Transient cleavage failure in dividing cells is not sufficient to establish stable populations of cells with extra centrosomes.
Abstract
We tested whether cleavage failure as a transient event establishes an incidence of centrosome amplification in cell populations. Five rounds of ∼30% cytochalasin-induced cleavage failure in untransformed human cell cultures did not establish centrosome amplification in the short or long terms. The progeny of binucleate cells progressively dropped out of the cell cycle and expressed p53/p21, and none divided a fourth time. We also tested whether cleavage failure established centrosome amplification in transformed cell populations. Tetraploid HCT116 p53−/− cells eventually all failed cleavage repeatedly and ceased proliferating. HeLa cells all died or arrested within four cell cycles. Chinese hamster ovary cells proliferated after cleavage failure, but five rounds of induced cleavage failure produced a modest increase in the incidence of centrosome amplification in the short term, which did not rise with more cycles of cleavage failure. This incidence dropped to close to control values in the long term despite a 2–6% rate of spontaneous cleavage failure in the progeny of tetraploid cells.
Introduction
Centrosome amplification, the presence of extra centrosomes, is found in many preinvasive carcinomas and most late-stage human solid tumor cells (Lingle and Salisbury, 2000; Pihan et al., 2003; Sagona and Stenmark, 2010). Supernumerary centrosomes generate chromosomal instability by increasing the incidence of unequal chromosome distribution on multipolar spindles (Brinkley, 2001; Nigg, 2002) or by generating merotelically attached chromosomes that are prone to missegregate (Cimini et al., 2001) even if the spindle becomes bipolar because of centrosome bundling (Ganem et al., 2009; Silkworth et al., 2009). Consequent whole-chromosome losses/gains lead to genetic imbalances that may promote unregulated growth, loss of heterozygosity for tumor suppressor genes, and resistance to chemotherapeutic agents (Lengauer et al., 1997; Orr-Weaver and Weinberg 1998; Pihan et al., 1998; Nigg, 2002, 2006). Chromosome instability is thought to be a major driver of multistep carcinogenesis (Pihan et al., 2001; D’Assoro et al., 2002; Goepfert et al., 2002; Krämer et al., 2002; Lingle et al., 2002; Weaver et al., 2007; Basto et al., 2008; Chandhok and Pellman, 2009).
How an incidence of centrosome amplification is established and maintained in tumor cell populations is not well understood. Possibilities include centriole reduplication (Balczon et al., 1995), centriole overduplication (Kleylein-Sohn et al., 2007; Duensing et al., 2009), de novo centriole assembly, and cleavage failure (and equivalent cell–cell fusion) particularly if they were ongoing events (Brinkley, 2001; Krämer et al., 2002; Meraldi et al., 2002; Nigg, 2002, 2006; Storchova and Pellman, 2004; Sagona and Stenmark, 2010). Overexpression of SAK/PLK4 or expression of the high risk papillomavirus protein E7 causes centriole overduplication and is implicated in tumor development (Ko et al., 2005; Duensing et al., 2009). Centrosome amplification from de novo centriole assembly would require cooperating defects because this phenomenon has been observed only after the resident centrioles have been removed (La Terra et al., 2005; Uetake et al., 2007).
Cleavage failure is another direct route to the establishment of an incidence of centrosome amplification in cell populations. For untransformed cells, it might be the only avenue to centrosome amplification because these cells do not show centriole reduplication/overduplication or de novo centriole assembly. Failure to divide immediately doubles centrosome number, and centrosome bundling at mitosis could maintain elevated centrosome content by allowing cells to undergo bipolar divisions (Borel et al., 2002; Sluder and Nordberg, 2004; Uetake and Sluder, 2004; Ganem et al., 2009). Importantly, doubling of the genome after cleavage failure increases the probability that some daughters of multipolar divisions will have enough chromosomes to remain viable. The importance of cleavage failure in the evolution of cellular transformation in vivo is supported by observations that tetraploidization often precedes aneuploidy in solid tumors (Shackney et al., 1989; Levine et al., 1991; Galipeau et al., 1996; Reid et al., 1996; Ganem et al., 2007). Also, the injection of tetraploid p53−/− mouse embryo fibroblasts into nude mice produces tumors, whereas the injection of diploid cells does not (Fujiwara et al., 2005).
Nevertheless, the ability of cleavage failure as a transient event to establish centrosome amplification in proliferating cell populations has not been directly examined. We tested whether repeated rounds of cleavage failure can establish centrosome amplification in populations of untransformed human cells. We also tested whether cooperating defects, such as a compromised p53 pathway, can enable cleavage failure to establish centrosome amplification in populations of transformed cells.
Results and discussion
Untransformed cells
We used human telomerase reverse transcriptase (hTERT)–immortalized RPE1 cells stably expressing low levels of centrin1/GFP to tag the centrioles. The centriole number per cell was determined by the number of bright focal GFP spots and/or immunostaining with a γ-tubulin monoclonal antibody, showing a consistent 1:1 colocalization between GFP spots and γ-tubulin–reactive spots (Fig. S1 A). Time-lapse recordings revealed that cytochalasin-induced binucleate cells assembled a single spindle at mitosis (86% segregate chromosomes and divide in a bipolar fashion; the remainder divide in a tripolar fashion; n = 121). All daughter cells were mononucleate after bipolar and tripolar mitoses. We never observed binucleate cells producing binucleate daughters. Centriole duplication in binucleate cells was normal, and in mitosis, centriole distribution to daughter cells could be equal or unequal (Fig. S1, B and C).
