Abstract
A major role for c-Myc in the proliferation of normal cells is attributed to its ability to promote progression through G1 and into S phase of the cell cycle. The absolute requirement of c-Myc for cell cycle progression in human tumor cells has not been comprehensively addressed. In the present work, we used a lentiviral-based short hairpin RNA (shRNA) expression vector to stably reduce c-Myc expression in a large number of human tumor cell lines and in three different types of normal human cells. In all cases, cell proliferation was severely inhibited, with normal cells ultimately undergoing G0/G1 growth arrest. In contrast, tumor cells demonstrated a much more variable cell cycle response with cells from several lines accumulating in S or G2/M phases. Moreover, in some tumor lines, the phase of cell cycle arrest caused by inhibition of c-Myc could be altered by depleting tumor suppressor protein p53 or its transcriptional target p21CIP/WAF. Our data suggest that, as in the case of normal cells, c-Myc is essential for sustaining proliferation of human tumor cells. However its rate-limiting role in cell cycle control is variable and is reliant upon the status of other cell cycle regulators.
Keywords: Myc, cell cycle, shRNA
Introduction
By virtue of its central role as a general transcription factor, the c-Myc oncoprotein regulates a number of key normal cellular processes such as growth, proliferation, cell cycle progression, apoptosis and differentiation (Grandori et al., 2000; Adhikary and Eilers, 2005; Meyer et al., 2006). The importance of c-Myc is underscored by the fact that its inhibition leads to cell cycle arrest in vitro and early embryonic lethality in vivo (Prochownik et al., 1988; Trumpp et al., 2001; Zhou and Hurlin, 2001; Lutz et al., 2002). Furthermore, deregulated overexpression of c-Myc, either in vitro or in vivo, leads to malignant transformation (Grandori et al., 2000; Lutz et al., 2002). Taken together, these data suggest that the accumulation of c-Myc mRNA and protein in numerous and diverse human tumors plays a role in the pathogenesis of these neoplasms (Nesbit et al., 1999).
The critical importance of c-Myc as a transforming oncogene has led to a number of strategies designed to inhibit its expression or function (Prochownik, 2004; Ponzielli et al., 2005; Vita and Henriksson, 2006). These efforts have been fueled, in part, by experimental systems showing that tumor cell proliferation and/or survival rely upon sustained expression of c-Myc (Adhikary and Eilers, 2005). Despite these findings, the idea that all cells have an obligate requirement for c-Myc has recently been challenged. For example, somatic knockout of both alleles of c-myc in spontaneously immortalized rat fibroblasts resulted in a several-fold reduction of proliferation but not in its complete inhibition (Mateyak et al., 1997). In liver, the conditional inactivation of c-myc impairs hepatocyte proliferation in neonatal but not adult mice (Baena et al., 2005), although another report suggested that proliferation of adult hepatocytes is also c-myc-independent (Li et al., 2006). Accordingly, conditional inactivation c-myc did not affect self-renewal of intestinal mucosa in juvenile and adult mice (Bettess et al., 2005). Additionally, c-myc was found to be dispensable for proliferation of mouse skin keratinocytes in vivo, but not for their transformation by activated H-Ras (Oskarsson et al., 2006). Based on these results, it is conceivable that the proliferation of certain types of cells occurs via c-Myc-independent pathways. In accord with this suggestion, we and others have demonstrated that resumption of normal proliferation as well as other aspects of c-Myc phenotype can be imparted to c-myc knockout fibroblasts by overexpression of individual downstream c-Myc target genes (Hermeking et al., 2000; Nikiforov et al., 2002; Rothermund et al., 2005). Together, these results raise the formal possibility that, due to their genetic heterogeneity, some tumor cells may overcome their dependency on c-Myc by expressing the proper combination of surrogate genes. Clearly, determining the extent to which this is true is critical before serious consideration can be given to the use of c-Myc inhibitors in the clinical setting.
In normal cells, inhibition of c-Myc invariably results in a G0/G1 cell cycle arrest (Prochownik et al., 1988; Mateyak et al., 1997; de Alboran et al., 2001; Prathapam et al., 2006). The type of cell cycle arrest caused by c-Myc depletion in tumor cells has not been adequately studied. Data available to date on this subject were collected on small number of lines (Skorski et al., 1995; McGuffie et al., 2000; Chen et al., 2001; Hermeking, 2003) and the methods utilized for inhibiting c-Myc were not consistent (D’Agnano et al., 2001; Citro, 1998). Therefore, the extent to which tumor cells resemble normal cells with regard to the type of cell cycle arrest, if any, caused by c-Myc depletion remains unclear.
In the current report, we have for the first time undertaken a comprehensive study of the phenotypes caused by c-Myc depletion in a large number of human tumor cell lines (22) and in three reference normal human cell types (fibroblasts, keratinocytes and melanocytes). For this purpose, we utilized a standardized and specific method of reducing endogenous c-Myc levels, for example, lentivirus-mediated delivery of short hairpin c-Myc RNAs (Myc-shRNAs). This approach was able to provide stable inhibition of c-Myc in nearly 100% of cells. As a result of these studies, we have demonstrated that the requirement for c-Myc to maintain proliferation is absolute in all examined cells, suggesting that this could be generally true. However, unlike normal cells, which invariably arrest in G0/G1 phase of the cell cycle in response to c-Myc depletion, tumor cells exhibit significant heterogeneity with regard to the positioning of cell cycle arrest. As demonstrated here, this heterogeneity could be dependent upon the intactness and relative importance of other cell cycle control pathways.
Results
Selection of human c-Myc-specific shRNA
To determine whether tumor cell proliferation depends on c-Myc, we depleted its endogenous levels using lentivirus-delivered shRNAs. As a prelude, several candidate Myc-shRNAs were cloned into lentiviral vector H1, which also encoded enhanced green fluorescent protein (eGFP) (Ivanova et al., 2006). In addition, a set of commercially available human c-Myc-specific shRNAs in the pLKO1 vector was also tested. The specificity and effectiveness of these shRNAs were first tested in immortalized c-myc-null rat fibroblasts (HO15.19) (Mateyak et al., 1997) reconstituted with human c-Myc cDNA (HO15.19-h-c-Myc). The ‘myc-null’ phenotype has been extensively characterized in HO15.19 cells, and therefore these cells serve as an excellent reference line in which to study shRNA-mediated inhibition of c-Myc expression. Supplementary Figure S1 summarizes features of HO15.19-h-c-Myc cells expressing the most effective Myc-shRNAs (M1, M2, M3). In each case we observed efficient inhibition of c-Myc as evidenced by western blotting, characteristic changes in cell morphology of Myc-depleted HO15.19-h-c-Myc cells and an increase in the fraction of cells in G0/G1 phase of the cell cycle. We therefore used these validated shRNAs for inhibition of c-Myc in human cells.
