APC/CCdh1-dependent degradation of USP1 allows for PCNA monoubiquitination and subsequent recruitment of trans-lesion synthesis polymerase to UV repair sites.
Abstract
Targeted protein destruction of critical cellular regulators during the G1 phase of the cell cycle is achieved by anaphase-promoting complex/cyclosomeCdh1 (APC/CCdh1), a multisubunit E3 ubiquitin ligase. Cells lacking Cdh1 have been shown to accumulate deoxyribonucleic acid (DNA) damage, suggesting that it may play a previously unrecognized role in maintaining genomic stability. The ubiquitin-specific protease 1 (USP1) is a known critical regulator of DNA repair and genomic stability. In this paper, we report that USP1 was degraded in G1 via APC/CCdh1. USP1 levels were kept low in G1 to provide a permissive condition for inducing proliferating cell nuclear antigen (PCNA) monoubiquitination in response to ultraviolet (UV) damage before DNA replication. Importantly, expression of a USP1 mutant that cannot be degraded via APC/CCdh1 inhibited PCNA monoubiquitination during G1, likely compromising the recruitment of trans-lesion synthesis polymerase to UV repair sites. Thus, we propose a role for APC/CCdh1 in modulating the status of PCNA monoubiquitination and UV DNA repair before S phase entry.
Introduction
Genotoxic stress, such as exposure to UV light, induces the accumulation of different types of DNA lesions and a wide range of cellular responses (Stokes and Comb, 2008). To maintain cell viability, replication forks that encounter damaged DNA must efficiently bypass these lesions to complete replication of the genome in a timely manner during S phase. DNA synthesis across damaged DNA is achieved by specialized DNA polymerases that incorporate nucleotides opposite to damaged bases in a process known as trans-lesion synthesis (TLS; Lehmann et al., 2007). However, overuse of TLS polymerases can increase mutagenesis because of their highly accommodating active sites and lack of proofreading activity (Waters et al., 2009). The recruitment of TLS polymerases for lesion bypass requires the monoubiquitination of proliferating cell nuclear antigen (PCNA; Kannouche et al., 2004; Watanabe et al., 2004; Bienko et al., 2005; Garg and Burgers, 2005; Plosky et al., 2006). Regulatory factors that control the level of PCNA ubiquitination, such as ubiquitin ligases and proteases, are important to promote an optimal balance between TLS-associated cell survival and TLS-associated mutagenesis (Prives and Gottifredi, 2008). This has been previously described in a series of elegant work that suggest the recruitment of Pol-η (a member of the Y family of TLS polymerases) to UV lesions by the Rad18-dependent monoubiquitination of PCNA (Kannouche et al., 2004; Watanabe et al., 2004). The model predicts that the blockage of replicative polymerases activates PCNA monoubiquitination at replication-stalled sites (Hoege et al., 2002; Friedberg et al., 2005). This event in turn promotes the recruitment of TLS polymerases to bypass the lesion and allow continuation of DNA replication. It has been suggested that the switch back from TLS to normal processive polymerases is regulated by ubiquitin-specific protease 1 (USP1), the deubiquitinating enzyme for PCNA (Huang and D’Andrea, 2006; Huang et al., 2006; Ulrich, 2006).
However, recent studies have shown that PCNA monoubiquitination can occur outside of S phase both in mammalian and yeast systems (Frampton et al., 2006; Soria et al., 2006, 2009; Daigaku et al., 2010; Karras and Jentsch, 2010; Ogi et al., 2010). Moreover, Pol-η and other TLS polymerases can be recruited to sites of UV lesions in quiescent or noncycling cells, which is in line with possible roles in gap-filling of DNA damage tracks left behind the replication forks (Lehmann and Fuchs, 2006; Lopes et al., 2006). As such, the gap-filling pathway is likely not limited to S phase but also occurs in G0, G1, and G2/M phases. PCNA monoubiquitination and TLS polymerase recruitment to UV lesions have also been recently implicated in nucleotide excision repair (NER), a DNA repair process that can take place outside of S phase (Ogi et al., 2010). It is currently unclear whether specific cell cycle phases, such as G1 or S phase, can dictate the mechanism of how the cell responds to UV-mediated DNA damage through the activation of PCNA monoubiquitination and subsequent DNA repair.
The anaphase-promoting complex/cyclosome (APC/C) is a multisubunit ubiquitin ligase that targets many key cell cycle regulators for proteolysis (Qiao et al., 2010). The activation of APC/C is dependent on two WD-40 repeat domain–containing proteins, Cdc20 and Cdh1. Whereas APC/CCdc20 principally regulates mitotic progression, APC/CCdh1 shows a broad spectrum of G1-specific substrates, including proteins that function beyond cell cycle control (Skaar and Pagano, 2008; Qiao et al., 2010). Previous studies have shown that Cdh1 knockout or depletion in mammalian cells can cause genomic instability, but the precise cause of this instability is unclear (Engelbert et al., 2008; García-Higuera et al., 2008). Recently, however, the stability of several proteins involved in the DNA damage checkpoint response and DNA repair, such as Claspin, Rad17, thymidine kinase 1, and the ribonucleotide reductase subunit RRM2, has been shown to be regulated by APC/CCdh1 (Chabes et al., 2003; Ke et al., 2005; Bassermann et al., 2008; Gao et al., 2009; Zhang et al., 2010). Moreover, although APC/CCdh1 is active only at the very end of mitosis and during G1, it is reactivated in G2 in response to DNA damage to target the mitotic kinase Plk1 for proteasomal degradation (Bassermann et al., 2008). It is likely that more Cdh1 substrates will be identified with roles in the DNA damage response (DDR) pathways, further underscoring the relevance and importance of APC/C in maintaining cellular genomic integrity.
In humans, protein deubiquitination is controlled by a family of nearly 100 deubiquitinases (DUBs), but the function of many of these ubiquitin proteases is unknown (Nijman et al., 2005b; Reyes-Turcu et al., 2009). The majority of DUBs are cysteine proteases that cleave ubiquitin from specific mono- and polyubiquitinated substrates or from linear ubiquitin polypeptides. The DUB USP1 is a critical regulator of genomic stability in mammalian cells (Kim et al., 2009). USP1 has been shown to regulate the monoubiquitination levels of three protein substrates involved in DNA repair: FANCD2 and FANCI (the Fanconi anemia [FA] effector proteins involved in DNA cross-link repair) and PCNA (the DNA replication processivity factor whose function is important for the recruitment of specialized TLS polymerases to sites of UV DNA lesions; Nijman et al., 2005a; Huang et al., 2006; Sims et al., 2007; Smogorzewska et al., 2007). However, it is unclear how USP1 negatively regulates the monoubiquitination of substrates involved in two distinct DDR pathways.
In this study, we explore whether the control of USP1 levels during cell cycle progression modulates the cellular response to UV DNA damage. We show that during G1, USP1 is targeted for degradation by APC/CCdh1 to ensure that USP1 levels are kept in check before S phase entry. Low levels of USP1 enable robust UV-induced PCNA monoubiquitination during G1, which is likely to allow the recruitment of TLS polymerases to UV lesions. These findings suggest that APC/CCdh1 plays a direct role in modulating the DNA repair choice in G1 and further solidify the link between cell cycle regulation and DNA repair.