Asynchronous cultures were treated with 0.5 µM cytochalasin D for 10 h to induce a 26–38% incidence of binucleate cells. After drug removal, we cultured the mixture of diploid and binucleate cells for 100 h, at which time the culture was passaged and split. One culture was treated again with cytochalasin, and the other was passaged seven times (∼50 doublings). This protocol was repeated four more times for a total of five rounds of cleavage failure (Fig. 1 A). For each round, we determined the incidence of centrosome amplification 100 h after cleavage failure and at passages 1, 3, and 7. The centrosome number per cell was determined by counting centrioles. Cells containing more than four centrin/γ-tubulin spots were scored as cases of centrosome amplification. Binucleate cells were not scored because time-lapse recordings (described in the second following paragraph) revealed that these were cells that arrested in first interphase. Data from two experiments with closely similar results are pooled for presentation.
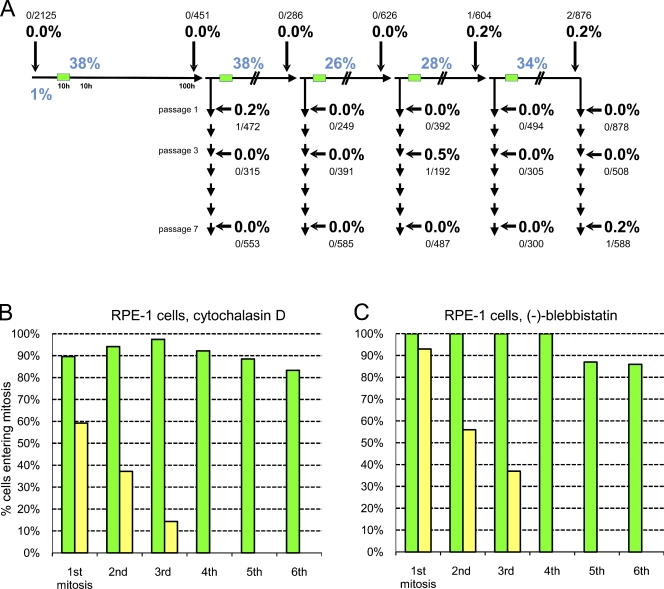
Figure 1.
Centrosome amplification and proliferation of untransformed cells after cleavage failure. (A) Centrosome amplification after repeated cleavage failure in RPE1 cell populations. Asynchronous cultures were treated with 0.5 µM cytochalasin D for 10 h (green bars) and split 100 h later. One culture was passaged seven times; the other was treated again with cytochalasin. This was repeated four more times. The percentages of binuclear cells after cytochalasin treatments are shown in blue. The percentages of centrosome amplification (more than four centrioles) are shown in bold with the number of cells counted shown above or below. (B) Proliferative capacity of binuclear cells and their progeny (yellow bars) and same-preparation control cells (green bars) after cytochalasin-induced cleavage failure. Bars show the percentage of cells that enter mitosis after completing the prior mitosis for each cell cycle; cells arresting in a previous cell cycle are not included. Percentages were calculated from lineages of multiple individual cells followed in one experiment. (C) Proliferative capacity of binuclear cells (yellow bars) and same-preparation control cells (green bars) after blebbistatin-induced cleavage failure. One experiment is shown on multiple cells.
Control cultures exhibited no centrosome amplification (n = 2,125). For all rounds of cleavage failure and subsequent passages collectively, we found only six cells of 9,552 scored that contained five to six centrioles (Fig. 1 A). For reasons outlined in the following paragraph, these cells were probably not proliferative. Importantly, there was no correlation between rounds of cleavage failure and the incidence of centrosome amplification. Thus, repeated cleavage failure is not sufficient to establish an incidence of centrosome amplification in untransformed cell populations.
To gain insight into this, individual binucleate cells and control cells in the same preparations were continuously followed for 100–168 h by video microscopy starting shortly after cytochalasin treatment. 48 control cells and their progeny propagated through six mitoses (Fig. 1 B, green bars) with a mean cell cycle duration of 17.3 h (n = 277) with no slowing of the cell cycle at later times in the film runs. For 120 binucleate cells, we found that progressively fewer and fewer of the progeny entered mitosis at each cell cycle (Fig. 1 B, yellow bars), and none divided a fourth time. For as long as the tetraploid cells propagated, their cell cycle duration was on average 18.5 h (n = 117). The gradual interphase arrest of the progeny of binucleate cells was not simply caused by catastrophic chromosome loss through multipolar divisions because 52 of the starting binucleate cells showed strictly bipolar lineages yielding 87 progeny remaining in the microscope fields, and none divided a fourth time.
These results are not particular to RPE1 cells. We followed 14 control and 44 same-preparation binucleate primary human fibroblasts for 100 h. The control cells proliferated through six mitoses at a 90–100% rate before the film recordings were terminated because of confluency (Fig. S2, green bars). The progeny of the binucleate cells, however, progressively dropped out of the cell cycle, and none entered a fourth mitosis even though 79% of the mitoses were bipolar (Fig. S2, yellow bars; and Table S1).
We note that 40–50% of the RPE1 cells and primary fibroblasts arrested as binucleates after cytochalasin-induced cleavage failure. Given that G1 progression in untransformed cells is extremely sensitive to the presence of cytochalasin (Lohez et al., 2003; Uetake and Sluder, 2004), we blocked cleavage in RPE1 cells with the myosin II inhibitor (−)-blebbistatin and followed binucleate cells and their same-preparation controls for 70–118 h. Nine control cells propagated through up to six mitoses before the film runs were terminated. For cells that failed cleavage, 28 of 30 progressed through the first postcleavage-failure mitosis. After that, their progeny progressively dropped out of the cell cycle with none dividing a fourth time (Fig. 1 C). Thus, the immediate postcleavage-failure arrest we observed for some tetraploid cells appears to be largely caused by residual cytochalasin, not tetraploidy. Also, the progressive withdrawal of doubled cells from the cell cycle is not specific to cytochalasin-induced cleavage failure.