Inhibition of proliferation of normal and tumor cells by Myc-shRNAs
We studied 22 human tumor cell lines originating from a wide variety of adult and pediatric cancers (Table 1). Normal control cells consisted of primary human fore-skin keratinocytes, melanocytes and fibroblasts. All cells were infected with control shRNA or Myc-shRNAs as described in Materials and methods. The efficiency of infection was quantified by counting GFP-positive cells (which was always ≥95%, not shown). When possible, the degree of c-Myc inhibition was monitored within 48–96 h after infection by western blotting (Figure 1a, Supplementary Figure S2 and data not shown). Our analysis revealed that Myc-suppressing activities of M1, M2 and M3 varied significantly among different lines (data not shown). Therefore, based on preliminary experiments, we utilized Myc-shRNAs, which provided the most efficient inhibition of c-Myc in a given cell line.
Table 1.
Normal and tumor cells utilized in the study
| # | Cell line | Type of tissue/tumor | Cell cycle arresta |
|---|---|---|---|
| 1 | Melanocytes | Skin | G0/G1 |
| 2 | Keratinocytes | Skin | G0/G1 |
| 3 | Fibroblasts | Skin | G0/G1 |
| 4 | Mel-1 9 | Metastatic melanoma | G0/G1 |
| 5 | SK-Mel-28 | Metastatic melanoma | G2/M |
| 6 | SK-Mel-29 | Metastatic melanoma | G0/G1 |
| 7 | SK-Mel-94Metasta | Metasta tic melanoma | G0/G1+G2/M |
| 8 | SK-Mel-103 | Metastatic melanoma | G0/G1 |
| 9 | SK-Mel-147 | Metastatic melanoma | G0/G1 |
| 10b | UACC62 | Metastatic melanoma | S |
| 11b | SK-Mel-173 | Metastatic melanoma | G0/G1 |
| 12 | Malme 3M | Metastatic melanoma | G0/G1+G2/M |
| 13b | UACC257 | Metastatic melanoma | S, G2/M |
| 14 | SAOS-2 | Osteosarcoma | G0/G1 |
| 15 | HT-1080 | Osteosarcoma | G2/M |
| 16 | A549 | Lung carcinoma | G0/G1+G2/M |
| 17 | OVCAR-8 | Ovary adenocarcinoma | G2/M |
| 18 | PC3 | Prostate adenocarcinoma | G2/M |
| 19 | U251 | Glioma | G2/M |
| 20 | RDES | Ewing’s sarcoma | G1/S |
| 21 | HT-29 | Colon adenocarcinoma | G2/M |
| 22 | CACO-2 | Colon adenocarcinoma | G2/M |
| 23 | HCT-116 | Colon adenocarcinoma | N/Ac |
| 24 | MDA-MB2 31 | Breast adenocarcinoma | S, G2/M |
| 25 | NCI-H460 | Large cell lung cancer | G2/M |
Predominant type of the cell cycle arrest.
Lines in which MYC-depletion causes apoptosis in more than 20% of cells at day 5 post infection.
No apparent changes in the cell cycle parameters.
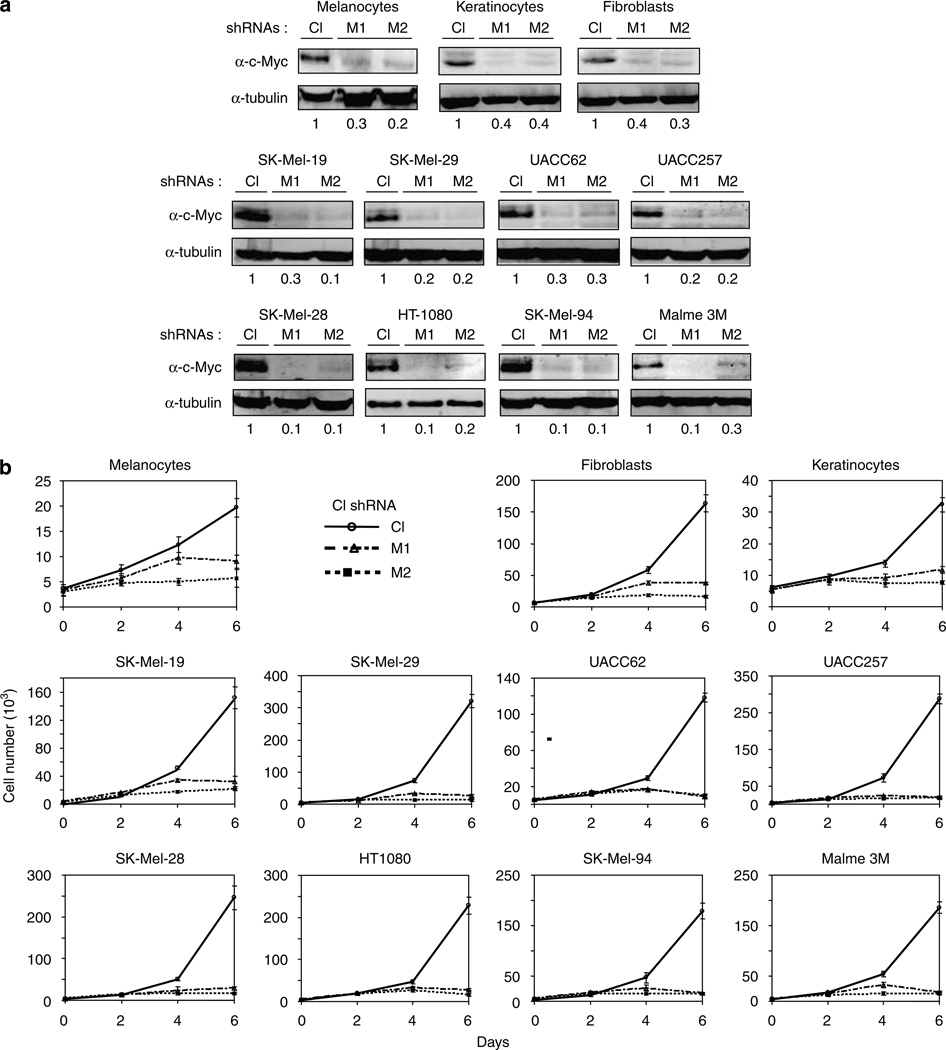
Figure 1.
Inhibition of c-Myc expression in normal and tumor human cells leads to arrest of proliferation. Each of the indicated tumor cell lines or populations was infected during log-phase growth with lentiviral vectors encoding a control shRNA (Cl) or two different c-Myc-shRNAs M1 or M2. (a) Cells were harvested at 4days post infection in the SDS-containing buffer, resolved on 8% polyacrylamide gel, transferred to PVDF membranes and probed in western blotting with the antibodies designated on the right. Numbers below the panels represent c-Myc amounts normalized by the amounts of tubulin and amounts of c-Myc in control samples. (b) Tumor cells were plated in triplicates in 24-well plates (2 × 103 cells per well), normal cells were plated in triplicates in 24-well plates (0.5–2 × 104 cells per well). Cells were counted every other day for 6 days. Numbers below the graph indicate days post infection.
In order to evaluate the impact of Myc-shRNAs expression on cell proliferation, cells were plated at low density 2 days after infection and were counted and/or photographed during the ensuing 6 days. As seen in Figure 1b and Supplementary Figure S3, all cells infected with control lentiviral continued to proliferate logarithmically. In contrast, cells infected with the Myc-shRNAs showed an abrupt cessation of proliferation starting at day 3 or 4 post infection. In some cases, cells underwent apoptotic death starting as early as day 4 post infection (Supplementary Figure S4).
