Abstract
We demonstrate a general strategy to determine structures from showers of microcrystals. It uses acoustic droplet ejection (ADE) to transfer 2.5 nanoliter droplets from the surface of microcrystal slurries, through the air, and onto mounting micromesh pins. Individual microcrystals are located by raster-scanning a several micron X-ray beam across the cryocooled micromeshes. X-ray diffraction datasets merged from several micron-sized crystals are used to solve 1.8 Å resolution crystal structures.
Microcrystals measuring only a few microns along an edge are often easy to obtain, even from viruses, membrane proteins, protein-protein complexes, or protein-nucleic acid complexes. They are ubiquitous, but difficult to use because they are too small to yield a suitable diffraction pattern with conventional macromolecular crystallography (MX). Fortunately, advances in X-ray sources at third generation synchrotrons and free electron lasers (FEL) (1, 2) are rapidly reducing the sample size (3–5) and exposure time required for atomic level crystal structure determination.(6, 7) As an example, consider that an MX beamline at the National Synchrotron Light Source II (NSLS-II) currently under construction at the Brookhaven National Laboratory will produce a 1 μm diameter X-ray beam with sufficient intensity that a similarly sized protein crystal is projected to reach its maximum dose limit in as little as 5 ms.(8) However, as the crystal size is reduced, so is the signal relative to the noise in the X-ray diffraction data. Consequently, an essential strategy to improve the signal to noise ratio is to reduce the background scattering, especially from the mother liquor surrounding a micron-sized crystal. Unfortunately, removing all the excess mother liquor is very difficult as the crystal size approaches a few cubic microns. Therefore, to exploit the full capabilities of third generation synchrotrons and free electron lasers, robust new strategies must be developed to manipulate microcrystals for structure determination.
To address this critical gap, we are developing acoustic droplet ejection (ADE) methods to accurately and gently transfer small protein crystals (roughly 10 μm on each side) within microdroplets of mother liquor from the crystallization well, through a short air column (1–10 cm), to a standard X-ray diffraction mounting mesh. The acoustic droplet ejection instrumentation uses sound energy to transfer nanoliter to picoliter volumes from the surface of liquids. The principle that sound waves of great intensity near the surface of a liquid will eject droplets was demonstrated in 1927.(9) If sufficient energy from the transducer is focused near the surface, then a controlled ejection occurs. This is because the sound waves carry energy from a transducer to the focal point, where it produces a displacement of the surface that ejects a small droplet (Fig. 1A). Potential energy transfers from a periodic compression wave to kinetic energy of an ejected particle. The wavelength of the sound governs the volume into which the energy is deposited. (10, 11) Therefore, the ejected droplet diameter is proportional to the wavelength; consequently, smaller volume droplets are produced by higher frequency (shorter wavelength) sound waves. The proportionality also yields very accurate ejected volumes (σ < 4%) that scale to 12 microns in diameter (1 pL in volume) or below.(10, 12) If the hydrodynamic characteristics of the fluid are suitable and the sound wave is well focused, then the initial velocity of the ejected droplet is proportional to the amplitude of the focused wave.(13) Consequently, the transfer momentum of the droplet over a range of destination distances can be precisely controlled.
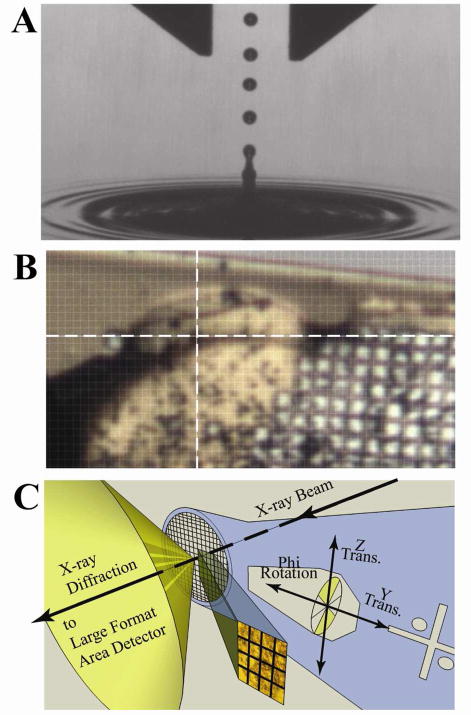
Figure 1.