Results and discussion
USP1 levels are low during the G1 phase of the cell cycle
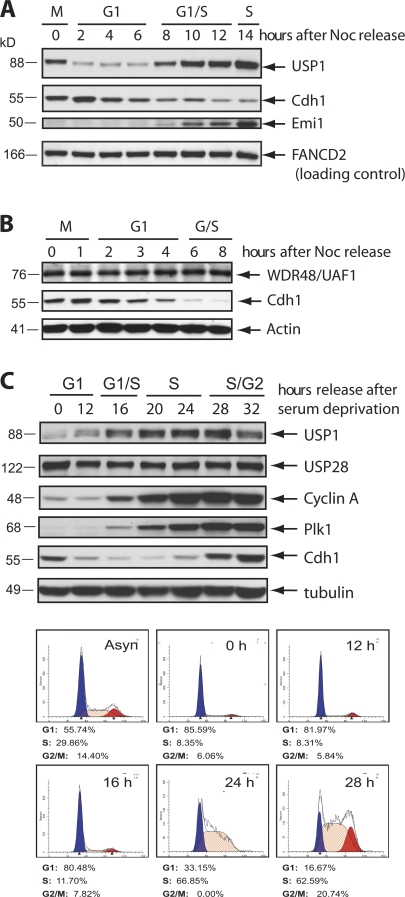
To investigate whether USP1 activity is cell cycle regulated, we examined whether USP1 protein levels fluctuate in cells progressing through the cell cycle. We synchronized both U2OS and HeLa cells in prometaphase and then let them progress from mitosis into the next cell cycle. We found that USP1 levels are low during G1 and high during S phase (Fig. 1, A and B). We also observed low levels of USP1 in serum-starved and G1 T98G cells, similar to other proteins known to be degraded in G0/G1, such as Plk1 and cyclin A (Fig. 1 C). USP1 DUB activity depends on the interaction with its catalytic cofactor WDR48 (also called UAF1; Cohn et al., 2007). In contrast to USP1, WDR48 levels did not change throughout the cell cycle (Fig. 1 B and not depicted).
Figure 1.
USP1 levels are low during the G1 phase of the cell cycle. (A and B) HeLa (A) and U2OS (B) cells were synchronized in M phase by treating cells with nocodazole (Noc) for 16 h, washed, released, and collected for the indicated time points. (C) T98G human glioblastoma cells were synchronized in G0/G1 by serum deprivation for 72 h and then refed with serum and harvested at the indicated time points. (A–C) Western blot analyses were performed and probed with the indicated antibodies. Separate samples were also collected, fixed, and stained with PI for FACS analysis according to procedures outlined in the Materials and methods section.
APC/CCdh1 regulates USP1 levels in G1
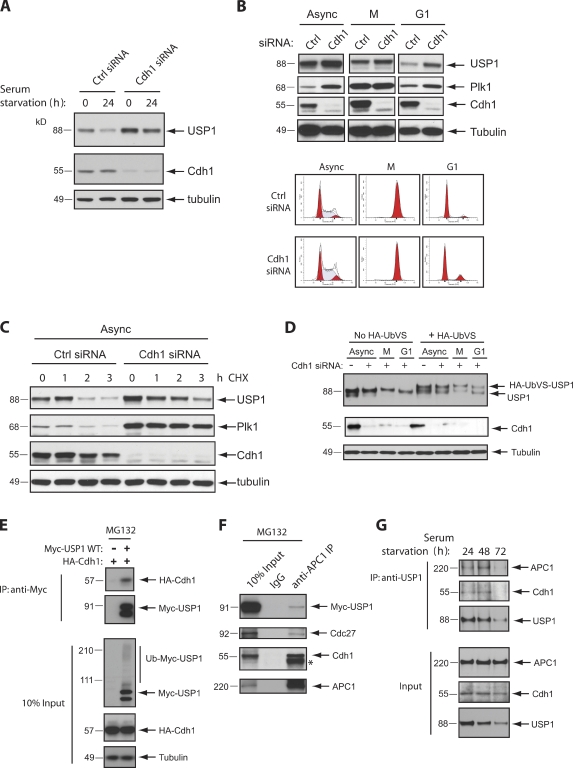
The down-regulation of USP1 in the G1 phase of the cell cycle led us to ask whether a ubiquitin ligase may be responsible for the degradation of USP1. As the APC/CCdh1 complex is critical for promoting the degradation of cell cycle regulators in G1, we tested whether this ubiquitin ligase also regulates the G1 levels of USP1. To this end, we silenced Cdh1 expression in both synchronized T98G and synchronized U2OS cells using well-established siRNA oligonucleotides (Bashir et al., 2004). We found that USP1 accumulated in both asynchronous and G1 cells depleted of Cdh1 (Fig. 2, A and B). This accumulation appears to be caused by USP1 stabilization, as shown by measuring USP1 half-life (Fig. 2 C). As expected (APC/CCdh1 is inactive during prometaphase), the accumulation of USP1 after Cdh1 silencing was not as prominent in M phase cells (Fig. 2 B). Similarly, upon Cdh1 silencing, Plk1, a known Cdh1 substrate, was also more accumulated in asynchronous and G1 cells than in M phase synchronized cells (Fig. 2 B).
Figure 2.
Cdh1 depletion stabilizes USP1 during the G1 phase of the cell cycle. (A) T98G cells were transfected for 48 h with a control (Ctrl) siRNA (AllStars Negative; QIAGEN) or Cdh1 siRNA. Cells were serum starved in culture media containing 0.05% FBS for 24 h to arrest them in G0/G1. 0 indicates cells grown in regular media. (B) U2OS cells were transfected with the indicated siRNAs, synchronized in M phase (M) by incubating with nocodazole for 16 h, washed, and released in fresh media for 3 h for G1 phase (G1). Separate samples were collected for FACS. (C) U2OS cells were transfected with the indicated siRNAs and treated for the indicated time points with cycloheximide (CHX) to inhibit protein synthesis. (D) U2OS cells were transfected with the indicated siRNAs and synchronized in G1 as in B. Samples were collected and lysed according to protocols described for the UbVS DUB activity assay (see Materials and methods). Higher shift in the USP1 protein band indicates active USP1 (covalently modified USP1 by HA-UbVS). (E) Expression constructs were cotransfected in U2OS cells and treated for 6 h with 10 µM of the proteasome inhibitor MG132 before termination. Samples were lysed and collected for immunoprecipitation (IP) or loaded for input. (F) U2OS cells were transfected with Myc-USP1 wild type (WT) for 48 h and treated with MG132 as in E. Samples were lysed and collected for immunoprecipitation. *, heavy chain band. (G) T98G cells were serum starved as in A for the indicated times. Samples were lysed and collected for immunoprecipitation and input (10%) and probed with the indicated antibodies for Western blot analysis.