In several cases, we observed the eventual emergence of proliferating cells from Myc-shRNA-infected cultures. Western blot analysis revealed that c-Myc expression, while initially low, had returned to the pre-infection levels of control cells (not shown). This suggested that these cells had somehow escaped Myc-shRNAs action or that the efficiency of infection of these cells was suboptimal, thus allowing for the outgrowth of cells with a proliferative advantage. We therefore conclude that the continuous suppression of c-Myc is mandatory for the proliferative arrest of all cell lines examined.
Cell line-specific effects of c-Myc depletion
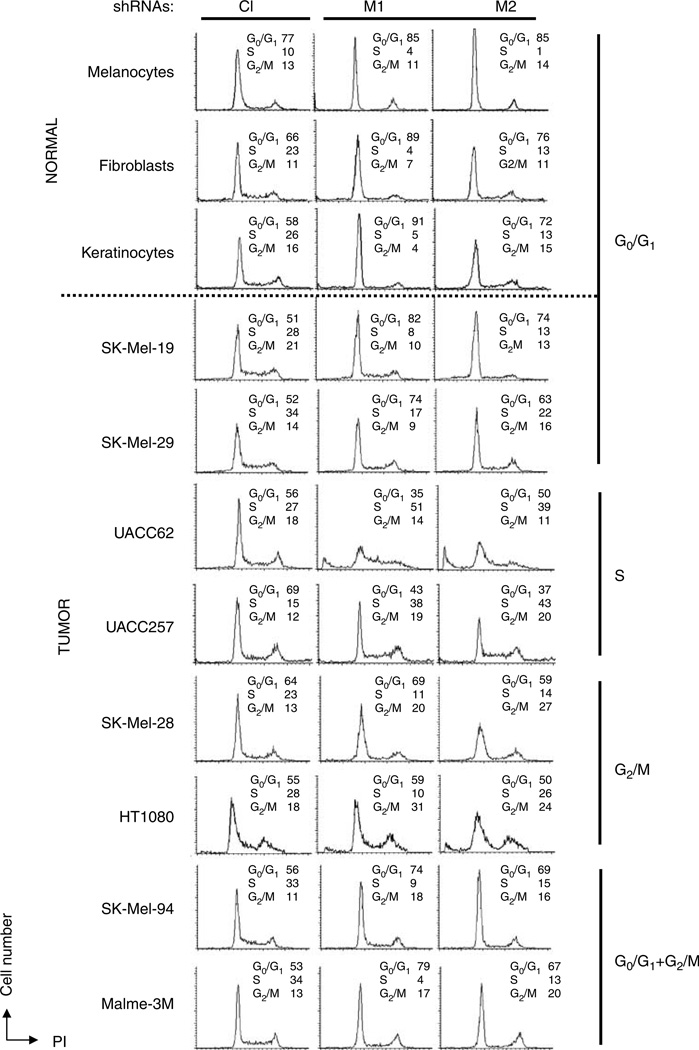
A large body of literature suggests that inhibition of c-Myc in primary cells or non-transformed cell lines generally leads to a strong proliferative arrest in the G0/G1 or G1/S phases of the cell cycle (Prochownik et al., 1988; Mateyak et al., 1997; de Alboran et al., 2001; Prathapam et al., 2006). In order to determine whether this is also the case for tumor cells, we used propidium iodide (PI) staining and flow cytometry to evaluate the cell cycle parameters of the above-described cells 4–5 days after infection (Figure 2, Supplementary Figure S5). In parallel, control and two Myc-shRNAs were introduced separately into populations of normal skin melanocytes, fibroblasts and keratinocytes. Overall, the cell cycle profiles of cells infected with control lentiviral vector did not differ significantly from those of their uninfected counterparts (not shown). In normal cells, depletion of c-Myc resulted in G0/G1 growth arrest, as previously reported (Prochownik et al., 1988; Mateyak et al., 1997; de Alboran et al., 2001; Prathapam et al., 2006). In contrast, tumor cells expressing Myc-shRNAs displayed several distinct types of cell cycle arrest as depicted in Figure 2 and Supplementary Figure S5. For example, similar to normal melanocytes, melanoma lines SK-Mel-19 and SK-Mel-29 arrested in G0/G1 in response to c-Myc depletion. In contrast, c-Myc depletion in melanoma cells UACC62 and UACC257 resulted in S-phase arrest accompanied by apoptosis at days 4–5 (Supplementary Figure S4), whereas SK-Mel-28 melanoma cells and HT1080 osteosarcoma cells arrested in G2/M without any discernible apoptosis. In further contrast, c-Myc inhibition in SK-Mel-94and Malme 3M melanoma cells resulted in cell cycle arrest concurrently in G0/G1 and G2/M phases. In colon cancer line HCT-116, cell cycle profiles of Myc-depleted cells were nearly identical to those of cells infected with the control vector. Since these cells demonstrated dramatically reduced proliferation rates (Supplementary Figure S3B), we conclude that c-Myc depletion blocks cell cycle progression in these cells equally in all phases. The cell cycle profiles of all other cancer lines were basically similar to one of those described above (Table 1, Supplementary Figure S5). Independent infection of a number of these lines with another c-Myc-shRNA lentivirus produced similar results thus indicating that the cell cycle arrest was truly a specific consequence of c-Myc depletion (not shown).
Figure 2.
c-Myc depletion causes different types of cell cycle arrest in normal and tumor cells. The indicated cell lines were infected during log phase growth with lentiviral vectors expressing either control (Cl) or c-Myc M1 or M2 shRNAs. Four days post infection the subconfluent cultures were harvested, stained with propidium iodide and analysed by flow cytometry. See Supplementary Figure S5 for the cell cycle profiles of other cell lines.
Our findings indicate that the cell cycle response to c-Myc depletion in normal cells is invariably associated with G0/G1 arrest. However, among tumor cell lines, considerable cell cycle heterogeneity is observed, even among lines derived from identical tumor types. These observations suggest that other factors, whose expression levels or mutational status differ between normal and tumor cells, determine the type of cell cycle arrest in c-Myc-depleted tumor cells.
