Summary
The zinc finger domain transcription factor prdm1a plays an integral role in the development of the neural plate border cell fates, including neural crest cells and Rohon-Beard (RB) sensory neurons. However, the mechanisms underlying prdm1a function cell fate specification is unknown. Here, we test more directly how prdm1a functions in this cell fate decision. Rather than affecting cell death or proliferation at the neural plate border, prdm1a acts explicitly on cell fate specification by counteracting olig4 expression in the neighboring interneuron domain. olig4 expression is expanded in prdm1a mutants and olig4 knockdown can rescue the reduced or abrogated neural crest and RB neuron phenotype in prdm1a mutants, suggesting a permissive role for prdm1a in neural plate border-derived cell fates. In addition, prdm1a expression is upregulated in the absence of Notch function, and inhibiting Notch signaling fails to rescue prdm1a mutants. This suggests that prdm1a functions downstream of Notch in the regulation of cell fate at the neural plate border and that Notch regulates the total number of progenitor cells at the neural plate border.
Keywords: Blimp1, neural crest, Rohon-Beard sensory neurons HD050698 to K.B.A
Introduction
The development of the nervous system involves a complex series of inductive interactions followed by the combinatory action of transcription factors to specify explicit cell fates. The embryonic neural plate is induced from the ectoderm in the presence of low levels of BMP signaling with opposing action from Shh required for ventral neural tube patterning. The neural plate border (NPB), which lies between the neural and non-neural ectoderm, requires an intermediate level of BMP signaling for its formation (Ahrens and Schlosser, 2005; Mancilla and Mayor, 1996; Rossi et al., 2008). Rohon-Beard sensory neurons, neural crest cells (NCCs), and placodal cell populations form at this junction in a medial to lateral orientation, respectively. Rohon-Beard (RB) sensory neurons are primary sensory neurons localized in the dorsal spinal cord that mediate proprioceptive mechanosensory information and are required for the touch response in zebrafish and Xenopus embryos (Lamborghini, 1980). Neural crest cells are a transient embryonic cell population that give rise to neurons and glia of the peripheral nervous system, melanocytes, and cartilage of the face. Placodal populations form at the lateralmost edge of the NPB giving rise to neurons, and along with neural crest cells, contribute to the cranial ganglia (Ahrens and Schlosser, 2005; Schlosser, 2006). NPB cell fate specification requires the combinatory action of many transcription factors including prdm1a, zic genes, pax3/7, msx1/2, and dlx family members (reviewed in Meulemans and Bronner-Fraser, 2004). Further, specification of the different types of cells that form at the NPB requires action of a different set of transcription factors, including foxd3, snail1, slug (snail2), twist, ap-2s, and sox9/10 for neural crest cells (Meulemans and Bronner-Fraser, 2004); dlx genes, msx genes, neurog1, neuroD and islet1 for RB neurons (Rossi et al., 2009); and dlx genes, six1, eya1 and pax genes for placodal cells (Schlosser et al., 2008). Expression analysis of these factors suggests that early in development, there is significant overlap between the NPB and placodal domain (marked by dlx3b), while there is little to no overlap between the NPB and the neural plate itself. Previous lineage tracing data suggests that neural crest cells and RB sensory neurons arise within the same domain at the NPB, while placodal cells arise from a domain lateral to the NPB (Cornell and Eisen, 2002; Schlosser, 2006).
In addition to BMP signaling, Delta/Notch signaling also regulates the specification of neural crest and RB neuron precursors. Notch receptors are expressed in a broad domain within the neural plate and non-neural ectoderm, while the delta ligand genes are expressed in primary neuron domains (Appel and Eisen, 1998; Hsiao et al., 2007). In the absence of Notch signaling, as occurs in the zebrafish mind bomb (mib) and deltaA mutants, there is an excess of all primary neurons, including RB sensory neurons and interneurons, and a concomitant reduction in the number of trunk neural crest cells (Cornell and Eisen, 2000; Itoh et al., 2003; Jiang et al., 1996). In Xenopus, overexpression of the Notch intracellular domain (NICD), which constitutively activates Notch signaling, at the end of gastrulation increases the expression of the neural crest markers Xslug and foxd3, suggesting that Notch signaling is required for NCC induction (Glavic et al., 2004). Blocking Notch with DAPT, a gamma-secretase inhibitor, produces a neurogenic phenotype with an increase in the number of primary neurons (Geling et al., 2002). These findings suggest that lateral inhibition between neurons and NCCs is perturbed such that neuronal fate is promoted at the expense of neural crest fate, at least in the trunk region (Cornell, 2005). Notch signaling is also required for neurogenesis within the neural plate itself. As the neural plate domain eventually folds into the neural tube, Notch is required in the neural epithelium for V2 and dIL interneuron populations (Batista et al., 2008; Mizuguchi et al., 2006).
Prdm1a is a PR/SET domain, zinc finger domain transcription factor that functions as a master cell fate regulator in many cell types. Prdm1a (Blimp-1 in mouse) has been implicated in differentiation of plasma cells from B-cells (Shaffer et al., 2002; Shapiro-Shelef et al., 2003) by regulating cell proliferation (Lin et al., 2002; Lin et al., 1997). A conditional knockout of Blimp-1 in mouse using Sox2-cre demonstrates that prdm1a is required for the development of the posterior forelimb, caudal pharyngeal arches, secondary heart field, and sensory vibrissae (Robertson et al, 2007). In zebrafish, prdm1a functions at multiple stages during development, including gastrulation, formation of head structures and fin development (Mercader et al., 2006; Wilm and Solnica-Krezel, 2005), and in a Hedgehog-regulated switch between slow twitch and fast twitch muscle development (Baxendale et al., 2004) (Liew et al., 2008; von Hofsten et al., 2008). In addition to these roles, prdm1a is required for neural crest and RB neuron cell fate. prdm1a mutant embryos have a decreased number of trunk neural crest cells, a complete loss of RB sensory neurons (Artinger et al., 1999; Roy and Ng, 2004)(Hernandez-Lagunas et al., 2005) and a loss of neural crest-derived ceratobranchial cartilage within the craniofacial skeleton (Birkholz et al., 2009). prdm1a is expressed at midgastrulation in the NPB and continues to be expressed until the 6-somite stage when expression is downregulated, but remains (Hernandez-Lagunas et al., 2005)(Wilm and Solnica-Krezel, 2005)(Birkholz et al., 2009).
olig4 (Olig3 in mouse) is a member of the basic helix-loop-helix family of transcription factors expressed within the interneuron domain of primary neurons, and has been shown to regulate interneuron fate in both mouse and zebrafish. Olig3 is expressed in the p1-p3 dorsal interneuron domain of the mammalian spinal cord and mutations in Olig3 eliminate p2-3 interneurons and severely reduce the d1 population (Muller et al., 2005). Interestingly, in contrast to olig4 mutants in zebrafish, neural crest cells are unaffected in these mouse mutants. Knock down of olig4 in zebrafish results in a loss of interneurons and expansion of the neighboring neural crest and RB neuron domain (Filippi et al., 2005)(Tiso et al., 2009), while overexpression reduces neural crest cell number. These data suggest that olig4 normally acts as a negative regulator of NPB cell fates. Interestingly, olig4 has been shown to be downstream of both BMP and Wnt signaling in the specification of dorsal interneurons (Filippi et al., 2005; Zechner et al., 2007). In addition, in zebrafish olig4 is downstream of Notch, as knockdown of olig4 in Notch-deficient embryos rescues the loss of neural crest cells. This suggests that olig4 expression must be reduced for Notch signaling to specify cell fate (Filippi et al., 2005).
Here, we have further dissected the function of prdm1a and olig4 in NPB cell fate decisions. We show that prdm1a acts to regulate cell fate at the NPB by counteracting olig4 expression, but not by regulating cell death or proliferation. In addition, prdm1a expression is upregulated in the absence of Notch function, and we are not able to rescue prdm1a mutants by blocking Notch with DAPT. This suggests that prdm1a functions downstream of Notch in the regulation of cell fate at the neural plate border.
Materials and Methods
Animals
The zebrafish were maintained according to Westerfield (1993) and staged by hours post fertilization (hpf) and morphology according to Kimmel (1995). The zebrafish prdm1am805, deltaA, and mind bomb mutants have been described previously (Artinger et al., 1999)(Hernandez-Lagunas et al., 2005)(Rossi et al., 2009)(Birkholz et al., 2009)(Itoh et al., 2003). Single embryo phenotyping and genotyping in prdm1a clutches was performed for cell death, proliferation and rescue experiments as previously described (Olesnicky et al, 2010)(Rossi et al., 2009). For deltaA genotyping, we used primers and protocols provided by ZIRC, mibm132 genenotyping is as described in (Itoh et al., 2003).