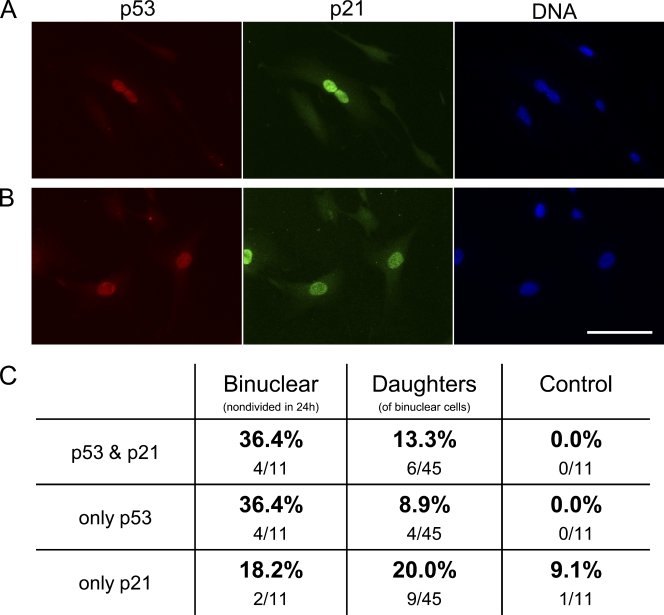
We next followed 46 binucleate RPE1 cells for 24 h after cytochalasin-induced cleavage failure, fixed them, and double labeled for p53 and p21 expression. 10 of the 11 that arrested in first interphase as binucleate cells expressed p53 and/or p21 (Fig. 2, A and C). For the 34 binucleates that divided, 45 of their daughters remained in the fields of view. 19 (42%) expressed p53 and/or p21, indicating that these were binucleate cell progeny that arrested after first mitosis (Fig. 2, B and C). Those not expressing p53 or p21 were presumably still cycling. Thus, cleavage failure can eventually lead to a p53 response and a p21-enforced cell cycle arrest.
Figure 2.
Expression of p53 and p21 in untransformed cells that stop cycling after cleavage failure. (A) Binuclear cell arrested after cleavage failure expressing p53 (red), p21 (green), and DNA (blue). (B) Daughters of a binuclear cell fixed 15 h after first tetraploid mitosis showing p53 (red) and p21 (green). (C) Percentages of cells expressing only p53, only p21, or both. The first column shows binucleates that arrested in first interphase, the second column shows daughters of binucleates, and the third column shows same-preparation control cells. Fluorescence micrographs are shown. Bar, 50 µm.
To test whether this p53 response is caused by DNA damage, we followed 36 binucleate cells and their progeny for 24 h, fixed them, and immunostained for nuclear phospho-H2AX foci, an indicator of DNA damage (Rogakou et al., 1998). Only 3 of the 68 daughter cells still in view showed phospho-H2AX foci (see Uetake and Sluder, 2010 for validation of the methodology). Thus, DNA damage is not the primary reason for tetraploid cells dropping out of the cell cycle.
We also examined whether doubled chromosome/centrosome content sufficiently prolonged prometaphase to trigger a p53-dependent G1 arrest of the daughter cells (Uetake and Sluder, 2010). For tetraploid RPE1 cells, prometaphase was on average 40.9 min (range of 12–75 min; n = 56) versus 20.8 min (range of 12–54 min; n = 38) for the same preparation controls, which is consistent with Yang et al. (2008). However, even the longest prometaphase duration in tetraploid cells was not long enough to stop daughter cell proliferation (Uetake and Sluder, 2010).
Previous work revealed that untransformed cells do not possess a tetraploidy checkpoint because most binucleate cells progressed through S phase and first mitosis (Uetake and Sluder, 2004). However, their progeny were not followed further. Our present longer term observations reveal that despite the lack of a classical checkpoint monitoring cleavage failure, the doubled condition is partially but poorly tolerated by untransformed cells. Though tetraploidy does not appear to greatly compromise proliferation in the short term, the progressive arrest of initially tetraploid cells after they had divided one or more times suggests that other factors are in play to cause a cell cycle arrest. What these might be is an intriguing mystery because the genome and cytoplasmic volumes, albeit doubled, are initially balanced and complete. Although doubled gene dosage might cause problems with the regulation of gene expression and growth control (Andalis et al., 2004; Upender et al., 2004), tetraploidy per se may not be the sole cause of the proliferation block. Proliferating tetraploid RPE1 cells with a normal centrosome complement can be selected through repeated FACS sorting (Ganem et al., 2009), and there are rare live births of tetraploid humans, albeit with lethal developmental defects (Nakamura et al., 2003; Storchova and Pellman, 2004). Perhaps chromosome missegregations caused by multipolar divisions or merotelically attached chromosomes on bipolar spindles (Ganem et al., 2009; Silkworth et al., 2009) contribute to the cell cycle arrest. Single-chromosome gains or losses in diploid RPE1 cells lead to a rapid p53-dependent cell cycle arrest (Thompson and Compton, 2008, 2010), but it is not known whether or not such single-chromosome gains or losses can induce a cell cycle arrest on an initially tetraploid background. Perhaps increased gene dosage, chromosome missegregations, and cytoskeletal alterations caused by extra centrosomes act additively to sufficiently stress the doubled cell to eventually trigger a p53 response (Ganem and Pellman, 2007; Ganem et al., 2007; Uetake et al., 2007). Regardless of why cells drop out of the cell cycle, our observations reveal that cleavage failure, as a transient event, is not a major driver of an incidence of centrosome amplification in proliferating populations of untransformed cells.
