuti SE, Cummings DJ. Nucleotide sequence of the mitochondrial genome of Paramecium. Nucleic Acids Res. 1990;18:173–180. doi: 10.1093/nar/18.1.173. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ricard G, de Graaf RM, Dutilh BE, et al. (13 co-authors) Macronuclear genome structure of the ciliate Nyctotherus ovalis: single-gene chromosomes and tiny introns. BMC Genomics. 2008;9:587. doi: 10.1186/1471-2164-9-587. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ricard G, McEwan NR, Dutilh BE, et al. (17 co-authors) Horizontal gene transfer from Bacteria to rumen Ciliates indicates adaptation to their anaerobic, carbohydrates-rich environment. BMC Genomics. 2006;7:22. doi: 10.1186/1471-2164-7-22. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Scheffler IE. Mitochondria make a come back. Adv Drug Deliv Rev. 2001;49:3–26. doi: 10.1016/s0169-409x(01)00123-5. [DOI] [PubMed] [Google Scholar]
- Sickmann A, Reinders J, Wagner Y, et al. (13 co-authors) The proteome of Saccharomyces cerevisiae mitochondria. Proc Natl Acad Sci U S A. 2003;100:13207–13212. doi: 10.1073/pnas.2135385100. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Smith DG, Gawryluk RM, Spencer DF, Pearlman RE, Siu KW, Gray MW. Exploring the mitochondrial proteome of the ciliate protozoon Tetrahymena thermophila: direct analysis by tandem mass spectrometry. J Mol Biol. 2007;374:837–863. doi: 10.1016/j.jmb.2007.09.051. [DOI] [PubMed] [Google Scholar]
- Smits P, Smeitink JAM, van den Heuvel LP, Huynen MA, Ettema TJG. Reconstructing the evolution of the mitochondrial ribosomal proteome. Nucleic Acids Res. 2007;35:4686–4703. doi: 10.1093/nar/gkm441. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Spinazzola A, Viscomi C, Fernandez-Vizarra E, et al. (20 co-authors) MPV17 encodes an inner mitochondrial membrane protein and is mutated in infantile hepatic mitochondrial DNA depletion. Nat Genet. 2006;38:570–575. doi: 10.1038/ng1765. [DOI] [PubMed] [Google Scholar]
- Stechmann A, Hamblin K, Perez-Brocal V, Gaston D, Richmond GS, van der Giezen M, Clark CG, Roger AJ. Organelles in Blastocystis that blur the distinction between mitochondria and hydrogenosomes. Curr Biol. 2008;18:580–585. doi: 10.1016/j.cub.2008.03.037. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Steinmetz LM, Scharfe C, Deutschbauer AM, et al. (11 co-authors) Systematic screen for human disease genes in yeast. Nat Genet. 2002;31:400–404. doi: 10.1038/ng929. [DOI] [PubMed] [Google Scholar]
- Szklarczyk R, Huynen MA. Expansion of the human mitochondrial proteome by intra- and inter-compartmental protein duplication. Genome Biol. 2009;10:R135. doi: 10.1186/gb-2009-10-11-r135. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tachezy J, Dolezal P. Iron-sulfur proteins and iron-sulfur cluster assembly in organisms with hydrogenosomes and mitosomes. In: Martin WF, Müller M, editors. Origin of mitochondria and hydrogenosomes. Berlin, Heidelberg (Germany): Springer Verlag; 2007. pp. 105–133. [Google Scholar]
- Tachezy J, Smid O. Mitosomes in parasitic protists. In: Tachezy J, editor. Hydrogenosomes and mitosomes: mitochondria of anaerobic eukaryotes. Berlin, Heidelberg (Germany): Springer Verlag; 2007. pp. 201–230. [Google Scholar]
- Thompson JD, Higgins DG, Gibson TJ. CLUSTAL W: improving the sensitivity of progressive multiple sequence alignment through sequence weighting, position-specific gap penalties and weight matrix choice. Nucleic Acids Res. 1994;22:4673–4680. doi: 10.1093/nar/22.22.4673. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tielens AG, Rotte C, van Hellemond JJ, Martin W. Mitochondria as we don't know them. Trends Biochem Sci. 2002;27:564–572. doi: 10.1016/s0968-0004(02)02193-x. [DOI] [PubMed] [Google Scholar]
- Tielens AG, van Grinsven KW, Henze K, van Hellemond JJ, Martin W. Acetate formation in the energy metabolism of parasitic helminths and protists. Int J Parasitol. 2010;40:387–397. doi: 10.1016/j.ijpara.2009.12.006. [DOI] [PubMed] [Google Scholar]
- Tielens AGM, van Hellemond JJ. Anaerobic mitochondria: properties and origin. In: Martin WF, Müller M, editors. Origin of mitochondria and hydrogenosomes. Berlin, Heidelberg (Germany): Springer Verlag; 2007. pp. 84–101. [Google Scholar]
- Ueda M, Fujimoto M, Arimura S, Tsutsumi N, Kadowaki K. Presence of a latent mitochondrial targeting signal in gene on mitochondrial genome. Mol Biol Evol. 2008;25:1791–1793. doi: 10.1093/molbev/msn139. [DOI] [PubMed] [Google Scholar]
- Wawrzyniak I, Roussel M, Diogon M, Couloux A, Texier C, Tan KS, Vivares CP, Delbac F, Wincker P, El Alaoui H. Complete circular DNA in the mitochondria-like organelles of Blastocystis hominis. Int J Parasitol. 2008;38:1377–1382. doi: 10.1016/j.ijpara.2008.06.001. [DOI] [PubMed] [Google Scholar]
- Yarlett N. Anaerobic protists and hidden mitochondria. Microbiology. 2004;150:1127–1129. doi: 10.1099/mic.0.27174-0. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.