Inhibition of p53 or p21 alters the cell cycle profile of c-Myc-depleted cells
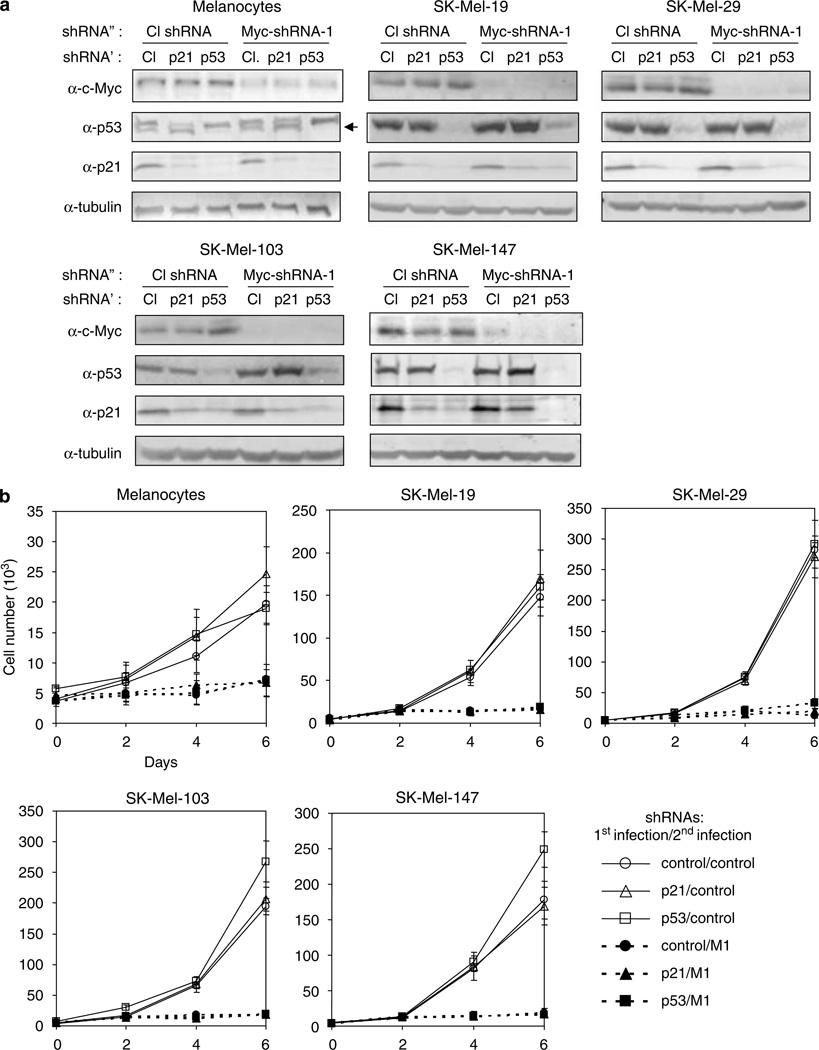
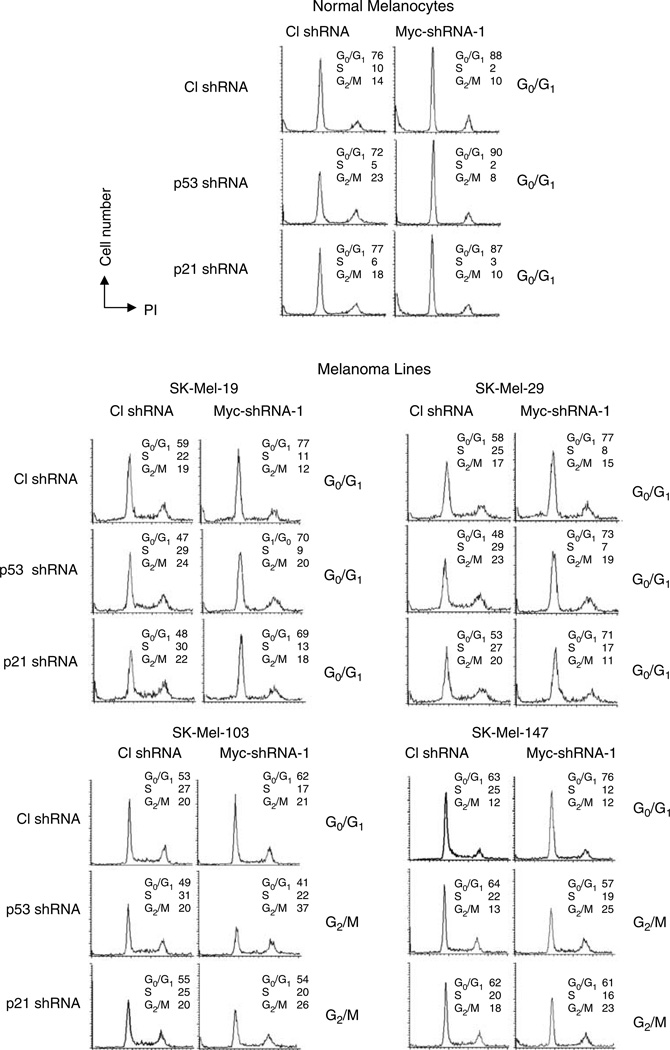
We were interested in understanding the molecular mechanisms that allow cells from several tumor lines to escape the G0/G1 growth arrest that characterizes c-Myc-depleted normal cells. It is well accepted that the expression of factors promoting G0/G1 arrest in response to different forms of stress can be altered in tumor cells (Hahn and Weinberg, 2002). Therefore, we hypothesized that qualitative or quantitative differences in such factors might underlie the different cell cycle profiles of c-Myc-depleted normal and tumor cells. To test this hypothesis, we first investigated the impact of inhibiting the p53 tumor suppressor protein on the cell cycle parameters of c-Myc-depleted cells undergoing G0/G1 arrest. We focused on p53, since it is involved in the G1/S checkpoint and because its inactivation or loss occurs frequently in tumor cells (Hanahan and Weinberg, 2000). To this end, we chose melanoma lines SK-Mel-19, -29, -103 and -147, which express wild-type p53 (Soengas et al., 2001), and which arrest in G0/G1 in response to c-Myc depletion (Figure 2). Primary melanocytes were also included in the experiment to compare cell cycle responses between normal and tumor cells of similar origin. As shown in Supplementary Figure S6, depletion of c-Myc resulted in slight increase in p53 levels in SK-Mel-19 and SK-Mel-29 cells and in moderate increase in p53 levels in SK-Mel-103 and SK-Mel-147 cells: 1.5–2 fold and 2.4–3.3 fold, respectively. Simultaneous suppression of p53 and c-Myc (by M1 shRNA) (Figure 3a) led to differences in the cell cycle arrest profiles among the studied cells (Figure 4). In normal melanocytes and cells from lines SK-Mel-19 and SK-Mel-29, G0/G1 phase was increased by 17, 30 and 32%, respectively, in response to c-Myc depletion only and by 25, 49 and 50%, respectively in cells depleted of both c-Myc and p53 (Figure 4). Depletion of c-Myc alone in melanoma cells SK-Mel-103 and SK-Mel-147 also resulted in corresponding increases of G0/G1 phase by 17 and 21%. In contrast, concurrent c-Myc and p53 depletion in these cells, rather than increasing the G0/G1 population actually caused modest declines of 17 and 11%, respectively and concomitant increases in the G2/M populations by 85 and 92%, respectively (Figures 3a and 4). Similar results were obtained with M2 shRNA (shown for SK-Mel-103 cells in Supplementary Figure S7). These data suggest that p53 levels could indeed influence the cell cycle consequences of c-Myc depletion in at least some tumor cell lines. Of note was that all c-Myc-depleted cells, irrespective of their p53 levels and wherein the cell cycle arrest occurred, showed total inhibition of proliferation (Figure 3b). This suggested that suppression of p53 is not sufficient to rescue the proliferative deficiencies caused by c-Myc depletion.