Transformed cells
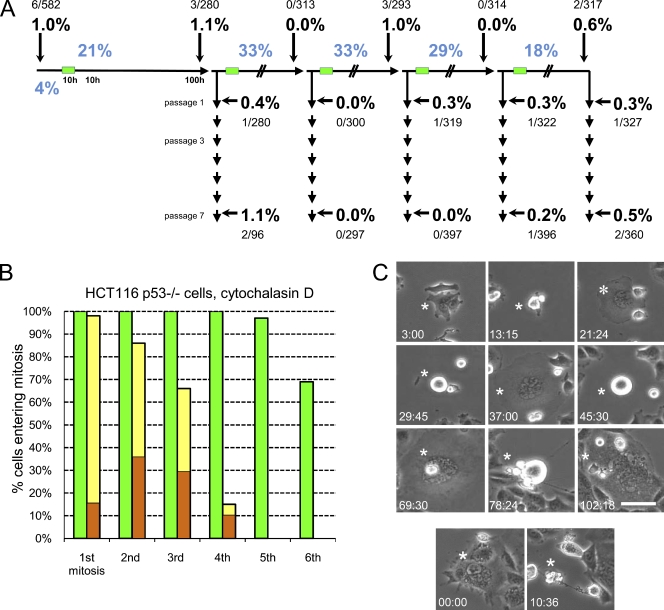
To determine whether cleavage failure can establish centrosome amplification in cell populations that continue proliferating, we repeated our experiments on three transformed cell lines with defective p53 pathways. For HCT116 p53−/− cells, we induced five rounds of 18–33% cleavage failure in asynchronous cultures using the same protocol used for RPE1 cells. Control populations showed a 1% incidence of centrosome amplification. Despite the five rounds of cleavage failure, we found that at 100 h, passage 1, and passage 7, the incidence of centrosome amplification was ∼1% or less in proliferating mononucleated cells (Fig. 3 A). Large multinucleated cells resulting from repeated cleavage failure (see following paragraph) had many centrosomes but were not counted because they were not proliferative.
Figure 3.
Centrosome amplification and proliferation of HCT116 p53−/− cells after cleavage failure. (A) Centrosome amplification in proliferative HCT116 p53−/− cell populations after repeated cleavage failure. The experimental protocol and display of the results are the same as those shown in Fig. 1 A. Large multinucleated cells are not scored because they do not proliferate. The percentages of binuclear cells after cytochalasin treatments are shown in blue. The percentages of centrosome amplification (more than four centrioles) are shown in bold with the number of cells counted shown above or below. (B) Proliferative capacity of binuclear cells (yellow bars) and same-preparation control cells (green bars). Orange portions of the bars indicate the percentages of cells that spontaneously fail cleavage at mitosis. Such cells cycle but repeatedly fail cleavage and cease proliferating. One experiment is shown on multiple cells. (C, top) Binuclear cell (asterisks) exhibiting multiple rounds of cleavage failure, resulting in a large multinucleated cell. (bottom) Death of another large multinuclear cell. Times are in hours and minutes. Phase-contrast microscopy is shown. Bar, 50 µm.
We followed individual binucleate cells and same-preparation controls for up to 125 h. 10 control cells and their progeny divided up to seven times with a mean cell cycle duration of 14 h (range of 11–17 h) with no slowing of the cell cycle later in the film runs (Fig. 3 B, green bars). All mitoses appeared normal with the exception of one cell that failed to cleave (Table S1). For 39 binucleate cells, 8–39% of their progeny failed cleavage at mitosis (Fig. 3 B, orange portions of yellow bars). Cells that spontaneously failed cleavage continued to cycle but repeatedly failed cleavage, yielding large multinucleated cells that eventually stopped cycling or died (Fig. 3 C).
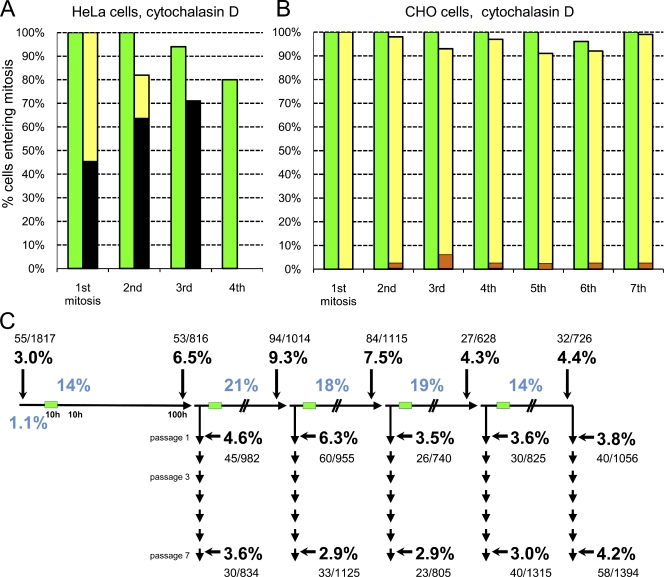
For HeLa cells, we followed binucleate cells and their same-preparation control cells for up to 83 h when the cultures became confluent. Eight control cells and their progeny divided four times (Fig. 4 A, green bars) with a mean cell cycle duration of 22.3 h (range of 18–29 h). All 51 mitoses were bipolar, and one cell failed cleavage (Table S1). For 41 binucleate cells and their progeny, all mitoses showed evidence of spindle multipolarity; 58% of the cells cleaved in a multipolar fashion or in a bipolar but unequal fashion as judged by cell and nuclear size after mitosis. Their cell cycle duration averaged 24.2 h (range of 17.5–33 h). After the first two tetraploid mitoses, a portion of the daughter cells died, and all died after the third mitosis (Fig. 4 A, black portion of yellow bars).