Embryo manipulation and analysis
Whole-mount in situ hybridization was adapted from Thisse and Thisse (1998). Fluorescent ISH was performed using the protocol described in (Pineda et al., 2006), in which a DIG-conjugated probe was developed using a fast red kit (Sigma F4648) and a fluorescein-conjugated probe was developed using a TSA kit (Perkin Elmer NEL741). Immunohistochemistry was performed as described (Ungos et al., 2003) and the following antibodies were used: HNK-1 antibody (Sigma) at a 1:1000 dilution; islet1/2 (39.4D5) at 1:200 (Developmental Studies Hybridoma Bank); anti-phosphohistone-H3 (Upstate) at 1:500; and Alexa568 goat anti-mouse at 1:750. Confocal microscopy was performed on Leitz TCS SP5 II laser scanning confocal using LAS AF software. Apoptosis was determined by TUNEL labeling using fluorescein-dUTP (TMR-Red, Roche). Total cells expressing pH3 and TUNEL were counted and compared to wildtype. 6-10 ng of prdm1a Morpholino (add sequence) was injected into the 1 cell stage for knockdown and rescue as previously described (Hernandez-Lagunas et al., 2005). 6-10ng of olig4 Morpholino (MO) was injected into 1cell stage embryos as described in (Filippi et al., 2005). The standard control MO 5′-CCT CTT ACC TCA GTT ACA ATT TAT A 3′ was injected at 10ng. At least three experiments in separate clutches were done for each experimental condition.
Analysis of prdm1a mutant rescue with olig4 MO injections was done as follows: Neural crest cell rescue was defined as the presence of NCC in 7 or more somites, since prdm1a mutants rarely have NCC in that number of somites. RB sensory neurons were counted across 20 segments at both 24 hpf in the tg[neurog1∷gfp] line or at 48 hpf using HNK-1 immunohistochemistry. Pigment cells were scored as rescued if there was pigment on the yolk (which prdm1a mutants rarely have) and an increase in pigment cells on the dorsal aspect of the embryo.
DAPT treatments were performed on wildtype or prdm1a-/- or morphant embryos. 100μM, 200μM, or 1% DMSO in embryo media was applied to embryos at 60% epiboly stage with holes poked in the chorions. Embryos were fixed at the tailbud to 2 somite stage in 4% PFA at 4°C overnight. Chorions were then fully removed and embryos were dehydrated in MeOH.
Results
prdm1a expression overlaps with pax3 but not olig4 at the neural plate border
To investigate the relationship between prdm1a and other cell fate regulators at the NPB, we performed double fluorescent in situ hybridization with markers of the NPB and neural tissue between 90% epiboly and the 2 somite stage. Previous work has shown that prdm1a overlaps extensively with the non-neural ectoderm marker dlx3b at gastrulation stages and 90% epiboly, but not with the neural markers sox19a or sox3. Expression of prdm1a and dlx3b further resolves into individual domains beginning at tailbud stage (Rossi et al., 2009). pax3 and pax7 act to specify the NPB and are required for NC development in other vertebrates (Basch et al., 2006). We examined the expression of the NPB marker pax3 and the interneuron marker olig4 in comparison to prdm1a. At 90% epiboly through tailbud, we observe overlap of expression of prdm1a with the NPB marker pax3 in the anterior region of the embryo. By the 2-somite stage, expression of prdm1a and pax3 resolves into separate domains (Figure 1A-C). pax3 expression is not altered in prdm1a mutant embryos (see Figure 3), indicating that prdm1a does not regulate pax3 expression. This observation is important as it allows us to utilize pax3 in other experiments to identify the NPB domain in prdm1a mutant embryos.
Figure 1. Double fluorescent in situ hybridization of prdm1a with pax3 and olig4.
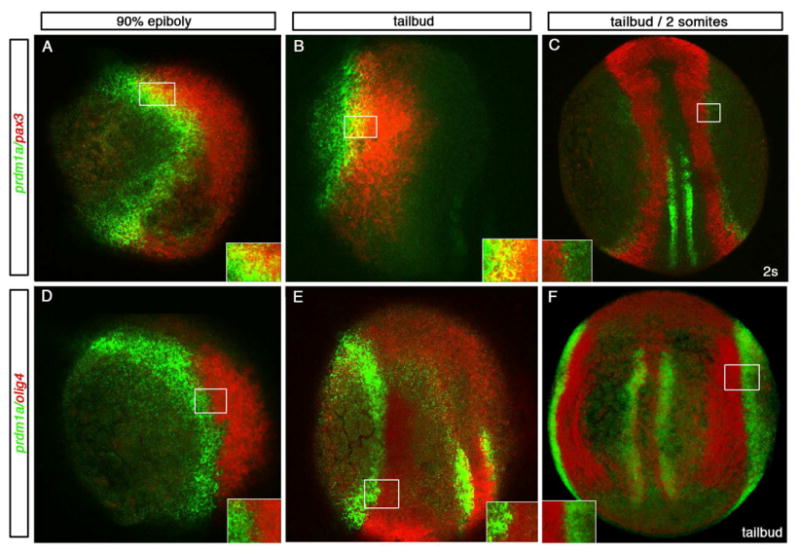
Dorsal and lateral views of 90% epiboly, tailbud and 2 somite stage (9 hpf-11 hpf) confocal micrographs in a single z-stack unless otherwise noted. Insets show higher resolution confocal images for each boxed area. (A-C) prdm1a in green and pax3 in red and (D-F) prdm1a in green and olig4 in red. A lateral view, dorsal to the right, of an embryo at the end of gastrulation (90% epiboly) and lateral view at tailbud exhibit overlap between prdm1a and pax3 (A, B; yellow). By 11hpf, dorsal view of a 2 somite stage embryo, projected image shows the domains are distinct (C). Even at the earliest stages examined, olig4 is distinct from prdm1a. Lateral view of 90% epiboly and dorsal lateral view of a tailbud stage embryo show no overlap in expression (D, E). At tailbud stages shown in a dorsal projected view (F), no overlap is seen in the dorsal domain or the medial adaxial domain.
Figure 3. Quantification of cell proliferation in prdm1a morphants.
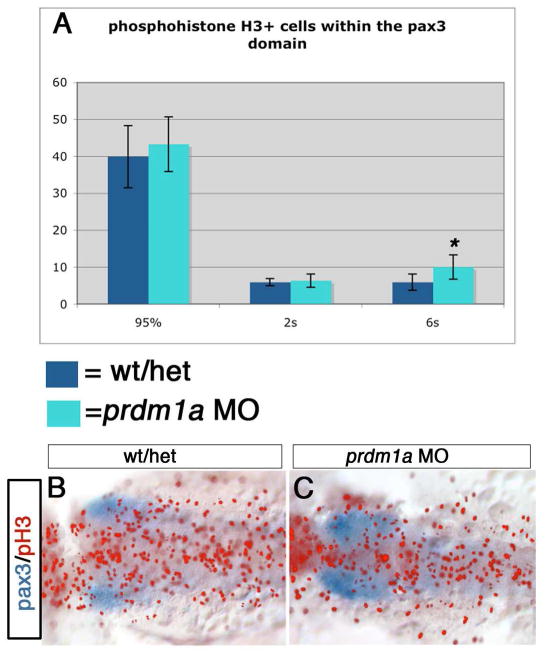
(A) Quantification of the pH3-expressing cells in wildtype and prdm1a morphant embryos at 95%, 2 somite stage and 6 somite stage: morphants in green, wildtype in blue. There is a significant increase in cell proliferation at the 6 somite stage, average of 5.9 pH3+ cells in wildtype (n=8) vs 10.0 in prdm1a mutants, n=10; p<0.003. (B, C) Dorsal view of pax3 expression in blue and pH3 pseudo-colored in red at the 6 somite stage in wildtype and prdm1a morphant embryos.
olig4 (previously named olig3) is expressed within the intermediate domain of primary neurons, and has been shown to regulate interneuron fate. Embryos in which Olig4 has been knocked down exhibit a loss of interneurons and expansion of the neighboring NC and RB neuron domain (Filippi et al., 2005)(Tiso et al., 2009). Since prdm1a is required in the neighboring neural crest and RB neuron domain, we sought to understand the relationship between these two transcription factors. To determine whether prdm1a expression overlaps with the interneuron domain, we examined the expression of olig4 and prdm1a using double fluorescent in situ hybridization. Previous work has shown no overlap in the expression of prdm1a with pan neural plate markers (Rossi et al., 2009). Consistently, prdm1a did not overlap with olig4 at any stage examined (Figure1D-F). Therefore, we conclude that prdm1a is expressed within the NPB but not within the neural plate/interneuron domain.