Next, we determined whether USP1 DUB activity is functional during the G1 phase. To this end, we measured the ability of USP1 to react with a chemically modified ubiquitin suicide substrate probe (ubiquitin vinyl sulfone [UbVS]; Borodovsky et al., 2002). We found that G1-stabilized USP1 was still catalytically active (Fig. 2 D). This suggests that USP1 proteolysis is likely a critical mechanism to inhibit USP1 activity during the G1 phase. Thus, we predict that a stable mutant of USP1 that could not be targeted by APC/CCdh1 during G1 would still interact with WDR48 (which is expressed in G1 as shown in Fig. 1 B) and deconjugate specific monoubiquitinated substrates.
To ensure that the Cdh1 knockdown effect on USP1 stability was direct, we addressed whether Cdh1 could interact with USP1 in vivo. We found that both exogenously and endogenously expressed USP1 can interact with members of the APC/CCdh1 complex (Fig. 2, E–G). In T98G cells that are accumulating in G0/G1 (serum starvation time course), we also show that endogenous USP1 can interact with Cdh1 before its degradation (Fig. 2 G).
Characterization of a USP1 mutant that cannot be degraded via APC/CCdh1
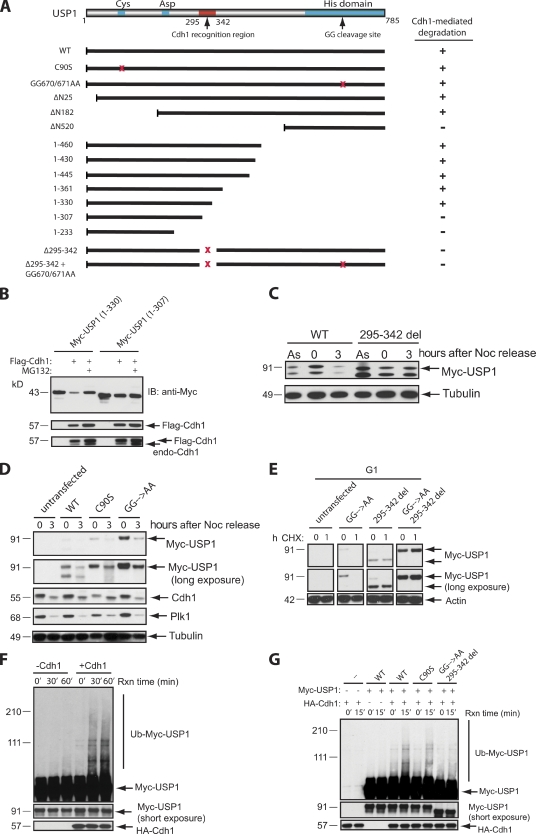
To search for a USP1 mutant that is stable in G1, we attempted to map the degradation motif (degron) in USP1. APC/CCdh1-mediated degradation typically requires the RXXL degron (known as destruction box or D box) in its target proteins (Glotzer et al., 1991). Although human USP1 contains three putative D box motifs and a KEN box (Fig. S1; Pfleger and Kirschner, 2000; Littlepage and Ruderman, 2002; Reis et al., 2006), their mutation, singly or in combination, had no appreciable effect on USP1 stability in G1 (not depicted). Thus, we suspected that USP1 possesses a noncanonical degron, which is similar to those found in Aurora A (Littlepage and Ruderman, 2002), Cdc20 (Reis et al., 2006), or Claspin (Bassermann et al., 2008). To map the USP1 degron, we generated a series of N- and C-terminal deletion mutants (Fig. 3 A) and tested them for their ability to resist degradation in response to Cdh1 overexpression. We found that a region surrounding amino acids 307–330 of human USP1 is required for Cdh1-dependent degradation of USP1 (Fig. 3 B). Accordingly, a near full-length USP1 construct containing an internal deletion of 295–342 amino acids was stable in U2OS cells progressing through G1 (Fig. 3 C), whereas wild-type USP1, a USP1 (C90S) mutant (which is catalytic inactive), or the USP1 (GG670/671AA) diglycine mutant (which is unable to cleave itself) was not (Fig. 3 D). These results show that the catalytic and autocleavage activities of USP1 are not necessary for USP1 to be targeted for degradation by APC/CCdh1 during G1. This strongly suggests that the mechanism for degrading USP1 in G1 is completely different from the UV-initiated autocleavage and degradation of USP1 during S phase, as the proteolysis of USP1 during G1 is independent of its own catalytic activity (Huang et al., 2006).
Figure 3.
Identification of a USP1 mutant that cannot be degraded via APC/CCdh1. (A) A schematic diagram of critical USP1 domains and Myc-USP1 expression constructs generated for testing of their stability when coexpressed with FLAG-Cdh1. +, degradation of Myc-USP1; −, stabilization of Myc-USP1; GG, diglycine residue for the USP1 autocleavage site; red x’s, deletion or point mutations of the region. (B) U2OS cells were transiently transfected with Myc-USP1 and/or Flag-Cdh1 constructs in the presence or absence of MG132 (3 h). IB, immunoblot. (C) U2OS cells were transfected with Myc-USP1 wild type (WT) or the 295–342 deletion (295–342 del) mutant and synchronized in M phase with nocodazole (Noc) for 16 h (0 h) and/or released into G1 (3 h after nocodazole release). As, asynchronous. (D) U2OS cells were transfected with Myc-USP1 wild type, C90S, or GG670/671AA and synchronized in M phase or G1 as in C. (E) U2OS cells were transfected with Myc-USP1 wild type, GG670/671AA, 295–342 deletion, or GG670/671AA plus the 295–342 deletion mutant, synchronized, released into G1 (3 h after nocodazole release), and treated with cycloheximide (CHX) for 1 h (1) or not (0). (F) In vitro ubiquitination assay of Myc-USP1 wild type in the presence or absence of HA-Cdh1. Reactions were performed as described in the Materials and methods section. (G) In vitro ubiquitination assay of Myc-USP1 wild type, C90S, and GG670/671AA plus the 295–342 deletion mutant in the presence or absence of HA-Cdh1. Rxn, reaction.
We also generated a double USP1 mutant that contained both the 295–342 deletion and the GG670/671AA mutation, which was also resistant to proteolysis by APC/CCdh1 during G1 (Fig. 3 E). To further support that USP1 is ubiquitinated via APC/CCdh1, we reconstituted the ubiquitination of USP1 in vitro. USP1 was efficiently ubiquitinated only when Cdh1 was present and a specific E2 combination was used (see Materials and methods section; Figs. 3 F and S2). Under our conditions, it is unclear why UBCH10-UBCH5 (Aristarkhov et al., 1996; King et al., 1996; Yu et al., 1996) works better than the canonical E2 pair UBCH10-UBE2S (Garnett et al., 2009; Williamson et al., 2009; Wu et al., 2010) in promoting USP1 polyubiquitination. Interestingly, both UBCH10 and UBE2S are also degraded in G1 by APC/CCdh1, suggesting that other E2 enzymes may be responsible for ubiquitinating G1 versus M phase substrates of APC/C (Williamson et al., 2009). Importantly, Cdh1-dependent ubiquitination was not observed in the USP1 degron mutant (Fig. 3 G). Unexpectedly, the degron mutant was still capable of binding to Cdh1 in vitro, suggesting that the degron region may be more critical for promoting efficient APC/CCdh1-dependent ubiquitination of USP1 (Fig. S3). We have not ruled out the possibility of multiple Cdh1 binding sites on USP1 that may contribute to its direct association with the APC/CCdh1 complex. Nevertheless, experiments in cell systems and in vitro show that Cdh1 promotes the ubiquitination and subsequent degradation of USP1 in a manner dependent on its degron region.