Figure 3.
Inhibition of p53 or p21CIP/WAF does not compensate for the depletion of c-Myc. Cells from the indicated lines were infected with lentiviral vectors expressing control − p53 −or p21CIP/WAF-shRNAs as described in Materials and methods. Two days later, cells were superinfected with control or c-Myc shRNA (M1). (a) Cells were harvested 4days after the second infection and examined by western blotting with the antibodies designated on the right. ShRNA′ and shRNA″ designate shRNAs used for primary and secondary infections, respectively. Note a background band in melanocyte cell extracts probed with p53 antibodies closely migrating with the correct band, designated with the arrow. (b) Cells were plated into 12-well plates and counted daily in triplicates in the presence of trypan blue.
Figure 4.
Cell cycle parameters of c-Myc-depleted cells are altered by inhibition of p53 or p21CIP/WAF. Cells from the indicated lines were infected with lentiviral vectors expressing control − p53 − or p21CIP/WAF-shRNAs as described in Materials and methods. Two days later, cells were superinfected with control or Myc shRNA (M1) lentiviral vectors. Cells were collected for cell cycle analysis 4days after the second infection.
One of the major mediators of p53-directed cell cycle arrest is the cyclin-dependent kinase inhibitor (CDKI) p21CIP/WAF (El-Deiry et al., 1993). We therefore assessed its role in mediating G0/G1 growth arrest induced by c-Myc depletion. As shown in Figures 3 and 4, inhibition of p21CIP/WAF in all cell lines resulted in patterns of cell cycle arrest similar to, although less pronounced than those observed in p53-depleted cells.
Taken together, our data suggest that c-Myc is essential for proliferation of all tested normal and tumor cells. However, the rate-limiting function of c-Myc in controlling cell cycle progression is cell line-dependent and is variably reliant upon the status of other cell cycle checkpoint regulators, even within tumors originating from the same cell type.
Discussion
The widespread deregulation of c-Myc in many tumor types together with its role in maintaining the transformed state in certain experimental settings (Arvanitis and Felsher, 2006) has provided the rationale for its consideration as a prime therapeutic target. Indeed, a number of novel strategies have already been employed to inhibit c-Myc expression or function in tumor cells (Hermeking, 2003; Prochownik, 2004; Ponzielli et al., 2005). In several transgenic mouse models of c-Myc-mediated tumorigenesis, abolishing c-Myc overexpression has been shown to lead to complete tumor regression (Felsher and Bishop, 1999; Pelengaris et al., 1999, 2002; Jain et al., 2002). In contrast to these reports, c-Myc appears to be dispensable for the maintenance of mouse mammary gland tumors (Boxer et al., 2004). It is also noteworthy that in all the above mouse models, c-myc was an initiating oncogene leading to transformation, whereas this is unlikely to be the case with the majority of spontaneously arising human cancers. Therefore, the absolute requirement of c-Myc for proliferation of tumor cells in general and human tumor cells in particular still remains an open question.
The studies reported here, performed with a variety of human tumor cell lines, indicate that they are all highly susceptible to Myc-shRNA-mediated growth inhibition. Indeed, some cell lines demonstrated marked growth arrest even when c-Myc ablation was less than complete (70–80% inhibition), thereby suggesting that human cancers could be sensitive to anti-c-Myc therapies that only partially reduce c-Myc levels or function. On the other hand, it was reported that loss of c-Myc did not inhibit proliferation of normal mouse hepatocytes (Baena et al., 2005; Li et al., 2006), keratinocytes (Oskarsson et al., 2006) and intestinal epithelium (Bettess et al., 2005) in vivo. There are several possible explanations for the difference between our data and those of Baena et al. (2005); Oskarsson et al. (2006) and Li et al. (2006). The first one is that in some cases proliferation of c-Myc-depleted cells could continue to depend on the expression of other Myc family members, including N-Myc, which has been shown to functionally compliment c-Myc in in vivo settings (Malynn et al., 2000). The second explanation is that expression of c-Myc downstream targets that are critical for the proliferation of the above cells in mouse could depend on the presence of cytokines or hormones (such as, for example, hedgehog protein for skin development (Athar et al., 2006) that are absent in the media routinely used in tissue culture. Finally, it is possible that proliferation of human tumor cells is more dependent on c-Myc than that of normal mouse cells, a phenomenon known as oncogene ‘addiction’ (Sharma et al., 2006).
The requirement of c-Myc for normal cell cycle progression has been extensively studied in primary or immortalized rodent cells where c-Myc inactivation results in severe or complete inhibition of proliferation and growth arrest predominantly in G0/G1 (Prochownik et al., 1988; Mateyak et al., 1997; de Alboran et al., 2001; Prathapam et al., 2006). Our data obtained in three different types of primary human cells are in good agreement with the majority of the above reports. On the other hand, while also achieving complete proliferative arrest in tumor cells following c-Myc depletion, we observed considerable variability in cell cycle consequences.
Tumor progression is often accompanied by mutations affecting genes whose products control G1/S transition (Hanahan and Weinberg, 2000). Therefore, the mechanisms involved in implementing G0/G1 growth arrest in response to c-Myc depletion present in normal cells could be impaired in tumor cells. Depending on the nature of these defects, c-Myc depletion would affect G1/S transition differently, even in tumors of the same type as shown in the current work. Consequently, due to the involvement of c-Myc in multiple cellular processes (Eisenman, 2001; Patel et al., 2004), its inhibition may result in activation of other cell cycle checkpoints downstream to G1/S, including those involving S (Gottifredi and Prives (2005) or G2/M (Stark and Taylor, 2006).
We demonstrated that interference with levels of p53 and its downstream CDKI target p21WAF/CIP in two melanoma lines (SK-Mel-103 and SK-Mel-147) permits them to transit G1/S and arrest instead in G2/M, without any apparent reversal of their proliferative defect. In contrast, suppression of p53 or p21CIP/WAF in normal melanocytes or in melanoma cells SK-Mel-19 and SK-Mel-29 resulted only in a more marked G0/G1 growth arrest. Accordingly, induction of p53 by c-Myc-depletion was more pronounced in SK-Mel-103 and SK-Mel-147 compared to SK-Mel-19 and SK-Mel-29 cells (Supplementary Figure S6). Interestingly, an observed increase in p53 levels (at least in SK-Mel-103 and SK-Mel-147 cells) is unlikely to be dependent on p14ARF since this gene is deleted in both of these cell lines (Paz et al., 2003). The simplest explanation for the difference in p53 or p21CIP/WAF dependency of G0/G1-growth arrest induced by c-Myc-depletion in studied cells is that, in addition to p53 and p21CIP/WAF, multiple proteins are involved in the implementation of G0/G1 arrest caused by c-Myc inhibition. According to our model, their mutational status and/or expression levels are not altered in normal melanocytes and in melanoma cells SK-Mel-19 and SKMel29 but are altered in SK-Mel-103 and SK-Mel-147 cells.