The olig4 domain is expanded in prdm1a mutant embryos
olig4 morphants have a phenotype opposite to that of prdm1a mutant embryos, exhibiting greater numbers of NCC and RB sensory neurons at the expense of interneurons, suggesting that both genes are required for establishing distinct cell fates within the NPB (Filippi et al., 2005). To assay the fate of cells after prdm1a loss, we examined the expression of both the non-neural ectoderm marker dlx3 and interneuron domain marker olig4 in prdm1a mutant embryos and prdm1a morphant embryos at 95% epiboly and tailbud stage. In wildtype embryos at both stages, a 2-4 cell gap exists between the olig4 and dlx3b expression domains, corresponding to the prdm1a expression domain (Figure 2A, D). Upon loss of prdm1a in the morphant or mutant embryos, this gap between olig4 and dlx3b expression domains is lost (Figure 2B, C, E, F, arrows as compared to wildtype). These data suggest that prdm1a normally acts to repress either dlx3b or olig4 to maintain a zone of cells fated to become NC and RB sensory neurons. Previous studies have shown that dlx3b expression is reduced and not expanded in the posterior domain (Artinger et al., 1999; Rossi et al., 2009). This suggests that prdm1a might normally repress olig4 expression and that olig4 expression may expand into the prdm1a domain in prdm1a mutant embryos. To quantify this increase, we examined olig4 expression at 2-somite stage in flat mounted embryos and counted the number of olig4 expressing cells along the anterior-posterior axis (Figure 2I). We observed a 2-3 cell increase in the anterior region and a slight increase in the posterior domain of olig4 expression in the prdm1a morphant embryos compared to wild type controls. Using confocal imaging to optically section in the Z-axis, we additionally show that this change in the olig4 expression domain is a change in the medial lateral domain of the neural plate and not an expansion into deeper cell layers (Figure 2J, K). Together, these data indicate that olig4 expands into the prdm1a domain in the absence of prdm1a function (Figure 2H; 2 somite uninjected, n=6, prdm1a MO, n=8). However, at earlier tailbud stages, we did not observe an increase in the size of the olig4 domain in prdm1a mutants, suggesting that while the initiation of olig4 expression is normal in prdm1a mutant embryos, the subsequent spatial refinement of olig4 expression requires Prdm1a function.
Figure 2. Loss of prdm1a results in expansion of the olig4 domain.
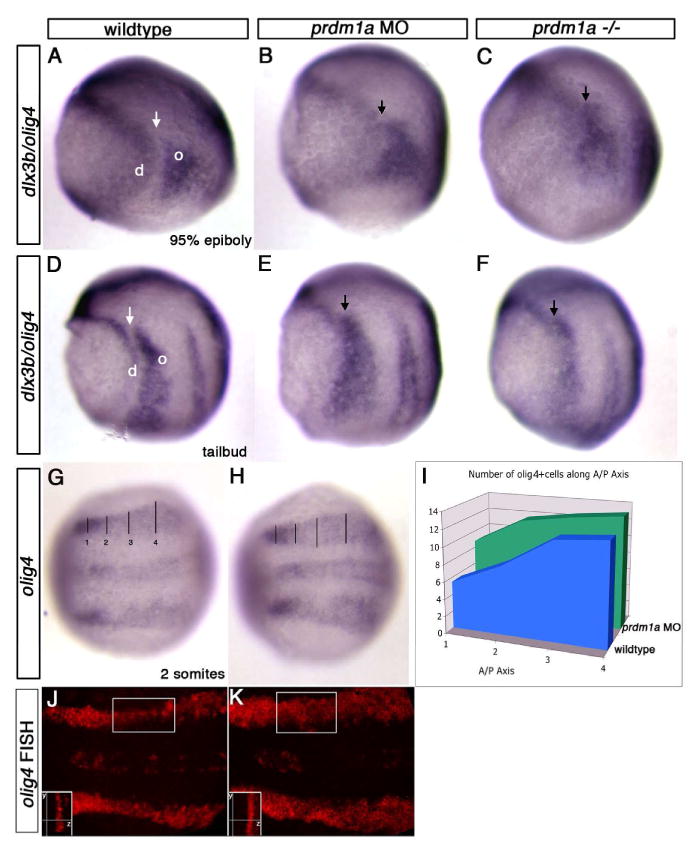
Dorsal and lateral views of wildtype, prdm1a morphant and prdm1a mutant embryos at 95% epiboly, tailbud and 2 somite stage. (A-C) Lateral view, dorsal to the right. There is a gap between the expression domains of dlx3b (d) and olig4 (o) in wildtype embryos that corresponds to the domain of prdm1a expression (arrow). When there is no prdm1a expression, the gap is lost (black arrow). Expression of dlx3b (left of arrow) in the non-neural ectoderm domain and olig4 (right of arrow) in the interneuron domain shifts to where there is no gap in prdm1a deficient embryos (arrows). The gap represents the position of the prdm1a domain. (D-F) Similar results are observed at the tailbud stage. (G-I) olig4 expression at the 2 somite stage in embryos that are mounted dorsally to count the number of cells in the olig4 domain. olig4 expression in the lateral interneuron domain and medial motor neuron domain (H) is expanded in prdm1a morphant embryos, as compared to wildtype embryos (G). (I) Cells were counted in four anterior/posterior (A/P) positions across the medial-lateral extent of the olig4 domain in wildtype (blue) and prdm1a morphant embryos (green) showing an expansion of the number of cells expressing olig4 especially in the anterior region (positions 1 and 2). (J,K) Fluorescent in situ hybridization (FISH) showing olig4 expression in prdm1a morphants is expanded in the medial lateral domain. However, expression does not expanded into the deeper cell layers, shown in a Z-plane confocal section (insets). Insets, boxed area observed at higher magnification in the Z-plane showing the same number of pixels in both uninjected and prdm1a morphants. y, y plane, anterior-posterior axis; z, z plane, dorsal-ventral axis
On the non-neural ectoderm side of the neural plate border, dlx3b expression is required for the development of RB sensory neurons and neurogenic placodes. Neurogenic placodes form as thickenings in the non-neural ectoderm next to the domain of NCCs, and form derivatives of the cranial and sensory organs (Nechiporuk et al., 2007). Previous work has shown that, in the absence of Prdm1a function, dlx3b is absent in the caudal domain, but does not expand into the NPB domain (Artinger et al., 1999). Since the neighboring interneuron domain marked by olig4 is affected in prdm1a mutants, we wanted to determine if the placodal domain or derivatives are also affected in prdm1a mutants. Placodal development was not changed in prdm1a mutant embryos as assessed by eya1 expression at the 2 somite stage (Supplemental Figure S1). There was also no change in the placodally-derived otic vesicle at the 10-somite stage, as demarked by pax2a expression (Supplemental Figure S1). Therefore, we conclude that the placodal domain is unaffected following prdm1a loss.
prdm1a acts to regulate cell fate, not cell proliferation or death, at the neural plate border
The results above indicate that prdm1a represses expression of the neighboring olig4 domain to maintain NPB cell fate. However, because prdm1a mutant embryos show decreased numbers of NPB derivatives, it is also possible that prdm1a regulates proliferation or death of NPB cells, similar to its role in the mouse immune system (Lin et al., 1997) and the zebrafish posterior pharyngeal arch region (Birkholz et al., 2009). To test the possibility that prdm1a mutants lack neural crest and RB neurons due to apoptosis within these cell types, we used TUNEL to assess rates of cell death at the NPB or dorsal neural tube of prdm1a mutants between 80% epiboly and the 6 somite stage within the pax3 expression domain. There is no change in the level of apoptosis at the NPB or in the dorsal neural tube of prdm1a mutant embryos compared to controls at either stage of development (data not shown). We found an average of 7.4 apototic cells at the 2 somite stage in both wildtype/heterozyogotes (n=29) and prdm1a-/- (n=12); and an average of 5.3 apototic cells at the 6 somite stage in wildtype/heterozyogotes (n=43) and 5.6 in prdm1a-/- (n=15). These results show that the loss of NPB derivatives in prdm1a mutants is not due to an increase in cell death.