Degradation of USP1 permits UV-induced monoubiquitination of PCNA during G1
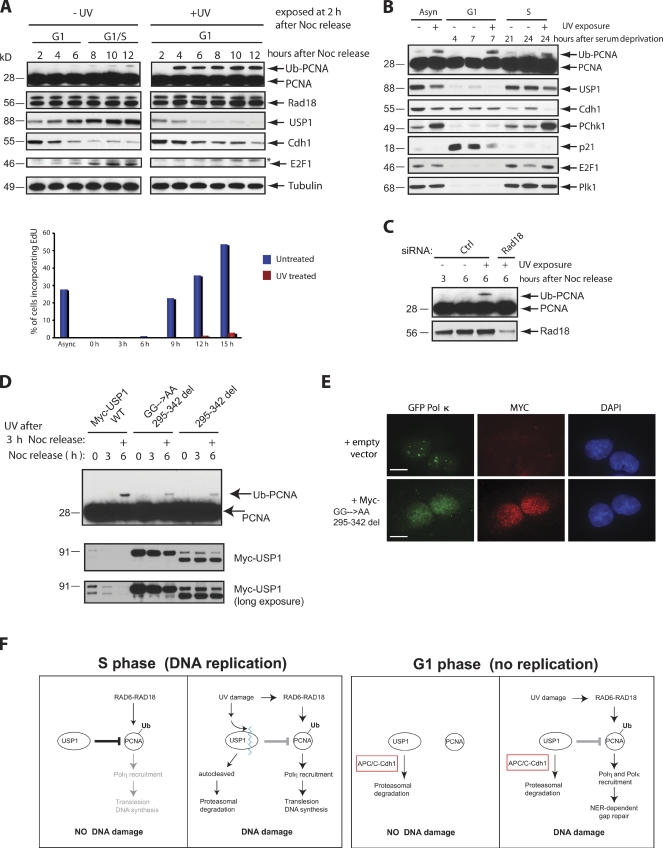
We then investigated the biological significance of USP1 degradation in G1. One possibility is that the repair of UV-induced lesions in G1 requires robust PCNA monoubiquitination. In fact, PCNA monoubiquitination is involved in the recruitment of TLS polymerases to sites of UV damage for repair synthesis (Ogi et al., 2010). To test whether UV-induced DNA damage during G1 induces PCNA monoubiquitination, U2OS cells were synchronized in G1 by releasing them from a prometaphase block. 2 h after release, cells were either left untreated or UV irradiated and followed for up to 10 h. After UV damage, USP1 levels decreased below the G1 levels, whereas in untreated cells, USP1 reaccumulated as they exited G1 and started DNA replication (Fig. 4 A). Accordingly, after UV damage, Cdh1 levels remained elevated, correlating with the loss of USP1 and a delay in S phase entry. Interestingly, robust monoubiquitination of PCNA already occurred by 2 h after UV damage and remained present for up to 10 h (Fig. 4 A). In contrast, detectable PCNA-monoubiquitinated species only started to appear when nonirradiated cells reached S phase. A strong UV-induced PCNA monoubiquitination response was also observed in T98G cells during G1 (Fig. 4 B). Interestingly, the monoubiquitination of FANCD2 and FANCI (FA pathway effector proteins) was not induced by UV damage in G1 (unpublished data). This is likely a result of low levels of UBE2T, the E2 conjugating enzyme for the FA pathway, in G1 that prevent activation of the FA pathway outside of S phase (unpublished data). We also noted that the DDR after UV damage is different in G1 versus S phase. For instance, UV irradiation targets the autocleavage and degradation of USP1 in S phase to promote PCNA monoubiquitination, as previously reported (Fig. 4 B; Huang et al., 2006). However, USP1 levels remain low and unchanged after UV damage in G1 (Fig. 4 B), suggesting that the environment in G1 was already conducive for UV-mediated activation of PCNA monoubiquitination. In addition, Cdh1 is degraded after UV damage in S phase cells (Liu et al., 2008) but not in G1. Also, the checkpoint kinase Chk1 is not activated by phosphorylation after UV damage in G1.
Figure 4.
UV-induced PCNA monoubiquitination during G1 requires prolonged degradation of USP1. (A) U2OS cells were synchronized in M phase with nocodazole (Noc) treatment and released for the indicated time points. Cells were also irradiated with 50 J/m2 UV in G1 (2 h after release from nocodazole treatment). Samples were then analyzed by Western blotting. Cells from corresponding samples were pulsed with EdU (BrdU analogue) to label cells undergoing DNA synthesis. Samples were processed according to procedures outlined in the Materials and methods section and were analyzed by FACS. The data displayed as a bar graph are representative of two separate experiments. (B) T98G cells were synchronized in G0/G1 by serum deprivation for 72 h, refed with fresh media, and collected at the indicated time points. Cells were either left untreated, UV exposed at 4 h after release from serum deprivation (G1 phase), or exposed after 21 h (early S phase). (C) U2OS cells were transfected with the indicated siRNAs, synchronized, released into G1 (3 h after nocodazole release), UV irradiated (50 J/m2), and collected 3 h after UV exposure (6 h after release). (D) U2OS cells were transfected with either Myc-USP1 wild type (WT), 295–342 deletion (295–342 del), or GG670/671AA plus the 295–342 deletion mutant, synchronized in M phase (0 h), released into G1 (3 h), UV irradiated (3 h after nocodazole release), and then collected 3 h after UV exposure (6 h). (E) U2OS cells stably expressing GFP–Pol-κ were selected in G418 for 10 d before isolation of GFP-positive cells through a FACS sorter (MoFlo; Dako). Stable U2OS GFP–Pol-κ–expressing cells were transiently transfected with Myc-tagged GG670/671AA plus the 295–342 deletion mutant, synchronized in M phase with nocodazole, and released for 3 h into the G1 phase. The cells were then irradiated with 50 J/m2 UV and fixed 2 h after UV exposure. Approximately 100 GFP-positive cells or GFP and Myc double-positive cells were analyzed. No Myc-expressing cells contained GFP–Pol-κ foci. A representative image of the cells analyzed is shown. GFP-positive nuclear foci formation was scored as the number of cells containing five or more foci. Bars, 20 µm. (F) A schematic representation of how USP1 is regulated by proteolysis in G1 versus S phase cells in the presence or absence of UV DNA damage. Black lines, active cells; gray lines, inactive cells; blue squiggly line, USP1 autocleavage site; Ub, ubiquitin.
We also found that the mechanism controlling PCNA monoubiquitination in G1 is surprisingly similar to the Rad18-dependent mechanism that governs its activation in S phase (Ogi et al., 2010). The knockdown of Rad18 resulted in the inhibition of UV-induced PCNA monoubiquitination during G1 (Fig. 4 C). The monoubiquitination of PCNA is dependent on lysine 164 (unpublished data). Likewise, expression of stable degron mutants of USP1 also reduced the levels of PCNA monoubiquitination and the recruitment of the TLS polymerase Pol-κ to sites of UV lesions in G1 after UV damage (Fig. 4, D and E). Thus, from our study, we provide a model suggesting that degradation of USP1 by the APC/CCdh1 during G1 is critical for cells to properly recruit TLS polymerases for UV-mediated DNA gap repair (Fig. 4 F).