The concepts of ADE and raster-scanning. A) A stroboscopic photomicrograph of a single 2.5 nL water droplet (~175 μm in diameter) launched via ADE from the liquid surface. The 1 mm width of the calipers at the top and the 400 microsecond strobe frequency are shown only to provide calibration, from which we estimate that the droplet was traveling at approximately 1 m/s. For droplets of about 2.5 nanoliters, the rate of drop ejection is typically every 2 ms. Higher rates are achieved for smaller drops; however, factors that impact this rate include the droplet size, solution hydrodynamic parameters for settling of the perturbed surface and the power load for the transducer duty cycle. B) An image from beamline X25 of the NSLS of the insulin microcrystal slurry supported on a micro mesh (25 μm × 25 μm grid pattern) used to solve the structure. The 20 μm × 20 μm X-ray beam is centered at the intersection of the white cross-hairs. C) An illustration of the concepts for raster scanning X-ray diffraction strategy with a micro-diffractometer and a several-micron sized X-ray beam (see also supplemental information section 3).
We and others have shown that ADE can rapidly transfer variable volumes of virtually any liquid with very high precision.(11, 14) Consequently, it is ideal for many applications, including drug discovery.(15) The successful use of ADE to transfer living cells and isolated DNA without inducing strand breaks suggests that it might be gentle enough for protein crystals. However, ADE must not impact the crystal lattice and consequently its diffraction quality, which is inherently more fragile than covalent bonds. Indeed, ADE has been used very recently for seeding protein crystallization trials,(16) but to our knowledge, not for X-ray diffraction and structure determination. Here, we report that ADE methods are well-suited for transferring 2.5 nanoliter droplets of microcrystal slurries of insulin or lysozyme from a 384-well plate to standard MiTeGen™ (Ithaca, NY) micromesh mounting pins. After cryocooling, the micron-sized crystals are located on the mesh with the X-ray beam via a raster-scan strategy by the presence or absence of diffraction. Once micro-crystals are located, partial datasets are collected and crystal structures solved to better than 2 Å resolution from merged datasets. Importantly, high resolution structures can be solved from slurries of microcrystals that traditionally would have been discarded as unsuitable for X-ray diffraction studies.
We prepared slurries of Sus scrofa (pig) Zn2+ insulin or Gallus gallus (hen egg-white) lysozyme microcrystals as detailed in the Supplemental Information sections (see also Figs. S2 – S4). Briefly, rhombohedral insulin microcrystals (space group R3) were obtained under low-salt conditions by slowly lowering the temperature from 313 K to 293 K.(17) By changing the rate of the temperature decrease, e.g. −100, −10, or −2 K per minute, we obtained slurry of uniformly-sized insulin microcrystals with largest dimensions of ~5 μm, ~10μm, or ~20 μm, respectively. Prior to ADE transfer, the insulin crystal slurries were concentrated by mild centrifugation and excess mother liquor removed until the volume of crystalline material to the total volume was ~50% for the 10 μm-sized and 25% for the 20 μm-sized microcrystals. Lysozyme microcrystals of tetragonal lattice (space group P43212) in the 5 – 20 μm sized-range were obtained at room temperature from highly saturated solution with sodium chloride and constant agitation using a reciprocating rocker.(18) The lysozyme microcrystal slurries, where ~10% of the total volume consists of crystalline material, were not concentrated and used as is for ADE transfer.
We used a modified Echo® Liquid Handling System at the Labcyte facility (Sunnyvale, CA) to transfer homogenous and heterogeneous slurries of microcrystals of insulin and lysozyme. Briefly, the critical characteristics important to crystallography include: i) a robotic system for loading/unloading trays from a storage rack into the ejection apparatus, ii) automatic capability to seal and unseal source trays, iii) x/y translation control for both the source and the destination plate, iv) two on-axis optical visualization cameras, v) a perpendicular off-axis optical system equipped with a stroboscopic camera to measure droplet velocity, and vi) either manual or computer controlled operation. We typically transferred one to four 2.5 nanoliter droplets from a source well containing 100 μl of a slurry of microcrystals suspended in mother liquor augmented with cryosolvents to a MiTeGen 400/25 micromesh, and then immediately plunged the mesh into liquid nitrogen to flash-cool the samples.
