Abstract
Background
Approximately 10% of pancreatic ductal adenocarcinomas have a familial basis. While a small portion of this familial clustering can be explained by inherited mutations in known genes (BRCA2, p16/CDKN2A, PRSS1, and STK11), the genetic basis for the majority of this familial clustering remains unknown. In addition, a pancreatic cancer susceptibility locus has been reported to be linked to chromosome 4q32-34 in a single family having a high penetrance of early-onset pancreatic ductal adenocarcinoma and pancreatic insufficiency. The goal of this study is to determine if linkage to chromosome 4q exists in our series of well-characterized families with idiopathic familial pancreatic cancer enrolled in the Pancreatic Cancer Genetic Epidemiology Consortium (PACGENE).
Methods
Parametric and nonparametric linkage analyses were performed using 21 microsatellite markers on chromosome 4 on affected individuals with pancreatic cancer from 42 familial pancreatic cancer kindreds.
Results
Markov Chain Monte Carlo parametric and nonparametric linkage analyses using SIMWALK2 as well as nonparametric linkage analysis using MERLIN did not provide strong evidence of linkage in this region (LOD < 1.0). Only one family provided a multipoint LOD score of >0.5 adjacent to the reported region.
Conclusions
Our results do not support linkage to the 4q32-34 region in the majority of our familial pancreatic cancer kindreds. However, because multiple pancreatic cancer susceptibility genes are likely to exist, it is possible that a subset of the families in this study may be linked to this region.
Keywords: pancreatic cancer, genetics, linkage analysis, adenocarcinoma, familial pancreatic cancer, hereditary, gastrointestinal cancer
INTRODUCTION
Pancreatic cancer is the fourth leading cause of cancer death in the United States. In 2006, an estimated 33,700 new pancreatic cancer cases will be diagnosed in the United States and 32,300 individuals will die from pancreatic cancer.1 Approximately 7–10% of individuals with pancreatic cancer have a family history of pancreatic cancer.2
The genetic basis of the aggregation of pancreatic cancer in families remains largely unknown. Genetic factors, including germline mutations in the BRCA2,3–5 p16/CDKN2A,6 PRSS17,8 and STK119 genes, have been demonstrated to increase the risk of pancreatic cancer. Germline BRCA2 mutations are the most prevalent germline mutations known to predispose to pancreatic cancer, with deleterious mutations reported in about 17% of families with three or more individuals affected with pancreatic cancer,5 12% in families with two individuals with pancreatic cancer4 and 7% in individuals with apparently sporadic pancreatic cancer.3 Mutations in other pancreatic cancer susceptibility genes (p16, STK11, PRSS1) have been reported in only a handful of familial pancreatic cancer families. Thus, less than twenty percent of families with multiple members with pancreatic cancers carry mutations in known susceptibility genes.
In 2002, Eberle et al.10 reported that pancreatic cancer in a single familial pancreatic cancer kindred, which they designated as “Family X,” appeared to be linked to chromosome 4q32-34. The family was remarkable because multiple members exhibited the phenotype of early onset pancreatic insufficiency and diabetes mellitus, which ultimately progressed in some to pancreatic cancer, typically before age forty. Affected kindred members who have undergone pancreatic resection also have multiple pancreatic intraepithelial neoplasia (PanIN) lesions.11 In contrast, we have demonstrated that in most cases of familial pancreatic cancer, pancreatic insufficiency does not occur, and affected members usually develop pancreatic cancer in their sixth to eighth decade of life.12,13 The marked clinical phenotype of Family X compared to more typical familial pancreatic cancer phenotype may suggest that the genes responsible may be be distinct. Indeed, a recent study of European familial pancreatic cancer kindreds did not replicate linkage to the 4q region.14
The objective of this study was to determine if linkage to chromosome 4q32-34 exists in families with the more common phenotype of familial pancreatic cancer in the absence of pancreatic insufficiency who have enrolled in the Pancreatic Cancer Genetic Epidemiology Consortium.13
METHODS
Data collection
The PACGENE Consortium was organized in 2002 with funding from the National Cancer Institute, and data collection is ongoing. This consortium has been described in detail elsewhere.13 In brief, familial pancreatic cancer kindreds are enrolled through seven sites, including National Familial Pancreas Tumor Registry at Johns Hopkins University (Baltimore, MD), Mayo Clinic (Rochester, MN), M.D. Anderson Cancer Center (Houston, TX), Karmanos Cancer Institute-Wayne State University (Detroit, MI), University of Toronto (Ontario, Canada), Dana-Farber Cancer Institute (Boston, MA), and Creighton University (Omaha, NE). Pathologic diagnoses are confirmed by review of available pathologic materials, hospital charts, and death certificates. We defined patients with “pancreatic cancer” as those with infiltrating ductal adenocarcinoma of the pancreas or a variant of ductal adenocarcinoma of the pancreas. Patients with pure endocrine neoplasms and neoplasms solely in other organs were not included in the analyses. Kindreds were evaluated for their suitability for linkage analysis by each site team and then at semiannual meetings of the PACGENE Steering Committee.