Conclusion
Proteolytic control of cell cycle regulators by the APC/CCdh1 ubiquitin ligase complex is responsible not only for a stable G1 phase after mitotic exit but also for allowing accurate preparation for DNA replication in the following S phase (Nakayama and Nakayama, 2006). It is known that some of these APC/CCdh1 target proteins are frequently up-regulated in tumor cells, and inactivation of human and mouse Cdh1 interferes with genome integrity (Engelbert et al., 2008; García-Higuera et al., 2008). However, it is still unclear what protein targets of APC/CCdh1 are critical for maintaining genomic stability in human cells. Here, we show that USP1 can be targeted for degradation by the APC/CCdh1 cell cycle regulator. This mainly occurs in the G1 phase before the start of DNA replication. USP1 is an important negative regulator of monoubiquitination for two major DNA repair pathways (FA and TLS) that control genomic stability. Thus, proper dynamic control of USP1 levels throughout the cell cycle and during DNA damage may strongly impact how the cell deals with certain types of DNA lesions. To further validate this point, experimental evidence has suggested that higher than normal levels of USP1 will inhibit the DNA damage–induced monoubiquitination of USP1 targets, such as FANCD2, FANCI, and PCNA (Huang et al., 2006; Oestergaard et al., 2007), whereas the loss of USP1 has been shown to cause chromosomal instability and elevated perinatal lethality in mice (Kim et al., 2009).
Previous works by several groups have hinted at a new role for TLS that is linked to PCNA monoubiquitination outside of S phase. For example, Soria et al. (2009) has suggested that the TLS polymerase Pol-η can be recruited to UV-induced DNA lesions in cells outside S phase, including cells permanently arrested in G1. Work in DT40 cells has shown that PCNA ubiquitination may not be required to maintain normal fork progression on damaged DNA template but is essential for filling postreplicative gaps (Edmunds et al., 2008). More recently, Ogi et al. (2010) has suggested that ubiquitinated PCNA is involved in the recruitment of the TLS polymerase Pol-κ to sites of UV lesions in the G1 phase. The function of ubiquitinated PCNA outside of S phase is tied to a gap-filling role or repair synthesis, which occurs downstream of NER. From our study, we believe that the cell prefers to maintain a low level of USP1 during G0/G1 to establish a permissive environment and allow for robust PCNA monoubiquitination if necessary (Fig. 4 F). Thus, in the absence of its negative regulator, PCNA can be rapidly activated after UV damage to allow repair synthesis to occur downstream of the NER pathway. In contrast, expression of a nondegradable form of USP1 during G1 will lead to less monoubiquitinated PCNA and defective recruitment and repair of UV lesions by the TLS polymerases. The reliance on excision and gap filling–based repair mechanisms becomes more crucial when cells are in quiescence or in the postmitotic phase because of the lack of available sister chromatid for homologous recombination repair (Hoeijmakers, 2001).
Materials and methods
Cell culture and cell cycle synchronization and cell images
T98G, HeLa, and U2OS cells were grown in DME with 10% FBS, 1% penicillin/streptomycin, and 1% glutamine. Cells were synchronized in G0/G1 by serum deprivation (0.05% FBS) for 72 h. After 72 h, cells were split, plated, and refed with normal cell culture media. Cells were synchronized in M phase by incubating cells with 0.1 µg/ml nocodazole for 12–16 h. After 12–16 h, mitotic cells were shaken off, collected, and washed twice with 1× PBS and then plated with fresh media. To verify the cell cycle phase for the cell synchronization experiments, cell samples were stained by propidium iodide (PI) according to the following staining procedure: cell pellets were washed and fixed in 70% ethanol overnight and stained with PI staining buffer (50 µg/ml PI and 10 µg/ml RNase in PBS). Flow cytometry analysis was performed using FACSCaliber and analyzed using CellQuest software (BD) and Modfit LT (V3.1; Verity Software House). Immunocytochemistry was performed according to protocol as previously described (Colnaghi et al., 2011). In brief, for detection of GFP–Pol-κ and Myc-USP1 proteins, U2OS cells were fixed in 100% methanol for 10 min, washed in PBS, blocked in 1% BSA in TBS–Tween 20, and incubated with mouse anti-GFP and rabbit anti-Myc antibodies (Santa Cruz Biotechnology, Inc.). Slides were mounted with Vectashield (Vector Laboratories) with DAPI and analyzed using 60× NA 1.42 objective lenses under oil immersion at 26°C using Alexa Fluor 488 and 546 fluorochromes. Microscope images were captured using a DeltaVision system (Applied Precision) on a base microscope (IX71; Olympus) and a charged-coupled device camera (CoolSnap HQ2; Photometrics). Images were deconvolved, and maximum intensity Quick projections were generated using SoftWorx Suite software (Applied Precision). The images were opened, sized, and placed into figures using ImageJ (National Institutes of Health) or Photoshop (7.0 Professional; Adobe) before transferring to Illustrator (CS3; Adobe).
Transfections, siRNA oligonucleotides, DNA constructs, and antibodies
Transfections with plasmids or siRNA oligonucleotides were performed using Fugene 6 (Roche) or Hiperfect (QIAGEN) reagent. The siRNA oligonucleotide sequences targeting human Cdh1 (5′-AATGAGAAGTCTCCCAGTCAG-3′) and human Rad18 (5′-CCCGAGGTTAATGTAGTTGTT-3′) were used. To generate human Myc-tagged USP1 truncation and deletion mutants, USP1 cDNA was PCR amplified and subcloned into modified pcDNA3 plasmid (Invitrogen) containing the 5′ sequence coding for 2× Myc epitope tag. Point mutations C90S or GG(670/671)AA were made by two-step PCR mutagenesis from the original USP1 template and verified by DNA sequencing as previously described (Huang et al., 2006). Truncation mutants were obtained by PCR amplification using specific 5′ or 3′ USP1 sequences. The internal deletion 295–342 amino acids of USP1 was generated by a site-directed mutagenesis kit (Agilent Technologies) using the primers forward 5′-CCAGTCATTGGAAGAGAAGTCTGCAACTAAGC-3′ and reverse 5′-GCTTAGTTGCAGACTTCTCTTCCAATGACTGG-3′. The following antibodies were used for Western blot analysis and/or immunoprecipitation: WDR48/UAF1 (Evoquest; Invitrogen), USP28 (Bethyl Laboratories, Inc.), Cdh1 (EMD), c-Myc (9E10; Santa Cruz Biotechnology, Inc.), HA (Covance), E2F1 (Bethyl Laboratories, Inc.), Emi1 (Invitrogen), PCNA (PC10; Santa Cruz Biotechnology, Inc.), tubulin (Abcam), Plk1 (Abcam), actin (Abcam), cyclin A (EMD), Rad18 (Abcam), pChk1-317 (Abcam), p21 (Abcam), and USP1, APC1, and Cdc27 (Bethyl Laboratories, Inc.). The GFP–Pol-κ expression construct was a gift from T. Ogi and A. Lehmann (University of Sussex, East Sussex, England, UK). Anti-USP1 and FANCD2 antibodies were a gift from A. D’Andrea (Dana-Farber Cancer Institute, Boston, MA).