X-ray diffraction data were collected at NSLS beamline X25 with a 20 μm × 20 μm X-ray beam (Fig. 1b), and at APS beamline 23ID-D with a 10 μm diameter X-ray beam. At each beamline, software designed for raster-scan searching (Fig. 1c), X-ray diffraction image evaluation, and meta-data handling were used to find and collect X-ray diffraction data from the microcrystals.(19, 20) Complete datasets were obtained by merging data from a small number of microcrystals (between 2 and 14) that were simultaneously ejected with one of the 2.5 nL droplets. Tables 1 and S1 summarizes our crystallographic results. Molecular replacement solutions were readily obtained for both insulin and lysozyme data using a polyalanine search model. The insulin crystals also yielded an anomalous signal in the data from two Zn2+ atoms located on the three fold symmetry axis. The small anomalous signal (~2.4%) and the phasing ambiguity that arises from anomalous scatterers located on a symmetry axis make insulin a challenging test case for SAD-based structure determination and have previously been used to evaluate new phasing methods.(21) The anomalous SAD data readily gave a correct heavy atom sub-structure The initial maps obtained after density modification improvement of SAD phases from the 20 μm insulin crystals were clearly interpretable (Fig 2) and led to a high quality SAD structure. All insulin structures determined from ADE transfer microcrystals revealed intact disulfide bridges (Fig S1) indicating that the microcrystals had not experienced significant X-ray induced radiation damage.(22) We conclude that the diffraction quality of microcrystals is not appreciably impacted by the sound waves inherent to ADE methods or other forces encountered by the droplet capture and cryocooling.
Table 1.
Data collection and model refinement statistics
| Insulin (10 μm) | Insulin (20 μm) | Lysozyme (20 μm) | |
|---|---|---|---|
| X-ray Diffraction Data Collection | |||
| # of crystals | 14 | 9 | 2 |
| X-ray source | NSLS X25 | NSLS X25 | APS 23ID-D |
| Wavelength (Å) | 1.280 | 1.280 | 0.9733 |
| Beam size (μm) | 20 × 20 | 20 × 20 | 10 × 10 |
| Resolution (Å) | 50 – 1.9 | 50 – 1.8 | 30 – 1.8 |
| Rsym or Rmerge | 24.0 (39.6) a | 11.4 (61.6) a | 10.9 (36.7) a |
| I/σI | 46.4 (3.4) a | 62.2 (2.1) a | 16.9 (1.2) a |
| Complete (%) | 93.5 (58.7) a | 99.8 (96.7) a | 94.9 (62.5) a |
| Redundancy | 6.5 | 10.5 | 1.8 |
|
| |||
| Model Refinement | |||
| # Reflections | 5917 | 7398 | 10280 |
| Rwork/Rfree | 16.7/20.3 | 18.3/22.0 | 16.8/21.2 |
| R.m.s. deviations in bond | |||
| Distances (Å) | 0.025 | 0.025 | 0.021 |
| Angles (°) | 2.244 | 2.159 | 1.938 |
Highest resolution shell
Figure 2.

High quality electron density maps are obtained from ADE-mounted, serial micro-crystallography. This experimental map, obtained from Zn SAD phases improved by solvent flattening (2.2 Å resolution, contoured at 1.5 σ), was generated using data merged from 9 different ADE-mounted insulin crystals. The anomalous signal, 2.4% of total diffraction, and the phase ambiguity introduced because the two zinc anomalous scatterers are located on a three-fold symmetry axis makes this case sensitive to noisy data. The interpretation of the map is consistent with two anti-parallel beta strands assigned as B23–B28 (shown with C, N, and O atoms rendered in green, blue and red, respectively) and the D23–D28 (not shown), which were manually built into density. This model was used as a seed by PHENIX, which was able to automatically build-in 74 out of the 102 total residues (supplementary section 3).