It is conceivable that G0/G1 arrest caused by c-Myc inhibition could occur via at least two non-mutually exclusive mechanisms. The first could involve deregulated expression of c-Myc-dependent checkpoint genes (CDKs and CDKIs, such as cdk4 and p27KIP1; Bernard and Eilers, 2006; Dang et al., 2006). Alternatively, c-Myc inactivation could lead to changes in the pools of specific cellular metabolites, since c-Myc directly regulates genes whose products play key roles in several metabolic pathways (Dang, 1999). The consequences of these alterations could be growth arrest at different stages of the cell cycle depending on the status of other key cell cycle regulators. In accord with the latter suggestion are the findings that depletion of the pyrimidine nucleotide pool in normal fibroblasts, achieved by chemical inhibition of the direct c-Myc target CAD (Carbamoyl-phosphate synthetase 2, Aspartate transcarbamylase, Dihydroorotase), results in G0/G1 growth arrest. Interestingly, similar inhibition of CAD in p53-null fibroblasts also results in growth arrest, although in S-phase rather than G0/G1 (Agarwal et al., 1998).
In conclusion, our findings regarding the absolute reliance of tumor cells on endogenous levels of c-Myc could be important when considering therapeutic approaches that aim to target this oncoprotein. The considerable differences in the cell cycle responses to c-Myc depletion noted here also point to yet additional heterogeneity at the level of other cell cycle regulators, even within tumors arising from the same tissues.
Materials and methods
Tumor cell lines
A list of the tumor cell lines used is provided in Table 1. Cell lines were originally obtained from the ATCC or from Memorial Sloan Kettering Cancer Center. Cells were cultured in either RPMI-1640 or Dulbecco’s modified Eagle’s essential minimal medium as recommended by the supplier. Supplements included fetal calf serum (10–20%), 2 mm glutamine and 100 U ml−1 penicillin G + 100 µg ml−1 streptomycin. All cell culture agents were purchased from Invitrogen Inc. (Carlsbad, CA, USA). Primary fibroblasts, melanocytes and keratinocytes were obtained from neonatal foreskin and maintained as described before (Fernandez et al., 2005; Denoyelle et al., 2006).
Plasmids and transfections
The lentiviral vector containing shRNA specific for p53 was described before (Boiko et al., 2006). The lentiviral vectors containing an shRNA specific for human p21CIP/WAF and for human c-Myc were purchased from Sigma. The lentiviral construct H1 (Ivanova et al., 2006) was used to express shRNAs directed against human c-Myc. The c-Myc targeting sequences consisted of nt. 1567–1585 (M1), 1341–1359 (M2) and 1919–1937 (M3) of the previously published sequence (GenBank Accession no. NM_002467.3). A control lentiviral vector contained an irrelevant sequence. Lentiviral packaging reactions were performed in the 293-FT cell line in the presence of packaging plasmids pVSV-G and pΔDR (Ivanova et al., 2006) using Superfect Transfection Reagent (Qiagen Inc., Chatsworth, CA). Viral supernatants were collected 48 h after transfection, filtered through disposable 0.45 µm cellulose acetate filters (VWR Scientific Inc. West Chester, PA, USA) and frozen in individual aliquots at −80 °C. For infection cells were plated in 60 or 100 mm tissue culture dishes (VWR) and allowed to achieve 40–50% confluence before adding viral supernatant in the presence of 8 µg ml−1 polybrene for 24h (Sigma, St Louis, MO, USA). Cells infected with H1 lentiviral vector expressing different shRNAs were examined by fluorescence microscopy for the expression of EGFP in 48 h. Cells were then split and cultured for an additional 2–4 days after infection before being utilized for further studies.
Immunoblotting, immunohistochemistry and flow cytometry
Total cell lysates were prepared and immunoblotting was performed as previously described using 50–80 µg of total protein (48). The following antibodies were used: 9E10 and C33 monoclonal antibody (mAb) for human c-Myc (SC-40, Santa Cruz Biotechnology, Inc., Santa Cruz, CA, USA), p53 (VP-P955, Vector Laboratories Ltd, Peterborough, UK) and p21CIP/WAF (SC-397), H2AX-Ser139-phospho-specific antibodies (Millipore Corporation, Billerica, MA, USA), D-10 mAb against tubulin (Santa Cruz Biotechnology). Membranes were developed using enhanced chemiluminescence kit (Super-Signal West Femto Maximum Sensitivity Substrate, Pierce, Rockland, IL, USA) or using alkaline phosphatase secondary antibodies and detection/quantification of the signal was performed using ImageJ 1.38 software or STORM Phosphor-Imager and ImageQuant 2.0 Program as a part of ImagePro Analysis Software package. Background was calculated from an equivalent area in each lane and subtracted from the value for c-Myc in that lane. Propidium iodide staining, flow cytometry and statistical evaluation of cell cycle data were performed as previously described (Nikiforov et al., 2002; Yin et al., 2002).
Supplementary Material
Acknowledgements
We are grateful to Dr Gary Fisher, Dr Andrzej Dlugosz and Dr Laure Rittie for critical reading of the manuscript. We thank John Lazo, David Hackham and Yatin Vyas for cell lines, Jie Lu for DNA sequencing, Josh Solomon for assistance in the initial optimization of lentiviral infection protocols and MaryBeth Ribblet and Suresh Patil for technical assistance. This work was supported by Dermatology Research Foundation Carrier Development Award to MAN, NIH grants R01- CA120244-A1 to MAN, RO1-CA105033, RO1-CA078259 to EVP and R01-CA107237 to MSS. MAN is a Melanoma Research Foundation Scholar. HW was supported by a postdoctoral fellowship award from the Research Advisory Committee of Children’s Hospital of Pittsburgh.
Footnotes
Supplementary Information accompanies the paper on the Oncogene website (http://www.nature.com/onc).