In vitro ubiquitination, DUB activity assay, and coimmunoprecipitation
USP1 constructs and Cdh1 were in vitro translated using the TNT T7–coupled wheat germ extract system (Promega) and TNT SP6–coupled reticulolysate system (Promega), respectively, following the manufacturer’s protocol. In vitro translation reactions were stopped using 1 mg/ml cycloheximide. Ubiquitination of in vitro–translated USP1 was performed at 30°C with 50 mM Tris, pH 7.6, 5 mM MgCl2, 2 mM ATP, 2.5 mg/ml ubiquitin, 1 µM ubiquitin aldehyde, 1.5 ng/µl UBE1, 25 ng/µl UBCH10, 5 ng/µl UBCH5 (Boston Biochem), and 10 µM MG132 (EMD) in the presence or absence of in vitro–translated HA-Cdh1. UBCH10 and UBCH5 were chosen as the E2 enzyme pair to use for the in vitro ubiquitination assay after screening several E2 pairs to determine the best E2 combination to use (Fig. S2 A). Cdc6 was used as a positive control for an APC/CCdh1-specific ubiquitination substrate (Fig. S2 B), and methyl ubiquitin was used to confirm chain elongation for the in vitro ubiquitination assay (Fig. S2 C). UbVS DUB activity assay was performed according to Borodovsky et al. (2002) with several modifications. Cells were lysed for 1 h on ice with gentle tapping (250 mM Tris, 150 mM NaCl, 3 mM EDTA, and 0.5% NP-40). Supernatants from cell lysis were collected after a 10-min spin of 14,000 rpm at 4°C. Cell extracts and 50 µM HA-UbVS probe (Boston Biochem) were incubated at 25°C for 1.5 h in DUB buffer (50 mM Tris, 50 mM NaCl, 10% glycerol, and 1 mM EDTA). Reactions were terminated with Laemmli buffer and boiled for Western blot analysis. Coimmunoprecipitation studies were performed overnight at 4°C with the indicated antibodies according to the protocol previously described (Sims et al., 2007). In brief, cell lysis, antibody incubation, and bead washes were performed using low immunoprecipitation buffer (25 mM Tris, pH 7.5, 150 mM NaCl, 2 mM EDTA, and 0.5% NP-40 with protease inhibitor cocktail [Roche]).
Induction of DNA damage and measurement of DNA synthesis
T98G and U2OS cells were treated and harvested between 50 and 80% visible cofluency. 254-nm UV-C irradiation was performed (Stratalinker 2400; Agilent Technologies) in the absence of DME media at 50 J/m2. Recovery after UV damage was in complete medium for the times indicated. Replicating cells were detected using a flow cytometry assay kit (Click-iT EDU; Invitrogen) according to the manufacturer’s instructions. Flow cytometric data were acquired on a flow cytometer (LSR II; BD) using the FACS DiVa (BD) and Modfit LT softwares.
Online supplemental material
Fig. S1 shows the sequence alignment of the USP1 degron region. Fig. S2 shows APC/CCdh1-dependent ubiquitination of USP1 using different E2 pairs and methyl ubiquitin. Fig. S3 shows that the degron mutant can still interact with Cdh1. Online supplemental material is available at http://www.jcb.org/cgi/content/full/jcb.201101062/DC1.
Acknowledgments
We especially would like to thank T. Ogi, A. Lehmann, and A. D’Andrea for valuable tools and reagents for this study and members of the T. Huang, M. Pagano, D. Bar-Sagi, and D. Reinberg laboratories for their reagents, technical assistance, equipment, and helpful discussions. We also thank Peter Lopez at the New York University Flow Cytometry Core Facility for helpful assistance with FACS analysis.
This work was supported by National Institutes of Health grants to T.T. Huang (R01-GM084244) and to M. Pagano (R01-GM057587, R37-CA076584, and R21-CA161108). M. Pagano is an Investigator with the Howard Hughes Medical Institute.
Footnotes
Abbreviations used in this paper:
- APC/C
- anaphase-promoting complex/cyclosome
- DDR
- DNA damage response
- DUB
- deubiquitinase
- FA
- Fanconi anemia
- NER
- nucleotide excision repair
- PCNA
- proliferating cell nuclear antigen
- PI
- propidium iodide
- TLS
- trans-lesion synthesis
- UbVS
- ubiquitin vinyl sulfone
References
- Aristarkhov A., Eytan E., Moghe A., Admon A., Hershko A., Ruderman J.V. 1996. E2-C, a cyclin-selective ubiquitin carrier protein required for the destruction of mitotic cyclins. Proc. Natl. Acad. Sci. USA. 93:4294–4299 10.1073/pnas.93.9.4294 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bashir T., Dorrello N.V., Amador V., Guardavaccaro D., Pagano M. 2004. Control of the SCF(Skp2-Cks1) ubiquitin ligase by the APC/C(Cdh1) ubiquitin ligase. Nature. 428:190–193 10.1038/nature02330 [DOI] [PubMed] [Google Scholar]
- Bassermann F., Frescas D., Guardavaccaro D., Busino L., Peschiaroli A., Pagano M. 2008. The Cdc14B-Cdh1-Plk1 axis controls the G2 DNA-damage-response checkpoint. Cell. 134:256–267 10.1016/j.cell.2008.05.043 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bienko M., Green C.M., Crosetto N., Rudolf F., Zapart G., Coull B., Kannouche P., Wider G., Peter M., Lehmann A.R., et al. 2005. Ubiquitin-binding domains in Y-family polymerases regulate translesion synthesis. Science. 310:1821–1824 10.1126/science.1120615 [DOI] [PubMed] [Google Scholar]
- Borodovsky A., Ovaa H., Kolli N., Gan-Erdene T., Wilkinson K.D., Ploegh H.L., Kessler B.M. 2002. Chemistry-based functional proteomics reveals novel members of the deubiquitinating enzyme family. Chem. Biol. 9:1149–1159 10.1016/S1074-5521(02)00248-X [DOI] [PubMed] [Google Scholar]
- Chabes A.L., Pfleger C.M., Kirschner M.W., Thelander L. 2003. Mouse ribonucleotide reductase R2 protein: a new target for anaphase-promoting complex-Cdh1-mediated proteolysis. Proc. Natl. Acad. Sci. USA. 100:3925–3929 10.1073/pnas.0330774100 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cohn M.A., Kowal P., Yang K., Haas W., Huang T.T., Gygi S.P., D’Andrea A.D. 2007. A UAF1-containing multisubunit protein complex regulates the Fanconi anemia pathway. Mol. Cell. 28:786–797 10.1016/j.molcel.2007.09.031 [DOI] [PubMed] [Google Scholar]