Figure 4.
Centrosome amplification and proliferation of HeLa and CHO cells after cleavage failure. (A) Proliferative capacity of binuclear HeLa cells (yellow bars) and same preparation control cells (green bars). Black portions of bars indicate the percentage of the cells that die during or just after that mitosis. (B) Proliferative capacity of binuclear CHO cells (yellow bars) and same-preparation control cells (green bars). The darker portions of the yellow bars denote the percentage of cells that fail cleavage at each mitosis. (A and B) One experiment is shown on multiple cells. (C) Centrosome amplification after repeated cleavage failure in CHO cell populations. The experimental protocol and display of the results are the same as those shown in Fig. 1 A. The percentages of binuclear cells after cytochalasin treatments are shown in blue. The percentages of centrosome amplification (more than four centrioles) are shown in bold with the number of cells counted shown above or below.
For CHO cells in 85-h film runs, five control cells divided in a bipolar fashion eight times with a mean cell cycle duration of 11.2 h (range of 9.5–16.5 h; n = 72; Fig. 4 B, green bars). The progeny of 21 same-preparation binucleate cells propagated for eight cycles with a mean cell cycle duration of 12 h (range of 9.5–23 h; n = 160; Fig. 4 B, yellow bars). 90% of all mitoses (n = 150) were bipolar and equal in appearance, 4% were unequal, and 2–6% failed cleavage (Fig. 4 B, orange portions of the yellow bars; and Table S1).
In three experiments, we induced five rounds of cleavage failure using the protocol we used for RPE1 cells. Control populations showed a 3.0% incidence of centrosome amplification (n = 1,817). At 100 h, taking all cycles of cleavage failure together, the incidence of centrosome amplification ranged from 4.3 to 9.3%. At passage 1, the incidence of centrosome amplification ranged from 3.5 to 6.3%, and at passage 7, it ranged from 2.9 to 4.2% (Fig. 4 C). The incidence of centrosome amplification did not systematically increase with more rounds of induced cleavage failure or with increasing passage number.
Together, these observations reveal that the response of transformed cells to cleavage failure is cell line specific. For many transformed cell types, cleavage failure can be lethal, as we found for HeLa and Ganem et al. (2009) found for several other transformed lines. Perhaps these cell lines are particularly sensitive to unequal chromosome distribution induced by spindle multipolarity. For others, such as HCT116 p53−/−, cleavage failure predisposes cells to additional rounds of cleavage failure, resulting in huge cells that cease proliferating or die. It appears that this cell type does not have a sufficiently robust cleavage apparatus to divide larger than normal cells with high fidelity. Perhaps HCT116 p53−/− cells after repeated cleavage failure are representative of cells from high grade human tumors that show extensive centrosome amplification (Lingle et al., 1998; Pihan et al., 1998; Sato et al., 1999), but whose proliferative capacity with many centrosomes is uncertain.
For CHO cells, repeated rounds of cleavage failure lead to a modest step up in the incidence of centrosome amplification at 100 h, and this incidence diminishes at later passages, though it sometimes remains slightly above the 3% control values. These observations suggest a source and sink situation for the incidence of proliferating cells with extra centrosomes (Nigg, 2006). The source of centrosome amplification is an ongoing 2–6% rate of cleavage failure for mitotic tetraploid cells. The sink may be that tetraploidy and/or extra centrosomes diminish the proliferative capacity of these cells under our culture conditions, as indicated by our finding that the incidence of centrosome amplification diminished at later passages and did not progressively increase with more cycles of cleavage failure. These observations suggest that if centrosome amplification is to become stably established in proliferating populations of cancer cells, there must be an ongoing incidence of an event that increases centrosome numbers, be it cleavage failure or centriole reduplication/overduplication.
Materials and methods
Cell culture and live-cell imaging
hTERT-RPE1 cells (Takara Bio, Inc.) stably expressing low levels of human centrin1-GFP were cultured in 1:1 DME and Ham’s F12 media. Human primary foreskin fibroblasts (BJ strain, used at doublings 25–35) were obtained from American Type Culture Collection and cultured in MEM media. HeLa cells (American Type Culture Collection) were grown in DME, and CHO-K1 cells (American Type Culture Collection) and CHO cells expressing centrin1-GFP (gift from A. Khodjakov, Wadsworth Center, New York State Department of Health, Albany, NY) were maintained in Ham’s F12 medium. HCT116 p53−/− cells (gift from W. Theurkauf, University of Massachusetts Medical School, Worcester, MA) were maintained in McCoy’s modified media. All media (Invitrogen) contained 12.5 mM Hepes, 10% FCS (Invitrogen), 100 U/ml penicillin G, and 100 µg/ml streptomycin (Invitrogen). 16–18 h before drug treatment, cells were seeded on 18 × 18– or 22 × 22–mm coverslips and pretreated with 1 ml of 20-µg/ml fibronectin solution (F1141; Sigma-Aldrich) for 1 h at 37°C on cleaned coverslips previously treated with poly-l-lysine (Sigma-Aldrich). Cytochalasin D (Sigma-Aldrich) was used at 0.5 µM by a dilution of 1,000× DMSO stock. Asynchronous cultures were treated with 100 µM (−)-blebbistatin (Sigma-Aldrich) for 50 min. Drug treatments were terminated by washing the coverslips with drug-free medium more than five times over a period of 30 min.