We next examined the levels of cell proliferation between 95% epiboly and the 6 somite stage to determine if prdm1a is required for proliferation of NPB progenitors. We used a phosphohistone H3 (pH3) antibody to identify proliferating cells and counted pH3+ cells within the pax3 expression domain observed by in situ hybridization, which overlaps with the prdm1a domain at the NPB until the 2 somite stage (Figure 1). At 95% epiboly through the 2 somite stage, there was no significant difference in the number of proliferating cells in control embryos compared to prdm1a mutant embryos (Figure 3). At 95% epiboly, uninjected embryos had an average of 39.9 proliferating cells (n=9) and prdm1a MO had an average of 43.3 (n=8) within the NPB. At the 2 somite stage, uninjected embryos had an average of 5.9 (n=10) and prdm1a MO had an average of 6.3 proliferating cells (n=12). However, at the 6 somite stage, there is a significant increase in the number of mitotic cells within the region of the cranial neural crest in prdm1a MO embryos; uninjected embryos averaged 5.9 pH3+ cells at the 6 somite stage (n=8), while prdm1a MO embryos had an average of 10.0 (n=10; Student's t test, p=0.003). This is consistent with the observation that while prdm1a mutants have reduced cranial neural crest cells early in development, the number of cranial NCCs recovers to near normal levels by the 5-10 somite stage, as assessed by snail2 and dlx2 expression (Artinger et al, 1999). This suggests that the remaining neural crest cells in the cranial region of prdm1a mutant embryos proliferate to compensate for the earlier reduction in cranial NCCs. To further assess the role of proliferation in the prdm1a phenotype, we inhibited proliferation with Aphidicolin and HydroxyUrea from 70% epiboly to 1 somite, but this did not phenocopy the prdm1a mutation with respect to loss of neural crest markers and RB-specific markers (not shown). We therefore conclude that prdm1a is important specifically in regulation of cell fate decisions at the NPB and does not function by controlling cell death or proliferation of NPB progentitors.
olig4 knockdown can rescue the neural crest and Rohon-Beard sensory neuron phenotype of prdm1a mutants
If prdm1a and olig4 interact to define the NPB region, we should be able to determine the interaction of these transcription factors by epistasis experiments. If prdm1a and olig4 repress each other in neighboring domains, thereby promoting NPB and interneuron fates respectively, we would expect that removing olig4 would promote NC and RB cell fate even in the absence of prdm1a. First, we confirmed the published Morpholino knockdown phenotype of olig4 alone on neural crest cell fates. We found that olig4 increased foxd3 and crestin expression and pigment cell number (Supplemental Figure 2). To determine if olig4 knockdown can restore the depleted NC in prdm1a mutants, we injected olig4 Morpholino into prdm1a mutant and wild-type embryos. Consistent with previous reports, olig4 knockdown promoted NC and RB fate in wildtype embryos (data not shown). In addition, control Morpholino injected at the same concentration shows no phenotype or restoration of neural crest cells (data not shown). Knockdown of olig4 in prdm1a mutant embryos rescued NCC specification as demonstrated by an increase in expression of crestin (26 of 29 embryos - 89% - express crestin) and sox10 (29 of 32 embryos − 90% - express sox10) in prdm1a mutants injected with olig4 MO (Figure 4A-C). Overall pigment cells also increased compared to uninjected control embryos 93% - 41/44 of prdm1a mutants injected with olig4 MO -have increased pigment; Figure 4D-I). RB sensory neurons are also partially rescued, albeit to a lesser extent, as shown by the partial induction of islet1/2 and HNK-1 (4 of 10 prdm1a mutants injected with olig4 MO express HNK-1 and islet1 compared to prdm1a mutants without olig4 MO; Figure 4M-O). Uninjected or control MO-injected prdm1a mutants exhibit no expression of either marker (Figure 4). These results show that olig4 knockdown promotes the formation of NPB derivatives in the absence of prdm1a function. Thus, prdm1a is dispensable for NC and RB neuron specification, but is necessary to define the precise spatial domain of the NPB. Consequently, prdm1a plays a permissive rather than an instructive role in NPB specification and, thereby, in the cell fate specification of NPB derivatives. Moreover, the mutual repression of prdm1a and olig4 is required for the establishment and refinement of distinct interneuron and NPB domains.
Figure 4. Injections of olig4 Morpholino rescues the prdm1a phenotype.
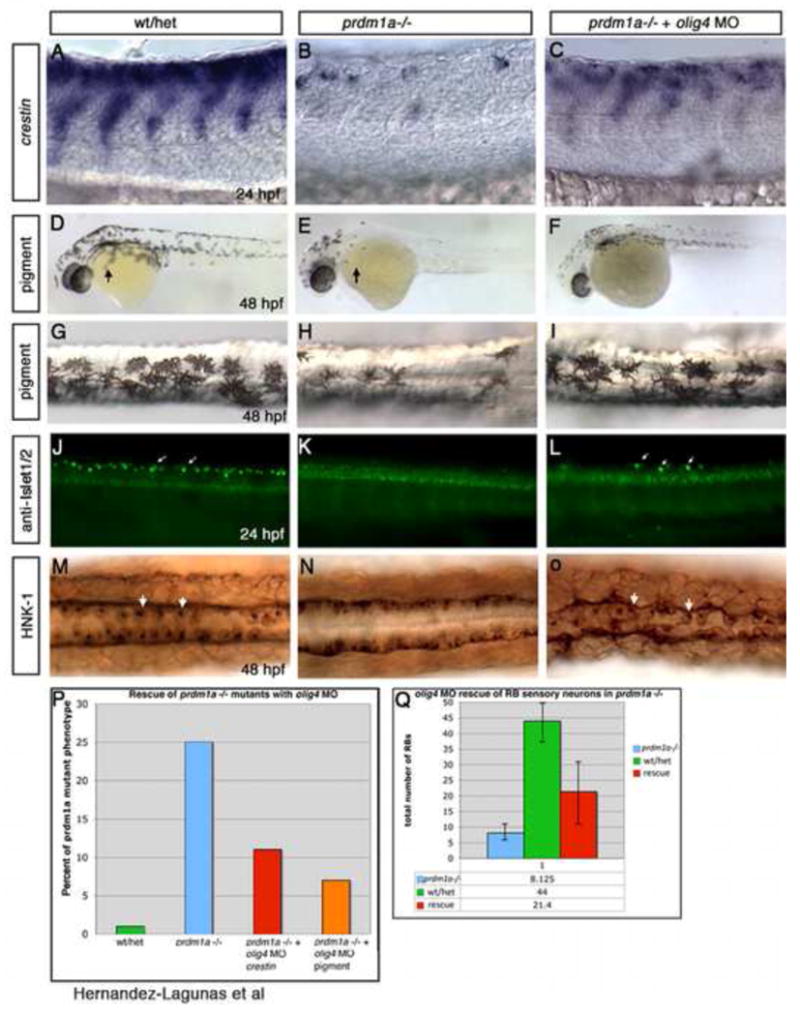
(A-F, J-L) Lateral views and (G-I, M-O) dorsal view, anterior to the left of 24 and 48 hpf embryos. (A) crestin expression in wildtype or heterozygous embryo show a wildtype pattern of neural crest migration. (B) prdm1a mutant embryos have few neural crest cells, while injection of olig4 MO rescues the neural crest cell deficit (C). (D, G) Low and high magnification of wildtype pigment pattern at 48 hpf compared to (E, H) prdm1a mutant embryos that have a reduction of pigment especially in the head and on the yolk (arrow). (F, I) olig4 MO injection shows an increase in pigment cells overall and some migrate over the yolk. (J, M) Rohon-Beard sensory neuron expression with Islet1/2 or HNK-1 antibody at 24 hpf and 48 hpf respectively in wildtype embryo. (K, N) prdm1a mutant embryos have no RB sensory neurons but still have ventral expressing interneurons and motor neurons. (L, O) Injection of olig4 MO into prdm1a mutant embryos increases the number of RB sensory neurons.