References
- Adhikary S, Eilers M. Transcriptional regulation and transformation by Myc proteins. Nat Rev Mol Cell Biol. 2005;6:635–645. doi: 10.1038/nrm1703. [DOI] [PubMed] [Google Scholar]
- Agarwal ML, Agarwal A, Taylor WR, Chernova O, Shama Y, Stark GR. A p53-dependent S-phase checkpoint helps to protect cells from DNA damage in response to starvation for pyrimidine nucleotides. Proc Natl Acad Sci USA. 1998;95:14775–14780. doi: 10.1073/pnas.95.25.14775. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Arvanitis C, Felsher DW. Conditional transgenic models define how MYC initiates and maintains tumorigenesis. Semin Cancer Biol. 2006;16:313–317. doi: 10.1016/j.semcancer.2006.07.012. [DOI] [PubMed] [Google Scholar]
- Athar M, Tang X, Lee JL, Kopelovich L, Kim AL. Hedgehog signalling in skin development and cancer. Exp Dermatol. 2006;15:667–677. doi: 10.1111/j.1600-0625.2006.00473.x. [DOI] [PubMed] [Google Scholar]
- Baena E, Gandarillas A, Vallespinos M, Zanet J, Bachs O, Redondo C, et al. c-Myc regulates cell size and ploidy but is not essential for postnatal proliferation in live. Proc Natl Acad Sci USA. 2005;102:7286–7291. doi: 10.1073/pnas.0409260102. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bernard S, Eilers M. Control of cell proliferation and growth by Myc proteins. Results Probl Cell Differ. 2006;42:329–342. doi: 10.1007/400_004. [DOI] [PubMed] [Google Scholar]
- Bettess MD, Dubois N, Murphy MJ, Dubey C, Roger C, Robine S, et al. c-Myc is required for the formation of intestinal crypts but dispensable for homeostasis of the adult intestinal epithelium. Mol Cell Biol. 2005;25:7868–7878. doi: 10.1128/MCB.25.17.7868-7878.2005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Boiko AD, Porteous S, Razorenova OV, Krivokrysenko VI, Williams BR, Gudkov AV, et al. A systematic search for downstream mediators of tumor suppressor function of p53 reveals a major role of BTG2 in suppression of Ras-induced transformation. Genes Dev. 2006;20:236–252. doi: 10.1101/gad.1372606. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Boxer RB, Jang JW, Sintasath L, Chodosh LA. Lack of sustained regression of c-MYC-induced mammary adenocarcinomas following brief or prolonged MYC inactivation. Cancer Cell. 2004;6:577–586. doi: 10.1016/j.ccr.2004.10.013. [DOI] [PubMed] [Google Scholar]
- Chen JP, Lin C, Xu CP, Zhang XY, Fu M, Deng YP. Molecular therapy with recombinant antisense c-myc adenovirus for human gastric carcinoma cells in vitro and in vivo. J Gastroenterol Hepatol. 2001;6:22–28. doi: 10.1046/j.1440-1746.2001.02361.x. [DOI] [PubMed] [Google Scholar]
- Citro G, D’Agnano I, Leonetti C, Perini R, Bucci B, Zon G, et al. c-myc antisense oligodeoxynucleotides enhance the efficacy of cisplatin in melanoma chemotherapy in vitro and in nude mice. Cancer Res. 1998;58:283–289. [PubMed] [Google Scholar]
- D’Agnano I, Valentini A, Fornari C, Bucci B, Starace G, Felsani A, et al. Myc down-regulation induces apoptosis in M14melanoma cells by increasing p27(kip1) levels. Oncogene. 2001;20:2814–2825. doi: 10.1038/sj.onc.1204392. [DOI] [PubMed] [Google Scholar]
- Dang CV. c-Myc target genes involved in cell growth, apoptosis, and metabolism. Mol Cell Biol. 1999;19:1–11. doi: 10.1128/mcb.19.1.1. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dang CV, O’Donnell KA, Zeller KI, Nguyen T, Osthus RC, Li F. The c-Myc target gene network. Semin Cancer Biol. 2006;16:253–264. doi: 10.1016/j.semcancer.2006.07.014. [DOI] [PubMed] [Google Scholar]
- de Alboran IM, O’Hagan RC, Gartner F, Malynn B, Davidson L, Rickert R, et al. Analysis of C-MYC function in normal cells via conditional gene-targeted mutation. Immunity. 2001;14:45–55. doi: 10.1016/s1074-7613(01)00088-7. [DOI] [PubMed] [Google Scholar]
- Denoyelle C, Abou-Rjaily G, Bezrookove V, Verhaegen M, Johnson TM, Fullen DR, et al. Anti-oncogenic role of the endoplasmic reticulum differentially activated by mutations in the MAPK pathway. Nat Cell Biol. 2006;8:1053–1063. doi: 10.1038/ncb1471. [DOI] [PubMed] [Google Scholar]
- Eisenman RN. Deconstructing myc. Genes Dev. 2001;15:2023–2030. doi: 10.1101/gad928101. [DOI] [PubMed] [Google Scholar]
- El-Deiry WS, Tokino T, Velculescu VE, Levy DB, Parsons R, Trent JM, et al. WAF1, a potential mediator of p53 tumor suppression. Cell. 1993;75:817–825. doi: 10.1016/0092-8674(93)90500-p. [DOI] [PubMed] [Google Scholar]
- Felsher DW, Bishop JM. Reversible tumorigenesis by MYC in hematopoietic lineages. Mol Cell. 1999;4:199–207. doi: 10.1016/s1097-2765(00)80367-6. [DOI] [PubMed] [Google Scholar]
- Fernandez Y, Verhaegen M, Miller TP, Rush JL, Steiner P, Opipari AW, et al. Differential regulation of noxa in normal melanocytes and melanoma cells by proteasome inhibition: therapeutic implications. Cancer Res. 2005;65:6294–6304. doi: 10.1158/0008-5472.CAN-05-0686. [DOI] [PubMed] [Google Scholar]
- Grandori C, Cowley SM, James LP, Eisenman RN. The Myc/Max/Mad network and the transcriptional control of cell behavior. Annu Rev Cell Dev Biol. 2000;16:653–659. doi: 10.1146/annurev.cellbio.16.1.653. [DOI] [PubMed] [Google Scholar]
- Gottifredi V, Prives C. The S phase checkpoint: when the crowd meets at the fork. Semin Cell Dev Biol. 2005;16:355–368. doi: 10.1016/j.semcdb.2005.02.011. [DOI] [PubMed] [Google Scholar]
- Hahn WC, Weinberg RA. Rules for making human tumor cells. N Engl J Med. 2002;347:1593–1603. doi: 10.1056/NEJMra021902. [DOI] [PubMed] [Google Scholar]
- Hanahan D, Weinberg RA. The hallmarks of cancer. Cell. 2000;100:57–70. doi: 10.1016/s0092-8674(00)81683-9. [DOI] [PubMed] [Google Scholar]
- Hermeking H, Rago C, Schuhmacher M, Li Q, Barrett JF, Obaya AJ, et al. Identification of CDK4as a target of c-Myc. Proc Natl Acad Sci USA. 2000;97:2229–2234. doi: 10.1073/pnas.050586197. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hermeking H. The MYC oncogene as a cancer drug target. Curr Cancer Drug Targets. 2003;3:163–175. doi: 10.2174/1568009033481949. [DOI] [PubMed] [Google Scholar]
- Ivanova N, Dobrin R, Lu R, Kotenko I, Levorse J, DeCoste C, et al. Dissecting self-renewal in stem cells with RNA interference. Nature. 2006;442:533–538. doi: 10.1038/nature04915. [DOI] [PubMed] [Google Scholar]