The X-ray brilliance and flux at new third-generation synchrotrons and X-ray FELs provide an opportunity to collect full diffraction datasets in much less than one second. Advances in technology and automation are required to support high-throughput use of micron-sized crystals to fully exploit these X-ray sources. Our results demonstrate the feasibility of using ADE and raster-scanning strategies in microcrystallography at sources like the NSLS-II. We also anticipate that ADE will match the 20 ms pulse rate for delivering crystal-containing droplets to the FEL X-ray beam. Moreover, by matching the frequency of the incident sound wave to the size of the crystals within the slurry, it is possible to mount a single microcrystal with minimal solvent. This is critical to obtain the signal to background noise ratio that is essential to solve structures from microcrystals. Therefore, ADE provides solutions to several challenges that must be overcome to realize the full potential of modern third generation synchrotron sources and FELs.
Supplementary Material
Acknowledgments
Insulin crystal data for this study were measured at beamline X25 of the National Synchrotron Light Source (NSLS) at the Brookhaven National Laboratory. We thank Annie Héroux and the PXRR staff for numerous discussions. Use of the NSLS was supported by the U.S. Department of Energy Office of Basic Energy Sciences, under Contract DE-AC02-98CH10886. Lysozyme crystal data for this study were measured at beamline 23ID-D of the Advanced Photon Source (APS) at Argonne National Laboratory. We thank Derek Yoder and the GMCA staff for their support. GM/CA CAT was funded in whole or in part with Federal funds from the National Cancer Institute (Y1-CO-1020) and the National Institute of General Medical Science (Y1-GM-1104). Use of the Advanced Photon Source was supported by the U.S. Department of Energy, Basic Energy Sciences, Office of Science, under contract No. DE-AC02-06CH11357.
Footnotes
This work was supported by the Brookhaven National Laboratory/U.S. Department of Energy, Laboratory Directed Research and Development grants 08-022 to AMO and 11-008 to ASS. Additional support was provided by the Office of Biological and Environmental Research, U.S. Department of Energy, the National Center for Research Resources (2 P41 RR012408, to ASS, JMS, and AMO) and the National Institute of General Medical Sciences (Y1 GM 0080-03, to MA) of the National Institutes of Health.
Atomic coordinates for the insulin structure solved using anomalous data and the corresponding structure factors have been deposited to the PDB with access code 3RTO.
SUPPORTING INFORMATION AVAILABLE: Sections describing insulin and lysozyme microcrystal preparations, crystal screening software, phasing and model refinement, ADE transfer of microcrystals, Table S1, and Figures S1 – S4. Electronic Supporting Information files are available without a subscription to ACS Web Editions (http://pubs.acs.org/journal/bichaw).
References
- 1.Seibert MM, Ekeberg T, Maia FR, Svenda M, Andreasson J, Jonsson O, Odic D, Iwan B, Rocker A, Westphal D, et al. Nature. 2011;470:78–81. doi: 10.1038/nature09748. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Chapman HN, Fromme P, Barty A, White TA, Kirian RA, Aquila A, Hunter MS, Schulz J, DePonte DP, Weierstall U, et al. Nature. 2011;470:73–77. [Google Scholar]
- 3.Sawaya MR, Sambashivan S, Nelson R, Ivanova MI, Sievers SA, Apostol MI, Thompson MJ, Balbirnie M, Wiltzius JJ, McFarlane HT, et al. Nature. 2007;447:453–457. doi: 10.1038/nature05695. [DOI] [PubMed] [Google Scholar]