prdm1a is downstream of Notch signaling in neural crest and RB cell fate specification
Notch signaling plays an important role during generation of cell fate in the nervous system and, more specifically, in the fate determination step that defines neural crest and RB sensory neurons. In zebrafish Notch mutants and Xenopus embryos in which Notch signaling is overexpressed or knocked down, there is an increase in the number of primary neurons at the expense of neural crest. Because prdm1a controls the fate of both of these populations, we wanted to determine how prdm1a expression is affected by Notch mutations and explore the epistatsis of prdm1a and Notch signaling. First, we examined prdm1a expression in mib and deltaA mutants. In mib mutant embryos, which have complete blockage of Notch signaling, prdm1a expression was increased throughout the neural plate border domain and significantly expanded in the anterior domain (Figure 5). In deltaA mutants, prdm1a expression was also increased within this domain but to a lesser degree (data not shown). As an alternate approach to genetically blocking Notch signaling, we also treated wildtype embryos with 100-200 μm DAPT at 60% epiboly until fixation at the 2 somite stage to inhibit Notch signaling specifically during the developmental window when neural crest and RB sensory neurons are being specified. We observed an increase in prdm1a expression in the NPB, but it was not statistically significant due to variability of treatment penetrance (Figure 5). It is also possible that inhibition of Notch signaling must occur at earlier developmental stages to affect prdm1a expression completely. These results suggest that Notch signaling negatively regulates prdm1a expression and by doing so increases the progenitors available to produce more neurons. To test this, we performed epistasis experiments with Notch and prdm1a. We used DAPT to treat wildtype and prdm1a mutants as described. Embryos were examined for border domain markers (olig4/dlx3b), neural crest (by foxd3 expression), and RB sensory neurons (by huC expression). We confirmed that treatment with DAPT in wildtype embryos increased the total number of primary neurons, as previously reported (Geling et al., 2002). However, even after application of DAPT to prdm1a-/- embryos, we did not observe a rescue of the border domain with restoration of the gap between olig4 and dlx3b expression (Figure 6A-D). In addition, RB sensory neurons were not recovered in prdm1a mutants (n=39) as none exhibited huC expression at the 2 somite stage (Figure 6E-H). All DAPT-treated prdm1a mutants (n=27) had foxd3 expression at similar levels to wildtype at 2 somite stage (Figure 6 I-L). However, there was no difference between the foxd3 expressing neural crest cells between DAPT- and vehicle-treated prdm1a mutant embryos, indicating that inhibition of Notch signaling was unable to restore the neural crest phenotype observed in prdm1a mutants. These results are consistent with Notch acting upstream of prdm1a.
Figure 5. prdm1a expression following Notch inhibition.
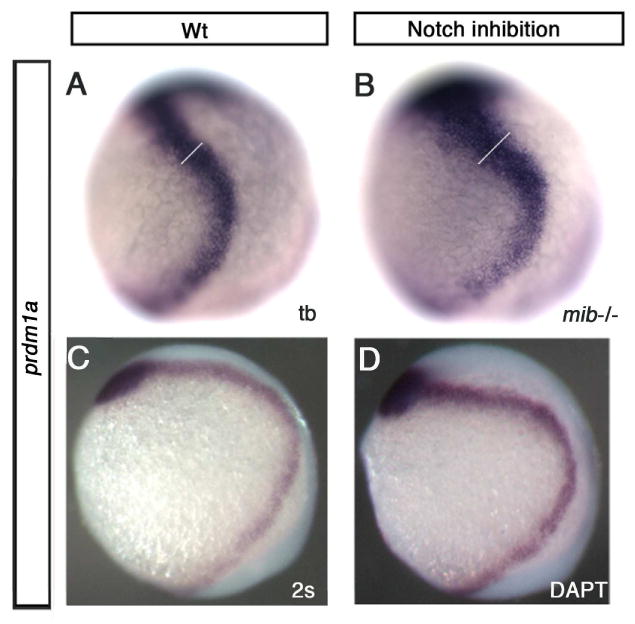
Lateral views, anterior to the top, dorsal to the right of tailbud and 2 somite stage embryos. (A) prdm1a expression at tailbud in a wildtype embryo (B) and in the mib-/-background. The overall level of expression is increased and the border domain was significantly expanded in mib mutant embryos (white line indicates where anterior prdm1a expression was measured using Photoshop; WT average 1.56 arbitrary pixel units, n=5; mib average 2.01 arbitrary pixel units, n=6; Student's t-test, p=0.03). (C) DMSO control treated wildtype and (D) embryo treated with DAPT a Notch inhibitor. prdm1a expression was increased overall when treated from 60% epiboly to 2 somite stage with DAPT.
Figure 6. Inhibition of Notch signaling cannot rescue the prdm1a mutant phenotype.
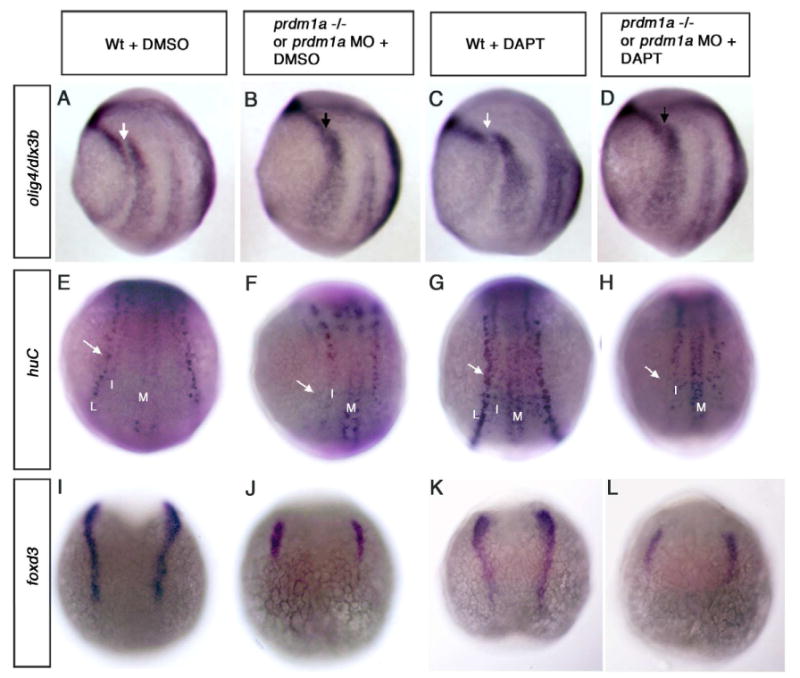
(A-D) Lateral views and (E-L) dorsal view, anterior to the top of tailbud and 2 somite stage embryos. (A) Wildtype embryo showing expression of olig4 and dlx3b and (B) prdm1a deficient embryos show no gap in expression of olig4 and dlx3b at tailbud stage as observed in wildtype. With the addition of DAPT from 60% epiboly to tailbud, wildtype embryos (C) have a slightly wider gap corresponding to the prdm1a expression domain, while prdm1a mutant embryos maintain the gap in olig4 and dlx3b expression after DAPT treatment (arrows). (E, G) Wildtype and prdm1a (F, H) mutant embryo at 2 somites; RB sensory neurons shown by the lateral expression of huc (arrows) were not recovered in prdm1a-/- embryos following DAPT treatment and look similar to DMSO control treated mutants. Wildtype and mutant embryos treated with DAPT show anincrease in the overall number of primary neurons (L, I, M) as previously reported. (I-L) Similarly foxd3 expressing neural crest cells are not increased in prdm1a mutant embryos following DAPT treatment. L, Lateral Rohon-Beard sensory neurons; I, intermediate interneurons, M, primary motor neurons.
Discussion
Here, we have further defined the function of prdm1a and olig4 in NPB cell fate decisions. We show that prdm1a regulates cell fate at the NPB by counteracting olig4 expression, not by regulating cell death or proliferation. In addition, prdm1a expression is upregulated in the absence of Notch function, and we are not able to rescue prdm1a mutants by blocking Notch signaling with DAPT. This suggests that prdm1a functions downstream of Notch in the regulation of cell fate at the neural plate border.