- Jain M, Arvanitis C, Chu K, Dewey W, Leonhardt E, Trinh M, et al. Sustained loss of a neoplastic phenotype by brief inactivation of MYC. Science. 2002;297:102–104. doi: 10.1126/science.1071489. [DOI] [PubMed] [Google Scholar]
- Li F, Xiang Y, Potter J, Dinavahi R, Dang CV, Lee LA. Conditional deletion of c-myc does not impair liver regeneration. Cancer Res. 2006;66:5608–5612. doi: 10.1158/0008-5472.CAN-05-4242. [DOI] [PubMed] [Google Scholar]
- Lutz W, Leon J, Eilers M. Contributions of Myc to tumorigenesis. Biochim Biophys Acta. 2002;1602:61–71. doi: 10.1016/s0304-419x(02)00036-7. [DOI] [PubMed] [Google Scholar]
- Malynn BA, de Alboran IM, O’Hagan RC, Bronson R, Davidson L, DePinho RA, et al. N-myc can functionally replace c-myc in murine development, cellular growth, and differentiation. Genes Dev. 2000;14:1390–1399. [PMC free article] [PubMed] [Google Scholar]
- Mateyak MK, Obaya AJ, Adachi S, Sedivy JM. Phenotypes of c-Myc-deficient rat fibroblasts isolated by targeted homologous recombination. Cell Growth Differ. 1997;8:1039–1048. [PubMed] [Google Scholar]
- McGuffie EM, Pacheco D, Carbone GM, Catapano CV. Antigene and antiproliferative effects of a c-myc-targeting phosphorothioate triple helix-forming oligonucleotide in human leukemia cells. Cancer Res. 2000;60:3790–3799. [PubMed] [Google Scholar]
- Meyer N, Kim SS, Penn LZ. The Oscar-worthy role of Myc in apoptosis. Semin Cancer Biol. 2006;16:275–277. doi: 10.1016/j.semcancer.2006.07.011. [DOI] [PubMed] [Google Scholar]
- Nesbit CE, Tersak JM, Prochownik EV. Myc oncogenes and human neoplastic disease. Oncogene. 1999;18:3004–3016. doi: 10.1038/sj.onc.1202746. [DOI] [PubMed] [Google Scholar]
- Nikiforov MA, Chandriani S, O’Connell B, Petrenko O, Kotenko I, Beavis A, et al. A functional screen for Myc-responsive genes reveals serine hydroxymethyltransferase, a major source of the one-carbon unit for cell metabolism. Mol Cell Biol. 2002;22:5793–5800. doi: 10.1128/MCB.22.16.5793-5800.2002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Oskarsson T, Essers MA, Dubois N, Offner S, Dubey C, Roger C, et al. Skin epidermis lacking the c-Myc gene is resistant to Ras-driven tumorigenesis but can reacquire sensitivity upon additional loss of the p21Cip1 gene. Genes Dev. 2006;20:2024–2029. doi: 10.1101/gad.381206. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Patel JH, Loboda AP, Showe MK, Showe LC, McMahon SB. Analysis of genomic targets reveals complex functions of MYC. Nat Rev Cancer. 2004;4:562–568. doi: 10.1038/nrc1393. [DOI] [PubMed] [Google Scholar]
- Paz MF, Fraga MF, Avila S, Guo M, Pollan M, Herman JG, et al. A systematic profile of DNA methylation in human cancer cell lines. Cancer Res. 2003;63:1114–1121. [PubMed] [Google Scholar]
- Pelengaris S, Littlewood T, Khan M, Elia G, Evan G. Reversible activation of c-Myc in skin: induction of a complex neoplastic phenotype by a single oncogenic lesion. Mol Cell. 1999;3:565–577. doi: 10.1016/s1097-2765(00)80350-0. [DOI] [PubMed] [Google Scholar]
- Pelengaris S, Khan M, Evan GI. Suppression of Myc-induced apoptosis in beta cells exposes multiple oncogenic properties of Myc and triggers carcinogenic progression. Cell. 2002;109:321–334. doi: 10.1016/s0092-8674(02)00738-9. [DOI] [PubMed] [Google Scholar]
- Ponzielli R, Katz S, Barsyte-Lovejoy D, Penn LZ. Cancer therapeutics: targeting the dark side of Myc. Eur J Cancer. 2005;41:2485–2501. doi: 10.1016/j.ejca.2005.08.017. [DOI] [PubMed] [Google Scholar]
- Prathapam T, Tegen S, Oskarsson T, Trumpp A, Martin GS. Activated Src abrogates the Myc requirement for the G0/G1 transition but not for the G1/S transition. Proc Natl Acad Sci USA. 2006;103:2695–2700. doi: 10.1073/pnas.0511186103. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Prochownik EV, Kukowska J, Rodgers C. c-myc antisense transcripts accelerate differentiation and inhibit G1 progression in murine erythroleukemia cells. Mol Cell Biol. 1988;8:3683–3695. doi: 10.1128/mcb.8.9.3683. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Prochownik EV. c-Myc as a therapeutic target in cancer. Expert Rev Anticancer Ther. 2004;4:289–302. doi: 10.1586/14737140.4.2.289. [DOI] [PubMed] [Google Scholar]
- Rothermund K, Rogulski K, Fernandes E, Whiting A, Sedivy J, Pu L, et al. c-Myc-independent restoration of multiple phenotypes by two C-Myc target genes with overlapping functions. Cancer Res. 2005;65:2097–2107. doi: 10.1158/0008-5472.CAN-04-2928. [DOI] [PubMed] [Google Scholar]
- Sharma SV, Fischbach MA, Haber DA, Settleman J. ‘Oncogenic shock’: explaining oncogene addiction through differential signal attenuation. Clin Cancer Res. 2006;12:4392s–4395s. doi: 10.1158/1078-0432.CCR-06-0096. [DOI] [PubMed] [Google Scholar]
- Skorski T, Nieborowska-Skorska M, Campbell K, Iozzo RV, Zon G, Darzynkiewicz Z, et al. Leukemia treatment in severe combined immunodeficiency mice by antisense oligodeoxynucleotides targeting cooperating oncogenes. J Exp Med. 1995;182:1645–1653. doi: 10.1084/jem.182.6.1645. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Soengas MS, Capodieci P, Polsky D, Mora J, Esteller M, Opitz-Araya X, et al. Inactivation of the apoptosis effector Apaf-1 in malignant melanoma. Nature. 2001;409:207–211. doi: 10.1038/35051606. [DOI] [PubMed] [Google Scholar]
- Stark GR, Taylor WR. Control of the G2/M transition. Mol Biotechnol. 2006;32:227–248. doi: 10.1385/MB:32:3:227. [DOI] [PubMed] [Google Scholar]
- Trumpp A, Refaeli Y, Oskarsson T, Gasser S, Murphy M, Martin GR, et al. c-Myc regulates mammalian body size by controlling cell number but not cell size. Nature. 2001;414:768–773. doi: 10.1038/414768a. [DOI] [PubMed] [Google Scholar]
- Vita M, Henriksson M. The Myc oncoprotein as a therapeutic target for human cancer. Semin Cancer Biol. 2006;16:318–330. doi: 10.1016/j.semcancer.2006.07.015. [DOI] [PubMed] [Google Scholar]
- Yin X, Grove L, Rogulski K, Prochownik EV. Myc target in myeloid cells-1, a novel c-Myc target, recapitulates multiple c-Myc phenotypes. J Biol Chem. 2002;277:19998–20010. doi: 10.1074/jbc.M200860200. [DOI] [PubMed] [Google Scholar]
- Zhou ZQ, Hurlin PJ. The interplay between Mad and Myc in proliferation and differentiation. Trends Cell Biol. 2001;11:S10–S14. doi: 10.1016/s0962-8924(01)02121-3. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.