Shortly after drug removal, binucleate cells and their same-preparation controls were continuously filmed for 70–118 h. For continuous live-cell film runs, coverslips bearing cells were assembled into chambers containing 1:1 DME and Ham’s F12 media as previously described (Sluder et al., 2005). The medium was changed every second day. Individual binucleate cells were circled on the coverslip with a diamond scribe and then followed at 37°C with microscopes (Universal [Carl Zeiss]; BH2 [Olympus]; or DMEXE [Leica]) equipped with phase-contrast optics using 10× objectives/0.3–0.32 NA. Image sequences were taken with cameras (Orca ER [Hamamatsu Photonics]; Retiga EX [Qimaging, Corp.]; or Retiga EXi Fast [Qimaging, Corp.]) in hoods enclosing the entire microscope. Images were acquired every 3 min with C-imaging software (Hamamatsu Photonics) and were exported as QuickTime videos (90% of IndeoVideo 5.1 compression mode or 100% of CinePak compression; Apple).
For GFP imaging of centrioles in living cells, single focal plane images or z stacks of three to eight images (40 ms each) were acquired every 4–12 h with a 100×/1.30 NA HCX PL Fluotar objective on a microscope (DMR; Leica). Images were acquired with a camera (Orca ER) using SlideBook software (Intelligent Imaging Innovations). Z-stack images were compiled to form maximum intensity point projection images.
Immunocytochemistry
Cells were fixed with −20°C methanol and immunostained as previously described (Uetake et al., 2007). The antibodies used in this study were monoclonal anti–γ-tubulin antibody at 1:1,000 (Sigma-Aldrich) or 1:100 (sc-51715; Santa Cruz Biotechnology, Inc.), p21 at 1:100 (7903; Abcam), p53 at 1:100 (sc-126; Santa Cruz Biotechnology, Inc.), phH2A.X at 1:1,000 (07–164; Millipore), anti–mouse IgG Alexa Fluor 488 at 1:1,000, or anti–rabbit IgG Alexa Fluor 594 at 1:1,000 (Invitrogen). Images were acquired with a 100×/1.30 NA oil HCX PL Fluotar or 63×/1.32 NA oil HCX Plan Apochromat objective on a microscope (DMR). Images were acquired with a camera (Retiga EXi Fast) using SlideBook software. In preparing the figures, images were cropped and assembled using Photoshop software (Adobe). Only contrast and/or brightness adjustments were made and performed uniformly across the entire images.
Online supplemental material
Fig. S1 shows hTERT-RPE1 cells expressing centrin1-GFP and its colocalization with γ-tubulin as well as images from live-cell imaging of binucleate RPE1 cells and centriole duplication and distribution in these cells. Fig. S2 displays the proliferative capacity of binucleate human primary fibroblasts after cytochalasin-induced cleavage failure. Table S1 summarizes mitotic outcomes for binucleate cells and same-preparation control cells for all cell lines used in this study. Online supplemental material is available at http://www.jcb.org/cgi/content/full/jcb.201101073/DC1.
Acknowledgments
We thank Josh Nordberg and Yumi Uetake for useful discussions and comments. We are grateful to Alexey Khodjakov and Bill Theurkauf for gifts of cell lines.
This work was supported by National Institutes of Health grant GM 30758 to G. Sluder.
Footnotes
Abbreviations used in this paper:
- hTERT
- human telomerase reverse transcriptase
References
- Andalis A.A., Storchova Z., Styles C., Galitski T., Pellman D., Fink G.R. 2004. Defects arising from whole-genome duplications in Saccharomyces cerevisiae. Genetics. 167:1109–1121 10.1534/genetics.104.029256 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Balczon R., Bao L., Zimmer W.E., Brown K., Zinkowski R.P., Brinkley B.R. 1995. Dissociation of centrosome replication events from cycles of DNA synthesis and mitotic division in hydroxyurea-arrested Chinese hamster ovary cells. J. Cell Biol. 130:105–115 10.1083/jcb.130.1.105 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Basto R., Brunk K., Vinadogrova T., Peel N., Franz A., Khodjakov A., Raff J.W. 2008. Centrosome amplification can initiate tumorigenesis in flies. Cell. 133:1032–1042 10.1016/j.cell.2008.05.039 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Borel F., Lohez O.D., Lacroix F.B., Margolis R.L. 2002. Multiple centrosomes arise from tetraploidy checkpoint failure and mitotic centrosome clusters in p53 and RB pocket protein-compromised cells. Proc. Natl. Acad. Sci. USA. 99:9819–9824 10.1073/pnas.152205299 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Brinkley B.R. 2001. Managing the centrosome numbers game: from chaos to stability in cancer cell division. Trends Cell Biol. 11:18–21 10.1016/S0962-8924(00)01872-9 [DOI] [PubMed] [Google Scholar]
- Chandhok N.S., Pellman D. 2009. A little CIN may cost a lot: revisiting aneuploidy and cancer. Curr. Opin. Genet. Dev. 19:74–81 10.1016/j.gde.2008.12.004 [DOI] [PubMed] [Google Scholar]
- Cimini D., Howell B., Maddox P., Khodjakov A., Degrassi F., Salmon E.D. 2001. Merotelic kinetochore orientation is a major mechanism of aneuploidy in mitotic mammalian tissue cells. J. Cell Biol. 153:517–527 10.1083/jcb.153.3.517 [DOI] [PMC free article] [PubMed] [Google Scholar]