Prdm1a has been shown to play a role as a cell fate switch in a variety of developmental paradigms, including neural crest/RB neurons and interneuron cell fate. In prdm1a mutant embryos, we previously observed an increase in islet1-expressing cells within a ventral interneuron domain, supporting the idea that prdm1a promotes formation of neural crest and RB sensory neurons by repressing interneuron cell fate (Olesnicky et al, 2010). This is supported further by the observation that overexpression of prdm1a expands sox10 and islet1 expression specifically in the neural crest and RB neuron domain, respectively (Olesnicky et al. 2010). Studies in zebrafish pharyngeal arch development and in the immune system of mice show that, in these systems, prdm1a plays a role in regulating cell proliferation (Birkholz et al., 2009; Lin et al., 1997). However, the data we present here shows that prdm1a does not regulate cell proliferation during the NPB stage, but instead plays a role in regulating NPB cell fate. At later neural plate stages (the 6 somite stage), there is a slight upregulation of cell proliferation within the pax3 domain. As the pax3 domain does not correlate completely with the prdm1a domain, we cannot rule out the possibility that the upregulation in cell proliferation is within the neural plate domain and not at the NPB. Thus, prdm1a may act non-cell autonomously to influence cell proliferation or cell cycle progression of the neighboring interneuron domain of neural plate cells. It is clear that these domains, which are often thought of as autonomous at the developing neural plate and NPB are not distinct at early stages, but become resolved as development proceeds via mutual repression and other complex genetic interactions. Double fluorescent in situ hybridization has revealed overlapping domains of non-neural ectoderm, NPB and neural plate during gastrulation. This overlap implies that during the process of resolving the progenitor domains of interneurons, neural crest, and RB sensory neurons, NPB genes interact extensively with genes expressed in overlapping and neighboring domains. While it is clear that the domains refine over time to produce specific neurons with specific functions, we do not yet know whether cells move in between the domains and if so whether these cells acquire the fate specified by their new environment or move back to their initial domain. Additional experiments are required to address these questions. Interestingly, prdm1a seems to play a similar role in other regions of the developing nervous system. Prdm1a appears to function in the mouse retina by simultaneously promoting photoreceptor fate and repressing bipolar interneuron cell fate. In conditional knockout studies, Prdm1a (Blimp1) mutants exhibited fewer photoreceptors and more bipolar interneuron cells (Brzezinski et al.2010; Katoh et al.2010).
Previous studies in zebrafish suggest that olig4 morphants have reduced interneuron cell number and increased numbers of neural crest cells and RB neurons (Filippi et al 2005). Because this is the opposite of the phenotype observed in prdm1a mutants, we were interested in understanding how these two transcription factors interact. Interestingly, olig4 expression does not overlap with prdm1a expression even at early gastrulation stages, and its expression domain is expanded in prdm1a mutants. In addition, olig4 Morpholino injection can restore NC development in prdm1a mutants. This suggests that prdm1a and olig4 are mutual repressors, creating a sharply delineated border by suppressing neighboring cell fate (see Model in Figure 7). This is reminiscent of what is observed in mouse spinal cord development where Olig3 is required for specification of dorsal class A interneurons (more dorsal dI1-3) and represses the formation of class B interneurons (intermediate dI3-6) (Muller et al., 2005). Class A interneurons are thought to migrate to a more ventral location in the spinal cord, and relay proprioceptive information (Ding et al., 2005; Muller et al., 2005). However, unlike the zebrafish Morpholino phenotype, Olig3-/- mice do not have a defect in FoxD3-and Sox10-expressing neural crest cells, suggesting that Olig3 is required to promote dI1-3 interneuron fate without having an effect on neural crest cell development. RB-like neurons have been characterized morphologically in mammalian embryos but not with molecular markers, thus it is unclear which cells, if any, in the developing mouse nervous system correspond to RB neurons (Humphrey, 1944; Humphrey, 1950). The difference between the function of Olig3/4 in mouse and zebrafish is intriguing and could provide clues to the cell fate relationships between neural crest, RB neurons and interneurons across species. While further studies are necessary, we speculate that RB neurons may have evolved into the more dorsal interneuron that relays proprioceptive information in mammalian embryos.
Figure 7. Model of prdm1a function at the neural plate border.
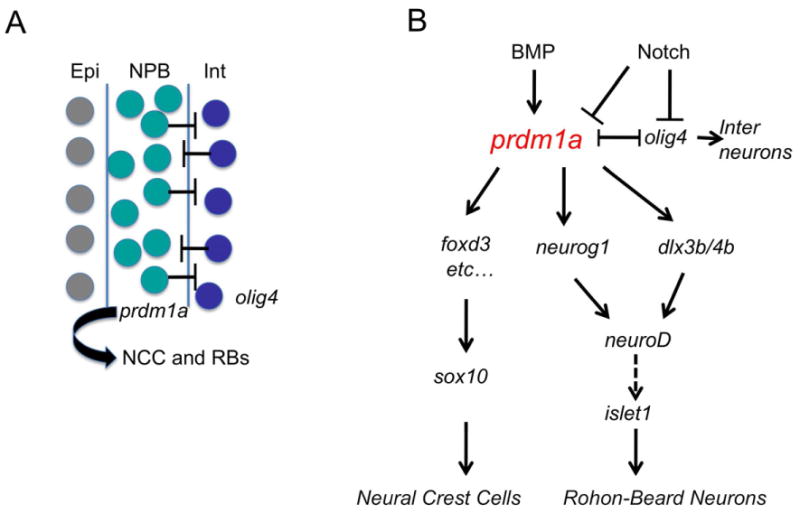
(A) Interneurons (Int; blue), Neural plate border (NPB, green) and Epidermis (Epi; grey). Mutual repression between prdm1a and olig4 is required to maintain NCC and RB sensory neuron cell fate. (B) Preliminary gene regulatory network for NCC and RB sensory neuron development.
Notch signaling is an important regulator of cell fate in the nervous system (Artavanis-Tsakonas et al., 1999; Cornell, 2005; Lewis, 1998). Its canonical role is in lateral inhibition, where cells within an equivalence domain (cells that are equivalent to each other in developmental potential) signal to each other, and by a stochastic process, one cell becomes a neuron and then inhibits its neighbors from becoming neurons. Notch receptors and delta ligands are expressed at the right time and place to be involved in segregation of cell fate between Rohon-Beard sensory neurons and neural crest cells. In mutations that affect Notch signaling in zebrafish, there is an excess of all primary neurons, including RB sensory neurons, at the expense of trunk neural crest cells (Cornell and Eisen, 2000; Itoh et al., 2003; Jiang et al., 1996). The transcription factors that mediate this process downstream of Notch include olig4 and prdm1a, which we have shown here to have opposing effects on NPB cell fate specification. While knockdown of olig4 increases neural crest cells and RB neurons (Filippi et al., 2005), loss of prdm1a reduces both cell fates. Because Notch mediates lateral inhibition between these two cell fates, perturbation of Notch signaling results in promotion of neuronal fate at the expense of neural crest fate (Cornell, 2005). We then examined what is required for mediating the positive and negative regulation of the Notch signaling pathway on neural crest development. We propose that the mutual repression between olig4 and prdm1a determines the fate of cells of the NPB region. Because olig4 knockdown can rescue prdm1a mutants, olig4 may be instructive while prdm1a is permissive in neural plate border fate. olig4 knockdown can rescue the neural crest phenotype seen in Notch inhibition, suggesting that olig4 functions downstream of Notch and normally inhibits neural crest development (Filippi et al., 2005). In the current study, we have shown that Notch can regulate prdm1a by normally downregulating its expression. This data is the first to support a role for Notch at two time points in zebrafish neural development: 1) At early stages, during neural induction, to regulate the number of progenitor cells at the neural plate border and, 2) At a later time point in lateral inhibition to promote neuronal fate at the expense of neural crest fate. Interestingly, inhibition of Notch cannot rescue any aspect of the prdm1a phenotype, suggesting that prdm1a acts downstream of Notch signaling at the neural plate border.
In conclusion, our studies provide further evidence that the transcription factor Prdm1a is a key cell fate regulator at the NPB. In particular, our data indicate that Prdm1a acts by specifically promoting neural crest cell and RB neuron fate, rather than by regulating cell death or proliferation of these cells or their progenitors. We have also shown that prdm1a acts downstream of Notch signaling in this cell fate pathway. Finally, prdm1a also inhibits olig4, thereby repressing the interneuron cell fate within the dorsal spinal cord, and it is the mutually suppressive function of olig4 and prdm1a that regulates neural crest and RB cell fate.
Supplementary Material
Dorsal and lateral views of 2 stage embryos. Placodal development was unchanged in prdm1a mutant embryos (B,D,F,H) compared to wildtype control embryos (A,C,E,G) as assessed by eya1 expression at tailbud and the 2 somite stage (A-D) and pax2a expression to label placodally derived otic vesicle at 10 somite stage (E-F).
Dorsal views of foxd3 expression at 12 hpf (A, B), and lateral views of crestin expression at 24 hpf (C,D) and pigmentation at 3 dpf (E,F) embryos. Neural crest formation is increased in the olig4 Morpholino injected embryos (B, D, F) as compared wildtype (A, C, E). We quantified the foxd3 expression domain at 12 hpf and show that the foxd3 domain is significantly expanded (p<0.02) into the neural plate in olig4 morphants (average of 1.38 pixel units, n=11) as compared to uninjected controls (1.17 pixel units, n=11).