- 4.Rasmussen SG, Choi HJ, Rosenbaum DM, Kobilka TS, Thian FS, Edwards PC, Burghammer M, Ratnala VR, Sanishvili R, Fischetti RF, et al. Nature. 2007;450:383–387. doi: 10.1038/nature06325. [DOI] [PubMed] [Google Scholar]
- 5.Cherezov V, Rosenbaum DM, Hanson MA, Rasmussen SG, Thian FS, Kobilka TS, Choi HJ, Kuhn P, Weis WI, Kobilka BK, et al. Science. 2007;318:1258–1265. doi: 10.1126/science.1150577. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Kirian RA, Wang X, Weierstall U, Schmidt KE, Spence JC, Hunter M, Fromme P, White T, Chapman HN, Holton J. Opt Express. 2010;18:5713–5723. doi: 10.1364/OE.18.005713. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Ozaki S, Bengtsson J, Kramer SL, Krinsky S, Litvinenko VN. Philosophy for NSLS-II design with subnanometer horizontal emittance. Particle Accelerator Conference, 2007. PAC. IEEE; 2007. pp. 77–79. [Google Scholar]
- 8.Hodgson KO, Anderson WF, Berman L, Fischetti R, Hendrickson WA, Kirz J, Makowski L, Phillips GNJ, Smith JL, Sweet RM, et al. Workshop of the National Institutes of Health National Center for Research Resources and the National Institute of General Medical Sciences on Plans for Support of Future Life Science Synchrotron Research at NSLS-II. National Institutes of Health; Bethesda MD: 2009. pp. 1–14. [Google Scholar]
- 9.Wood EW, Loomis AL. Philosophical Magazine Series. 1927;7(4):417–436. [Google Scholar]
- 10.Elrod SA, Hadimioglu B, Khuri x, Yakub BT, Rawson EG, Richley E, Quate CF, Mansour NN, Lundgren TS. Journal of Applied Physics. 1989;65:3441–3447. [Google Scholar]
- 11.Ellson R, Mutz M, Browning B, Lee L, Miller MF, Papen R. Journal of the Association for Laboratory Automation. 2003;8:29–34. [Google Scholar]
- 12.Comley J. Drug Discovery World Summer. 2004. pp. 43–54. [Google Scholar]
- 13.Stearns RG, Ellson RN. Acoustic ejection of fluids using large F-number focusing elements. Picoliter Inc; Mountain View, CA, United States: 2002. [Google Scholar]
- 14.David H, Mitchell M, Maria S, Richard S, Jean S, Siobhan P, Richard E, Joe O. Journal of the Association for Laboratory Automation. 2008;13:97–102. [Google Scholar]
- 15.Ellson R. Drug Discovery Today. 2002;7:S32–S34. [Google Scholar]
- 16.Villaseñor AG, Wong A, Shao A, Garg A, Kuglstatter A, Harris SF. Acta Crystallogr D Biol Crystallogr. 2010;66:568–576. doi: 10.1107/S0907444910005512. [DOI] [PubMed] [Google Scholar]
- 17.Soares AS, Caspar DL, Weckert E, Heroux A, Holzer K, Schroer K, Zellner J, Schneider D, Nolan W, Sweet RM. Acta Crystallogr D Biol Crystallogr. 2003;59:1716–1724. doi: 10.1107/s0907444903015403. [DOI] [PubMed] [Google Scholar]
- 18.Allaire M, Moiseeva N, Botez CE, Engel MA, Stephens PW. Acta Crystallogr D Biol Crystallogr. 2009;65:379–382. doi: 10.1107/S090744490900256X. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Cherezov V, Hanson MA, Griffith MT, Hilgart MC, Sanishvili R, Nagarajan V, Stepanov S, Fischetti RF, Kuhn P, Stevens RC. J R Soc Interface. 2009;6:S587–S597. doi: 10.1098/rsif.2009.0142.focus. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Skinner JM, Cowan M, Buono R, Nolan W, Bosshard H, Robinson HH, Heroux A, Soares AS, Schneider DK, Sweet RM. Acta Crystallogr D Biol Crystallogr. 2006;62:1340–1347. doi: 10.1107/S0907444906030162. [DOI] [PubMed] [Google Scholar]
- 21.Dauter Z, Dauter M, Dodson E. Acta Crystallogr D Biol Crystallogr. 2002;58:494–506. doi: 10.1107/s090744490200118x. [DOI] [PubMed] [Google Scholar]
- 22.Garman EF. Acta Crystallogr D Biol Crystallogr. 2010;66:339–351. doi: 10.1107/S0907444910008656. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.