- D’Assoro A.B., Lingle W.L., Salisbury J.L. 2002. Centrosome amplification and the development of cancer. Oncogene. 21:6146–6153 10.1038/sj.onc.1205772 [DOI] [PubMed] [Google Scholar]
- Duensing A., Spardy N., Chatterjee P., Zheng L., Parry J., Cuevas R., Korzeniewski N., Duensing S. 2009. Centrosome overduplication, chromosomal instability, and human papillomavirus oncoproteins. Environ. Mol. Mutagen. 50:741–747 10.1002/em.20478 [DOI] [PubMed] [Google Scholar]
- Fujiwara T., Bandi M., Nitta M., Ivanova E.V., Bronson R.T., Pellman D. 2005. Cytokinesis failure generating tetraploids promotes tumorigenesis in p53-null cells. Nature. 437:1043–1047 10.1038/nature04217 [DOI] [PubMed] [Google Scholar]
- Galipeau P.C., Cowan D.S., Sanchez C.A., Barrett M.T., Emond M.J., Levine D.S., Rabinovitch P.S., Reid B.J. 1996. 17p (p53) allelic losses, 4N (G2/tetraploid) populations, and progression to aneuploidy in Barrett’s esophagus. Proc. Natl. Acad. Sci. USA. 93:7081–7084 10.1073/pnas.93.14.7081 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ganem N.J., Pellman D. 2007. Limiting the proliferation of polyploid cells. Cell. 131:437–440 10.1016/j.cell.2007.10.024 [DOI] [PubMed] [Google Scholar]
- Ganem N.J., Storchova Z., Pellman D. 2007. Tetraploidy, aneuploidy and cancer. Curr. Opin. Genet. Dev. 17:157–162 10.1016/j.gde.2007.02.011 [DOI] [PubMed] [Google Scholar]
- Ganem N.J., Godinho S.A., Pellman D. 2009. A mechanism linking extra centrosomes to chromosomal instability. Nature. 460:278–282 10.1038/nature08136 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Goepfert T.M., Adigun Y.E., Zhong L., Gay J., Medina D., Brinkley W.R. 2002. Centrosome amplification and overexpression of aurora A are early events in rat mammary carcinogenesis. Cancer Res. 62:4115–4122 [PubMed] [Google Scholar]
- Kleylein-Sohn J., Westendorf J., Le Clech M., Habedanck R., Stierhof Y.D., Nigg E.A. 2007. Plk4-induced centriole biogenesis in human cells. Dev. Cell. 13:190–202 10.1016/j.devcel.2007.07.002 [DOI] [PubMed] [Google Scholar]
- Ko M.A., Rosario C.O., Hudson J.W., Kulkarni S., Pollett A., Dennis J.W., Swallow C.J. 2005. Plk4 haploinsufficiency causes mitotic infidelity and carcinogenesis. Nat. Genet. 37:883–888 10.1038/ng1605 [DOI] [PubMed] [Google Scholar]
- Krämer A., Neben K., Ho A.D. 2002. Centrosome replication, genomic instability and cancer. Leukemia. 16:767–775 10.1038/sj.leu.2402454 [DOI] [PubMed] [Google Scholar]
- La Terra S., English C.N., Hergert P., McEwen B.F., Sluder G., Khodjakov A. 2005. The de novo centriole assembly pathway in HeLa cells: cell cycle progression and centriole assembly/maturation. J. Cell Biol. 168:713–722 10.1083/jcb.200411126 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lengauer C., Kinzler K.W., Vogelstein B. 1997. Genetic instability in colorectal cancers. Nature. 386:623–627 10.1038/386623a0 [DOI] [PubMed] [Google Scholar]
- Levine D.S., Rabinovitch P.S., Haggitt R.C., Blount P.L., Dean P.J., Rubin C.E., Reid B.J. 1991. Distribution of aneuploid cell populations in ulcerative colitis with dysplasia or cancer. Gastroenterology. 101:1198–1210 [DOI] [PubMed] [Google Scholar]
- Lingle W.L., Salisbury J.L. 2000. The role of the centrosome in the development of malignant tumors. Curr. Top. Dev. Biol. 49:313–329 10.1016/S0070-2153(99)49015-5 [DOI] [PubMed] [Google Scholar]
- Lingle W.L., Lutz W.H., Ingle J.N., Maihle N.J., Salisbury J.L. 1998. Centrosome hypertrophy in human breast tumors: implications for genomic stability and cell polarity. Proc. Natl. Acad. Sci. USA. 95:2950–2955 10.1073/pnas.95.6.2950 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lingle W.L., Barrett S.L., Negron V.C., D’Assoro A.B., Boeneman K., Liu W., Whitehead C.M., Reynolds C., Salisbury J.L. 2002. Centrosome amplification drives chromosomal instability in breast tumor development. Proc. Natl. Acad. Sci. USA. 99:1978–1983 10.1073/pnas.032479999 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lohez O.D., Reynaud C., Borel F., Andreassen P.R., Margolis R.L. 2003. Arrest of mammalian fibroblasts in G1 in response to actin inhibition is dependent on retinoblastoma pocket proteins but not on p53. J. Cell Biol. 161:67–77 10.1083/jcb.200208140 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Meraldi P., Honda R., Nigg E.A. 2002. Aurora-A overexpression reveals tetraploidization as a major route to centrosome amplification in p53-/- cells. EMBO J. 21:483–492 10.1093/emboj/21.4.483 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nakamura Y., Takaira M., Sato E., Kawano K., Miyoshi O., Niikawa N. 2003. A tetraploid liveborn neonate: cytogenetic and autopsy findings. Arch. Pathol. Lab. Med. 127:1612–1614 [DOI] [PubMed] [Google Scholar]
- Nigg E.A. 2002. Centrosome aberrations: cause or consequence of cancer progression? Nat. Rev. Cancer. 2:815–825 10.1038/nrc924 [DOI] [PubMed] [Google Scholar]