Research Highlights.
prdm1a acts explicitly on cell fate specification by counteracting olig4 expression in the neighboring interneuron domain.
prdm1a in neural plate border-derived cell fates.
prdm1a expression is upregulated in the absence of Notch function, and inhibiting Notch signaling fails to rescue prdm1a mutants.
prdm1a functions downstream of Notch in the regulation of cell fate at the neural plate border and that Notch regulates the total number of progenitor cells at the neural plate border.
Acknowledgments
We would like to thank ZIRC (P40 RR012546-NIH-NCRR) for reagents; Bruce Appel, Linda Barlow, Lee Niswander and Christy Rossi for helpful comments on the manuscript; Morgan Singleton for extraordinary fish care; Keith Anderson for discussion; David McKean for the initial observation of prdm1a expression in mindbomb mutants; Ajay Chitnis for mind bomb mutants and discussion. We gratefully acknowledge the support of NIH P30 NS04815 zebrafish and imaging core grant, and NIH HD050698 to K.B.A.
Footnotes
Publisher's Disclaimer: This is a PDF file of an unedited manuscript that has been accepted for publication. As a service to our customers we are providing this early version of the manuscript. The manuscript will undergo copyediting, typesetting, and review of the resulting proof before it is published in its final citable form. Please note that during the production process errors may be discovered which could affect the content, and all legal disclaimers that apply to the journal pertain.
References
- Ahrens K, Schlosser G. Tissues and signals involved in the induction of placodal Six1 expression in Xenopus laevis. Dev Biol. 2005;288:40–59. doi: 10.1016/j.ydbio.2005.07.022. [DOI] [PubMed] [Google Scholar]
- Appel B, Eisen JS. Regulation of neuronal specification in the zebrafish spinal cord by Delta function. Development. 1998;125:371–80. doi: 10.1242/dev.125.3.371. [DOI] [PubMed] [Google Scholar]
- Artavanis-Tsakonas S, Rand MD, Lake RJ. Notch signaling: cell fate control and signal integration in development. Science. 1999;284:770–6. doi: 10.1126/science.284.5415.770. [DOI] [PubMed] [Google Scholar]
- Artinger KB, Chitnis AB, Mercola M, Driever W. Zebrafish narrowminded suggests a genetic link between formation of neural crest and primary sensory neurons. Development. 1999;126:3969–79. doi: 10.1242/dev.126.18.3969. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Basch ML, Bronner-Fraser M, Garcia-Castro MI. Specification of the neural crest occurs during gastrulation and requires Pax7. Nature. 2006;441:218–22. doi: 10.1038/nature04684. [DOI] [PubMed] [Google Scholar]
- Batista MF, Jacobstein J, Lewis KE. Zebrafish V2 cells develop into excitatory CiD and Notch signalling dependent inhibitory VeLD interneurons. Dev Biol. 2008;322:263–75. doi: 10.1016/j.ydbio.2008.07.015. [DOI] [PubMed] [Google Scholar]
- Baxendale S, Davison C, Muxworthy C, Wolff C, Ingham PW, Roy S. The B-cell maturation factor Blimp-1 specifies vertebrate slow-twitch muscle fiber identity in response to Hedgehog signaling. Nat Genet. 2004;36:88–93. doi: 10.1038/ng1280. [DOI] [PubMed] [Google Scholar]
- Birkholz DA, Killian EC, George KM, Artinger KB. Prdm1a is necessary for posterior pharyngeal arch development in zebrafish. Dev Dyn. 2009;238:2575–87. doi: 10.1002/dvdy.22090. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Brzezinski JAt, Lamba DA, Reh TA. Blimp1 controls photoreceptor versus bipolar cell fate choice during retinal development. Development. 137:619–29. doi: 10.1242/dev.043968. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cornell RA, Eisen JS. Delta signaling mediates segregation of neural crest and spinal sensory neurons from zebrafish lateral neural plate. Development. 2000;127:2873–82. doi: 10.1242/dev.127.13.2873. [DOI] [PubMed] [Google Scholar]
- Cornell RA, Eisen JS. Delta/Notch signaling promotes formation of zebrafish neural crest by repressing Neurogenin 1 function. Development. 2002;129:2639–48. doi: 10.1242/dev.129.11.2639. [DOI] [PubMed] [Google Scholar]
- Cornell RAa, E J. Notch in the pathway: The roles of Notch signaling in neural crest development. Semin Cell Dev Biol. 2005 Jul 27; doi: 10.1016/j.semcdb.2005.06.009. [DOI] [PubMed] [Google Scholar]
- Ding L, Takebayashi H, Watanabe K, Ohtsuki T, Tanaka KF, Nabeshima Y, Chisaka O, Ikenaka K, Ono K. Short-term lineage analysis of dorsally derived Olig3 cells in the developing spinal cord. Dev Dyn. 2005;234:622–32. doi: 10.1002/dvdy.20545. [DOI] [PubMed] [Google Scholar]
- Filippi A, Tiso N, Deflorian G, Zecchin E, Bortolussi M, Argenton F. The basic helix-loop-helix olig3 establishes the neural plate boundary of the trunk and is necessary for development of the dorsal spinal cord. Proc Natl Acad Sci U S A. 2005;102:4377–82. doi: 10.1073/pnas.0407284102. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Geling A, Steiner H, Willem M, Bally-Cuif L, Haass C. A gamma-secretase inhibitor blocks Notch signaling in vivo and causes a severe neurogenic phenotype in zebrafish. EMBO Rep. 2002;3:688–94. doi: 10.1093/embo-reports/kvf124. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Glavic A, Silva F, Aybar MJ, Bastidas F, Mayor R. Interplay between Notch signaling and the homeoprotein Xiro1 is required for neural crest induction in Xenopus embryos. Development. 2004;131:347–59. doi: 10.1242/dev.00945. [DOI] [PubMed] [Google Scholar]
- Hernandez-Lagunas L, Choi IF, Kaji T, Simpson P, Hershey C, Zhou Y, Zon L, Mercola M, Artinger KB. Zebrafish narrowminded disrupts the transcription factor prdm1 and is required for neural crest and sensory neuron specification. Dev Biol. 2005;278:347–57. doi: 10.1016/j.ydbio.2004.11.014. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hsiao CD, You MS, Guh YJ, Ma M, Jiang YJ, Hwang PP. A positive regulatory loop between foxi3a and foxi3b is essential for specification and differentiation of zebrafish epidermal ionocytes. PLoS One. 2007;2:e302. doi: 10.1371/journal.pone.0000302. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Humphrey Primative neurons in the embryonic human central nervous sytem. j comp neurol. 1944;81:1–45. [Google Scholar]
- Humphrey Intramedullary sensory gangion cells in the roof plate area of the embryonic human spinal cord. j comp neurol. 1950;92:333–399. doi: 10.1002/cne.900920304. [DOI] [PubMed] [Google Scholar]
- Itoh M, Kim CH, Palardy G, Oda T, Jiang YJ, Maust D, Yeo SY, Lorick K, Wright GJ, Ariza-McNaughton L, Weissman AM, Lewis J, Chandrasekharappa SC, Chitnis AB. Mind bomb is a ubiquitin ligase that is essential for efficient activation of Notch signaling by Delta. Dev Cell. 2003;4:67–82. doi: 10.1016/s1534-5807(02)00409-4. [DOI] [PubMed] [Google Scholar]
- Jiang YJ, Brand M, Heisenberg CP, Beuchle D, Furutani-Seiki M, Kelsh RN, Warga RM, Granato M, Haffter P, Hammerschmidt M, Kane DA, Mullins MC, Odenthal J, van Eeden FJ, Nusslein-Volhard C. Mutations affecting neurogenesis and brain morphology in the zebrafish, Danio rerio. Development. 1996;123:205–16. doi: 10.1242/dev.123.1.205. [DOI] [PubMed] [Google Scholar]