- Colnaghi L., Jones M.J., Cotto-Rios X.M., Schindler D., Hanenberg H., Huang T.T. 2011. Patient-derived C-terminal mutation of FANCI causes protein mislocalization and reveals putative EDGE motif function in DNA repair. Blood. 117:2247–2256 10.1182/blood-2010-07-295758 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Daigaku Y., Davies A.A., Ulrich H.D. 2010. Ubiquitin-dependent DNA damage bypass is separable from genome replication. Nature. 465:951–955 10.1038/nature09097 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Edmunds C.E., Simpson L.J., Sale J.E. 2008. PCNA ubiquitination and REV1 define temporally distinct mechanisms for controlling translesion synthesis in the avian cell line DT40. Mol. Cell. 30:519–529 10.1016/j.molcel.2008.03.024 [DOI] [PubMed] [Google Scholar]
- Engelbert D., Schnerch D., Baumgarten A., Wäsch R. 2008. The ubiquitin ligase APC(Cdh1) is required to maintain genome integrity in primary human cells. Oncogene. 27:907–917 10.1038/sj.onc.1210703 [DOI] [PubMed] [Google Scholar]
- Frampton J., Irmisch A., Green C.M., Neiss A., Trickey M., Ulrich H.D., Furuya K., Watts F.Z., Carr A.M., Lehmann A.R. 2006. Postreplication repair and PCNA modification in Schizosaccharomyces pombe. Mol. Biol. Cell. 17:2976–2985 10.1091/mbc.E05-11-1008 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Friedberg E.C., Lehmann A.R., Fuchs R.P. 2005. Trading places: how do DNA polymerases switch during translesion DNA synthesis? Mol. Cell. 18:499–505 10.1016/j.molcel.2005.03.032 [DOI] [PubMed] [Google Scholar]
- Gao D., Inuzuka H., Korenjak M., Tseng A., Wu T., Wan L., Kirschner M., Dyson N., Wei W. 2009. Cdh1 regulates cell cycle through modulating the claspin/Chk1 and the Rb/E2F1 pathways. Mol. Biol. Cell. 20:3305–3316 10.1091/mbc.E09-01-0092 [DOI] [PMC free article] [PubMed] [Google Scholar]
- García-Higuera I., Manchado E., Dubus P., Cañamero M., Méndez J., Moreno S., Malumbres M. 2008. Genomic stability and tumour suppression by the APC/C cofactor Cdh1. Nat. Cell Biol. 10:802–811 10.1038/ncb1742 [DOI] [PubMed] [Google Scholar]
- Garg P., Burgers P.M. 2005. Ubiquitinated proliferating cell nuclear antigen activates translesion DNA polymerases eta and REV1. Proc. Natl. Acad. Sci. USA. 102:18361–18366 10.1073/pnas.0505949102 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Garnett M.J., Mansfeld J., Godwin C., Matsusaka T., Wu J., Russell P., Pines J., Venkitaraman A.R. 2009. UBE2S elongates ubiquitin chains on APC/C substrates to promote mitotic exit. Nat. Cell Biol. 11:1363–1369 10.1038/ncb1983 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Glotzer M., Murray A.W., Kirschner M.W. 1991. Cyclin is degraded by the ubiquitin pathway. Nature. 349:132–138 10.1038/349132a0 [DOI] [PubMed] [Google Scholar]
- Hoege C., Pfander B., Moldovan G.L., Pyrowolakis G., Jentsch S. 2002. RAD6-dependent DNA repair is linked to modification of PCNA by ubiquitin and SUMO. Nature. 419:135–141 10.1038/nature00991 [DOI] [PubMed] [Google Scholar]
- Hoeijmakers J.H. 2001. Genome maintenance mechanisms for preventing cancer. Nature. 411:366–374 10.1038/35077232 [DOI] [PubMed] [Google Scholar]
- Huang T.T., D’Andrea A.D. 2006. Regulation of DNA repair by ubiquitylation. Nat. Rev. Mol. Cell Biol. 7:323–334 10.1038/nrm1908 [DOI] [PubMed] [Google Scholar]
- Huang T.T., Nijman S.M., Mirchandani K.D., Galardy P.J., Cohn M.A., Haas W., Gygi S.P., Ploegh H.L., Bernards R., D’Andrea A.D. 2006. Regulation of monoubiquitinated PCNA by DUB autocleavage. Nat. Cell Biol. 8:339–347 [DOI] [PubMed] [Google Scholar]
- Kannouche P.L., Wing J., Lehmann A.R. 2004. Interaction of human DNA polymerase eta with monoubiquitinated PCNA: a possible mechanism for the polymerase switch in response to DNA damage. Mol. Cell. 14:491–500 10.1016/S1097-2765(04)00259-X [DOI] [PubMed] [Google Scholar]
- Karras G.I., Jentsch S. 2010. The RAD6 DNA damage tolerance pathway operates uncoupled from the replication fork and is functional beyond S phase. Cell. 141:255–267 10.1016/j.cell.2010.02.028 [DOI] [PubMed] [Google Scholar]
- Ke P.Y., Kuo Y.Y., Hu C.M., Chang Z.F. 2005. Control of dTTP pool size by anaphase promoting complex/cyclosome is essential for the maintenance of genetic stability. Genes Dev. 19:1920–1933 10.1101/gad.1322905 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kim J.M., Parmar K., Huang M., Weinstock D.M., Ruit C.A., Kutok J.L., D’Andrea A.D. 2009. Inactivation of murine Usp1 results in genomic instability and a Fanconi anemia phenotype. Dev. Cell. 16:314–320 10.1016/j.devcel.2009.01.001 [DOI] [PMC free article] [PubMed] [Google Scholar]
- King R.W., Glotzer M., Kirschner M.W. 1996. Mutagenic analysis of the destruction signal of mitotic cyclins and structural characterization of ubiquitinated intermediates. Mol. Biol. Cell. 7:1343–1357 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lehmann A.R., Fuchs R.P. 2006. Gaps and forks in DNA replication: rediscovering old models. DNA Repair (Amst.). 5:1495–1498 10.1016/j.dnarep.2006.07.002 [DOI] [PubMed] [Google Scholar]
- Lehmann A.R., Niimi A., Ogi T., Brown S., Sabbioneda S., Wing J.F., Kannouche P.L., Green C.M. 2007. Translesion synthesis: Y-family polymerases and the polymerase switch. DNA Repair (Amst.). 6:891–899 10.1016/j.dnarep.2007.02.003 [DOI] [PubMed] [Google Scholar]
- Littlepage L.E., Ruderman J.V. 2002. Identification of a new APC/C recognition domain, the A box, which is required for the Cdh1-dependent destruction of the kinase Aurora-A during mitotic exit. Genes Dev. 16:2274–2285 10.1101/gad.1007302 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Liu W., Li W., Fujita T., Yang Q., Wan Y. 2008. Proteolysis of CDH1 enhances susceptibility to UV radiation-induced apoptosis. Carcinogenesis. 29:263–272 10.1093/carcin/bgm251 [DOI] [PubMed] [Google Scholar]
- Lopes M., Foiani M., Sogo J.M. 2006. Multiple mechanisms control chromosome integrity after replication fork uncoupling and restart at irreparable UV lesions. Mol. Cell. 21:15–27 10.1016/j.molcel.2005.11.015 [DOI] [PubMed] [Google Scholar]