- Nigg E.A. 2006. Origins and consequences of centrosome aberrations in human cancers. Int. J. Cancer. 119:2717–2723 10.1002/ijc.22245 [DOI] [PubMed] [Google Scholar]
- Orr-Weaver T.L., Weinberg R.A. 1998. A checkpoint on the road to cancer. Nature. 392:223–224 10.1038/32520 [DOI] [PubMed] [Google Scholar]
- Pihan G.A., Purohit A., Wallace J., Knecht H., Woda B., Quesenberry P., Doxsey S.J. 1998. Centrosome defects and genetic instability in malignant tumors. Cancer Res. 58:3974–3985 [PubMed] [Google Scholar]
- Pihan G.A., Purohit A., Wallace J., Malhotra R., Liotta L., Doxsey S.J. 2001. Centrosome defects can account for cellular and genetic changes that characterize prostate cancer progression. Cancer Res. 61:2212–2219 [PubMed] [Google Scholar]
- Pihan G.A., Wallace J., Zhou Y., Doxsey S.J. 2003. Centrosome abnormalities and chromosome instability occur together in pre-invasive carcinomas. Cancer Res. 63:1398–1404 [PubMed] [Google Scholar]
- Reid B.J., Barrett M.T., Galipeau P.C., Sanchez C.A., Neshat K., Cowan D.S., Levine D.S. 1996. Barrett’s esophagus: ordering the events that lead to cancer. Eur. J. Cancer Prev. 5(Suppl. 2):57–65 10.1097/00008469-199612002-00009 [DOI] [PubMed] [Google Scholar]
- Rogakou E.P., Pilch D.R., Orr A.H., Ivanova V.S., Bonner W.M. 1998. DNA double-stranded breaks induce histone H2AX phosphorylation on serine 139. J. Biol. Chem. 273:5858–5868 10.1074/jbc.273.10.5858 [DOI] [PubMed] [Google Scholar]
- Sagona A.P., Stenmark H. 2010. Cytokinesis and cancer. FEBS Lett. 584:2652–2661 10.1016/j.febslet.2010.03.044 [DOI] [PubMed] [Google Scholar]
- Sato N., Mizumoto K., Nakamura M., Nakamura K., Kusumoto M., Niiyama H., Ogawa T., Tanaka M. 1999. Centrosome abnormalities in pancreatic ductal carcinoma. Clin. Cancer Res. 5:963–970 [PubMed] [Google Scholar]
- Shackney S.E., Smith C.A., Miller B.W., Burholt D.R., Murtha K., Giles H.R., Ketterer D.M., Pollice A.A. 1989. Model for the genetic evolution of human solid tumors. Cancer Res. 49:3344–3354 [PubMed] [Google Scholar]
- Silkworth W.T., Nardi I.K., Scholl L.M., Cimini D. 2009. Multipolar spindle pole coalescence is a major source of kinetochore mis-attachment and chromosome mis-segregation in cancer cells. PLoS ONE. 4:e6564 10.1371/journal.pone.0006564 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sluder G., Nordberg J.J. 2004. The good, the bad and the ugly: the practical consequences of centrosome amplification. Curr. Opin. Cell Biol. 16:49–54 10.1016/j.ceb.2003.11.006 [DOI] [PubMed] [Google Scholar]
- Sluder G., Nordberg J.J., Miller F.J., Hinchcliffe E.H. 2005. A sealed preparation for long-term observations of cultured cells. Live Cell Imaging: A Laboratory Manual. Goldman R.D., Spector D.L., Cold Spring Harbor Laboratory Press, Cold Spring Harbor, New York: 345–349 [Google Scholar]
- Storchova Z., Pellman D. 2004. From polyploidy to aneuploidy, genome instability and cancer. Nat. Rev. Mol. Cell Biol. 5:45–54 10.1038/nrm1276 [DOI] [PubMed] [Google Scholar]
- Thompson S.L., Compton D.A. 2008. Examining the link between chromosomal instability and aneuploidy in human cells. J. Cell Biol. 180:665–672 10.1083/jcb.200712029 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Thompson S.L., Compton D.A. 2010. Proliferation of aneuploid human cells is limited by a p53-dependent mechanism. J. Cell Biol. 188:369–381 10.1083/jcb.200905057 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Uetake Y., Sluder G. 2004. Cell cycle progression after cleavage failure: mammalian somatic cells do not possess a “tetraploidy checkpoint”. J. Cell Biol. 165:609–615 10.1083/jcb.200403014 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Uetake Y., Sluder G. 2010. Prolonged prometaphase blocks daughter cell proliferation despite normal completion of mitosis. Curr. Biol. 20:1666–1671 10.1016/j.cub.2010.08.018 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Uetake Y., Loncarek J., Nordberg J.J., English C.N., La Terra S., Khodjakov A., Sluder G. 2007. Cell cycle progression and de novo centriole assembly after centrosomal removal in untransformed human cells. J. Cell Biol. 176:173–182 10.1083/jcb.200607073 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Upender M.B., Habermann J.K., McShane L.M., Korn E.L., Barrett J.C., Difilippantonio M.J., Ried T. 2004. Chromosome transfer induced aneuploidy results in complex dysregulation of the cellular transcriptome in immortalized and cancer cells. Cancer Res. 64:6941–6949 10.1158/0008-5472.CAN-04-0474 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Weaver B.A., Silk A.D., Montagna C., Verdier-Pinard P., Cleveland D.W. 2007. Aneuploidy acts both oncogenically and as a tumor suppressor. Cancer Cell. 11:25–36 10.1016/j.ccr.2006.12.003 [DOI] [PubMed] [Google Scholar]
- Yang Z., Loncarek J., Khodjakov A., Rieder C.L. 2008. Extra centrosomes and/or chromosomes prolong mitosis in human cells. Nat. Cell Biol. 10:748–751 10.1038/ncb1738 [DOI] [PMC free article] [PubMed] [Google Scholar]