- Katoh K, Omori Y, Onishi A, Sato S, Kondo M, Furukawa T. Blimp1 suppresses Chx10 expression in differentiating retinal photoreceptor precursors to ensure proper photoreceptor development. J Neurosci. 30:6515–26. doi: 10.1523/JNEUROSCI.0771-10.2010. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lamborghini JE. Rohon-beard cells and other large neurons in Xenopus embryos originate during gastrulation. J Comp Neurol. 1980;189:323–33. doi: 10.1002/cne.901890208. [DOI] [PubMed] [Google Scholar]
- Lewis J. Notch signalling and the control of cell fate choices in vertebrates. Semin Cell Dev Biol. 1998;9:583–9. doi: 10.1006/scdb.1998.0266. [DOI] [PubMed] [Google Scholar]
- Liew HP, Choksi SP, Wong KN, Roy S. Specification of vertebrate slow-twitch muscle fiber fate by the transcriptional regulator Blimp1. Dev Biol. 2008;324:226–35. doi: 10.1016/j.ydbio.2008.09.020. [DOI] [PubMed] [Google Scholar]
- Lin KI, Angelin-Duclos C, Kuo TC, Calame K. Blimp-1-dependent repression of Pax-5 is required for differentiation of B cells to immunoglobulin M-secreting plasma cells. Mol Cell Biol. 2002;22:4771–80. doi: 10.1128/MCB.22.13.4771-4780.2002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lin Y, Wong K, Calame K. Repression of c-myc transcription by Blimp-1, an inducer of terminal B cell differentiation. Science. 1997;276:596–9. doi: 10.1126/science.276.5312.596. [DOI] [PubMed] [Google Scholar]
- Mancilla A, Mayor R. Neural crest formation in Xenopus laevis: mechanisms of Xslug induction. Dev Biol. 1996;177:580–9. doi: 10.1006/dbio.1996.0187. [DOI] [PubMed] [Google Scholar]
- Mercader N, Fischer S, Neumann CJ. Prdm1 acts downstream of a sequential RA, Wnt and Fgf signaling cascade during zebrafish forelimb induction. Development. 2006;133:2805–15. doi: 10.1242/dev.02455. [DOI] [PubMed] [Google Scholar]
- Meulemans D, Bronner-Fraser M. Gene-regulatory interactions in neural crest evolution and development. Dev Cell. 2004;7:291–9. doi: 10.1016/j.devcel.2004.08.007. [DOI] [PubMed] [Google Scholar]
- Mizuguchi R, Kriks S, Cordes R, Gossler A, Ma Q, Goulding M. Ascl1 and Gsh1/2 control inhibitory and excitatory cell fate in spinal sensory interneurons. Nat Neurosci. 2006;9:770–8. doi: 10.1038/nn1706. [DOI] [PubMed] [Google Scholar]
- Muller T, Anlag K, Wildner H, Britsch S, Treier M, Birchmeier C. The bHLH factor Olig3 coordinates the specification of dorsal neurons in the spinal cord. Genes Dev. 2005;19:733–43. doi: 10.1101/gad.326105. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nechiporuk A, Linbo T, Poss KD, Raible DW. Specification of epibranchial placodes in zebrafish. Development. 2007;134:611–23. doi: 10.1242/dev.02749. [DOI] [PubMed] [Google Scholar]
- Olesnicky E, Hernandez-Lagunas L, Artinger KB. Prdm1a regulates sox10 and islet1 in the development of neural crest and Rohon-Beard sensory neurons. Genesis. 48(11):656–66. doi: 10.1002/dvg.20673. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pineda RH, Svoboda KR, Wright MA, Taylor AD, Novak AE, Gamse JT, Eisen JS, Ribera AB. Knockdown of Nav1.6a Na+ channels affects zebrafish motoneuron development. Development. 2006;133:3827–36. doi: 10.1242/dev.02559. [DOI] [PubMed] [Google Scholar]
- Robertson EJ, Charatsi Iphigenie, Joyner Clive J, Koonce Chad H, Morgan Marc, Islam Ayesha, Paterson Carol, L E, Arnold Sebastian J, Kallies Axel, Nutt Stephen L, Bikoff Elizabeth K. Blimp1 regulates development of the posterior forelimb, caudal pharyngeal arches, heart and sensory vibrissae in mice. Development. 2007;134:4335–4345. doi: 10.1242/dev.012047. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rossi CC, Hernandez-Lagunas L, Zhang C, Choi IF, Kwok L, Klymkowsky M, Artinger KB. Rohon-Beard sensory neurons are induced by BMP4 expressing non-neural ectoderm in Xenopus laevis. Dev Biol. 2008;314:351–61. doi: 10.1016/j.ydbio.2007.11.036. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rossi CC, Kaji T, Artinger KB. Transcriptional control of Rohon-Beard sensory neuron development at the neural plate border. Dev Dyn. 2009;238:931–43. doi: 10.1002/dvdy.21915. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Roy S, Ng T. Blimp-1 specifies neural crest and sensory neuron progenitors in the zebrafish embryo. Curr Biol. 2004;14:1772–7. doi: 10.1016/j.cub.2004.09.046. [DOI] [PubMed] [Google Scholar]
- Schlosser G. Induction and specification of cranial placodes. Dev Biol. 2006;294:303–51. doi: 10.1016/j.ydbio.2006.03.009. [DOI] [PubMed] [Google Scholar]
- Schlosser G, Awtry T, Brugmann SA, Jensen ED, Neilson K, Ruan G, Stammler A, Voelker D, Yan B, Zhang C, Klymkowsky MW, Moody SA. Eya1 and Six1 promote neurogenesis in the cranial placodes in a SoxB1-dependent fashion. Dev Biol. 2008;320:199–214. doi: 10.1016/j.ydbio.2008.05.523. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Shaffer AL, Lin KI, Kuo TC, Yu X, Hurt EM, Rosenwald A, Giltnane JM, Yang L, Zhao H, Calame K, Staudt LM. Blimp-1 orchestrates plasma cell differentiation by extinguishing the mature B cell gene expression program. Immunity. 2002;17:51–62. doi: 10.1016/s1074-7613(02)00335-7. [DOI] [PubMed] [Google Scholar]
- Shapiro-Shelef M, Lin KI, McHeyzer-Williams LJ, Liao J, McHeyzer-Williams MG, Calame K. Blimp-1 is required for the formation of immunoglobulin secreting plasma cells and pre-plasma memory B cells. Immunity. 2003;19:607–20. doi: 10.1016/s1074-7613(03)00267-x. [DOI] [PubMed] [Google Scholar]
- Tiso N, Filippi A, Benato F, Negrisolo E, Modena N, Vaccari E, Driever W, Argenton F. Differential expression and regulation of olig genes in zebrafish. J Comp Neurol. 2009;515:378–96. doi: 10.1002/cne.22054. [DOI] [PubMed] [Google Scholar]
- Ungos JM, Karlstrom RO, Raible DW. Hedgehog signaling is directly required for the development of zebrafish dorsal root ganglia neurons. Development. 2003;130:5351–62. doi: 10.1242/dev.00722. [DOI] [PubMed] [Google Scholar]
- von Hofsten J, Elworthy S, Gilchrist MJ, Smith JC, Wardle FC, Ingham PW. Prdm1- and Sox6-mediated transcriptional repression specifies muscle fibre type in the zebrafish embryo. EMBO Rep. 2008;9:683–9. doi: 10.1038/embor.2008.73. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wilm TP, Solnica-Krezel L. Essential roles of a zebrafish prdm1/blimp1 homolog in embryo patterning and organogenesis. Development. 2005;132:393–404. doi: 10.1242/dev.01572. [DOI] [PubMed] [Google Scholar]
- Zechner D, Muller T, Wende H, Walther I, Taketo MM, Crenshaw EB, 3rd, Treier M, Birchmeier W, Birchmeier C. Bmp and Wnt/beta-catenin signals control expression of the transcription factor Olig3 and the specification of spinal cord neurons. Dev Biol. 2007;303:181–90. doi: 10.1016/j.ydbio.2006.10.045. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Dorsal and lateral views of 2 stage embryos. Placodal development was unchanged in prdm1a mutant embryos (B,D,F,H) compared to wildtype control embryos (A,C,E,G) as assessed by eya1 expression at tailbud and the 2 somite stage (A-D) and pax2a expression to label placodally derived otic vesicle at 10 somite stage (E-F).
Dorsal views of foxd3 expression at 12 hpf (A, B), and lateral views of crestin expression at 24 hpf (C,D) and pigmentation at 3 dpf (E,F) embryos. Neural crest formation is increased in the olig4 Morpholino injected embryos (B, D, F) as compared wildtype (A, C, E). We quantified the foxd3 expression domain at 12 hpf and show that the foxd3 domain is significantly expanded (p<0.02) into the neural plate in olig4 morphants (average of 1.38 pixel units, n=11) as compared to uninjected controls (1.17 pixel units, n=11).