- Nakayama K.I., Nakayama K. 2006. Ubiquitin ligases: cell-cycle control and cancer. Nat. Rev. Cancer. 6:369–381 10.1038/nrc1881 [DOI] [PubMed] [Google Scholar]
- Nijman S.M., Huang T.T., Dirac A.M., Brummelkamp T.R., Kerkhoven R.M., D’Andrea A.D., Bernards R. 2005a. The deubiquitinating enzyme USP1 regulates the Fanconi anemia pathway. Mol. Cell. 17:331–339 10.1016/j.molcel.2005.01.008 [DOI] [PubMed] [Google Scholar]
- Nijman S.M., Luna-Vargas M.P., Velds A., Brummelkamp T.R., Dirac A.M., Sixma T.K., Bernards R. 2005b. A genomic and functional inventory of deubiquitinating enzymes. Cell. 123:773–786 10.1016/j.cell.2005.11.007 [DOI] [PubMed] [Google Scholar]
- Oestergaard V.H., Langevin F., Kuiken H.J., Pace P., Niedzwiedz W., Simpson L.J., Ohzeki M., Takata M., Sale J.E., Patel K.J. 2007. Deubiquitination of FANCD2 is required for DNA crosslink repair. Mol. Cell. 28:798–809 10.1016/j.molcel.2007.09.020 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ogi T., Limsirichaikul S., Overmeer R.M., Volker M., Takenaka K., Cloney R., Nakazawa Y., Niimi A., Miki Y., Jaspers N.G., et al. 2010. Three DNA polymerases, recruited by different mechanisms, carry out NER repair synthesis in human cells. Mol. Cell. 37:714–727 10.1016/j.molcel.2010.02.009 [DOI] [PubMed] [Google Scholar]
- Pfleger C.M., Kirschner M.W. 2000. The KEN box: an APC recognition signal distinct from the D box targeted by Cdh1. Genes Dev. 14:655–665 [PMC free article] [PubMed] [Google Scholar]
- Plosky B.S., Vidal A.E., Fernández de Henestrosa A.R., McLenigan M.P., McDonald J.P., Mead S., Woodgate R. 2006. Controlling the subcellular localization of DNA polymerases iota and eta via interactions with ubiquitin. EMBO J. 25:2847–2855 10.1038/sj.emboj.7601178 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Prives C., Gottifredi V. 2008. The p21 and PCNA partnership: a new twist for an old plot. Cell Cycle. 7:3840–3846 10.4161/cc.7.24.7243 [DOI] [PubMed] [Google Scholar]
- Qiao X., Zhang L., Gamper A.M., Fujita T., Wan Y. 2010. APC/C-Cdh1: from cell cycle to cellular differentiation and genomic integrity. Cell Cycle. 9:3904–3912 10.4161/cc.9.19.13585 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Reis A., Levasseur M., Chang H.Y., Elliott D.J., Jones K.T. 2006. The CRY box: a second APCcdh1-dependent degron in mammalian cdc20. EMBO Rep. 7:1040–1045 10.1038/sj.embor.7400772 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Reyes-Turcu F.E., Ventii K.H., Wilkinson K.D. 2009. Regulation and cellular roles of ubiquitin-specific deubiquitinating enzymes. Annu. Rev. Biochem. 78:363–397 10.1146/annurev.biochem.78.082307.091526 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sims A.E., Spiteri E., Sims R.J., III, Arita A.G., Lach F.P., Landers T., Wurm M., Freund M., Neveling K., Hanenberg H., et al. 2007. FANCI is a second monoubiquitinated member of the Fanconi anemia pathway. Nat. Struct. Mol. Biol. 14:564–567 10.1038/nsmb1252 [DOI] [PubMed] [Google Scholar]
- Skaar J.R., Pagano M. 2008. Cdh1: a master G0/G1 regulator. Nat. Cell Biol. 10:755–757 10.1038/ncb0708-755 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Smogorzewska A., Matsuoka S., Vinciguerra P., McDonald E.R., III, Hurov K.E., Luo J., Ballif B.A., Gygi S.P., Hofmann K., D’Andrea A.D., Elledge S.J. 2007. Identification of the FANCI protein, a monoubiquitinated FANCD2 paralog required for DNA repair. Cell. 129:289–301 10.1016/j.cell.2007.03.009 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Soria G., Podhajcer O., Prives C., Gottifredi V. 2006. P21Cip1/WAF1 downregulation is required for efficient PCNA ubiquitination after UV irradiation. Oncogene. 25:2829–2838 10.1038/sj.onc.1209315 [DOI] [PubMed] [Google Scholar]
- Soria G., Belluscio L., van Cappellen W.A., Kanaar R., Essers J., Gottifredi V. 2009. DNA damage induced Pol eta recruitment takes place independently of the cell cycle phase. Cell Cycle. 8:3340–3348 10.4161/cc.8.20.9836 [DOI] [PubMed] [Google Scholar]
- Stokes M.P., Comb M.J. 2008. A wide-ranging cellular response to UV damage of DNA. Cell Cycle. 7:2097–2099 10.4161/cc.7.14.6326 [DOI] [PubMed] [Google Scholar]
- Ulrich H.D. 2006. Deubiquitinating PCNA: a downside to DNA damage tolerance. Nat. Cell Biol. 8:303–305 10.1038/ncb0406-303 [DOI] [PubMed] [Google Scholar]
- Watanabe K., Tateishi S., Kawasuji M., Tsurimoto T., Inoue H., Yamaizumi M. 2004. Rad18 guides poleta to replication stalling sites through physical interaction and PCNA monoubiquitination. EMBO J. 23:3886–3896 10.1038/sj.emboj.7600383 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Waters L.S., Minesinger B.K., Wiltrout M.E., D’Souza S., Woodruff R.V., Walker G.C. 2009. Eukaryotic translesion polymerases and their roles and regulation in DNA damage tolerance. Microbiol. Mol. Biol. Rev. 73:134–154 10.1128/MMBR.00034-08 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Williamson A., Wickliffe K.E., Mellone B.G., Song L., Karpen G.H., Rape M. 2009. Identification of a physiological E2 module for the human anaphase-promoting complex. Proc. Natl. Acad. Sci. USA. 106:18213–18218 10.1073/pnas.0907887106 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wu T., Merbl Y., Huo Y., Gallop J.L., Tzur A., Kirschner M.W. 2010. UBE2S drives elongation of K11-linked ubiquitin chains by the anaphase-promoting complex. Proc. Natl. Acad. Sci. USA. 107:1355–1360 10.1073/pnas.0912802107 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yu H., King R.W., Peters J.M., Kirschner M.W. 1996. Identification of a novel ubiquitin-conjugating enzyme involved in mitotic cyclin degradation. Curr. Biol. 6:455–466 10.1016/S0960-9822(02)00513-4 [DOI] [PubMed] [Google Scholar]
- Zhang L., Park C.H., Wu J., Kim H., Liu W., Fujita T., Balasubramani M., Schreiber E.M., Wang X.F., Wan Y. 2010. Proteolysis of Rad17 by Cdh1/APC regulates checkpoint termination and recovery from genotoxic stress. EMBO J. 29:1726–1737 10.1038/emboj.2010.55 [DOI] [PMC free article] [PubMed] [Google Scholar]