In addition to clinical and family history questionnaires, all consenting probands and family members were asked to donate a blood sample for genetic studies. When a blood sample was not available, archival tissues (formalin-fixed paraffin-embedded tissues) were sought as a source of DNA. Fifty kindreds were selected to undergo genome-wide STRP genotyping by the Center for Inherited Disease Research (CIDR) in 2004 using ~400 microsatellite (STRP) markers, twenty-one of which were located on chromosome 4. For family members with lymphocyte samples, DNA was isolated from peripheral blood lymphocytes and subject to PCR amplification at CIDR as previously described.13 DNA was extracted from unstained 10-μm slides of formalin-fixed paraffin embedded tissue samples after deparaffinization as previously described.15 If an individual’s archival tissue contained cancer, the sections were microdissected to exclude cancer tissue. Tissue DNA samples were isolated and amplified in the Goggins lab. Tissue DNA samples were amplified using the CIDR marker panel using conditions adapted for archival DNA. DNA was first quantified by Quantifiler (ABI).16 Two ng of input DNA was used for each PCR reaction, each STRP marker was amplified singly for 45 PCR cycles using Platinum Taq polymerase and buffer. Markers were sized at CIDR using ABI3700 capillary sequencers. PCR products from the tissue DNA samples were run on the same ABI3700 sequencer as the peripheral blood DNA PCR products. PCR products from lymphocyte DNAs were sized in pools of multiple different PCR products, while PCR products from archival tissues were sized individually.
Relationship errors were identified in the genotype data using RELCHECK17 and Mendelian errors were identified using Pedcheck18 and SIB-PAIR.19
Statistical analysis
Parametric and nonparametric linkage analyses were performed using SIMWALK2, which utilizes Monte Carlo Markov Chain simulated annealing to perform multipoint linkage analysis on any size pedigree. For the nonparametric analysis we present the NPL-pairs statistic given this is the most appropriate statistic for when examining large pedigrees with multiple affected members such as those in this dataset.20 Nonparametric affected-only linkage analyses was also conducted using MERLIN (version 1.0.1) which computes exact linkage statistics using the Lander-Green algorithm, but is limited by pedigree size.21 In these analyses, nonparametric Kong and Cox LOD scores and associated p-values were calculated.22 This test statistic has been show to be less conservative than the NPL score when genotype data are not available on all pedigree members.22
For the Merlin analysis, allele frequency estimates were derived using maximum likelihood estimates; SIMWALK2 estimates were obtained using the LINKAGE program. The Marshfield genetic map, assuming a Haldane mapping function, was used in both sets of analyses. Due to the computational limitations of the Lander-Green algorithm, noninformative individuals were removed from the pedigrees for the MERLIN analysis only. Prior to this analysis, genotypes were imputed using SIB-PAIR.19 Genotypes were included in the analyses only when they could be assigned with certainty using SIB-PAIR.
Because genetic heterogeneity not only reduces power, but can also result in failure to replicate reported linkages,23 analysis was limited to the forty-two families reporting Caucasian ancestry.
RESULTS
The forty-two kindreds in the analyses included 128 individuals with pancreatic cancer, with a mean age of onset of 63 years (standard deviation ± 12.6 years). Four pancreatic cancer patients had unknown age at onset. There were twenty-three kindreds with two cases of pancreatic cancer, five kindreds with three cases of pancreatic cancer and fourteen kindreds with four or more cases of pancreatic cancer. Of the 126 pancreatic cancer cases, DNA from blood was available on fifty patients and DNA from paraffin embedded tissues was available on seventeen patients. Genotype data were available on 322 additional family members, which enabled us to reconstruct genotypes of some of the pancreatic cancer cases for whom DNA samples were unavailable. On average, genotype data was available on 9.3 individuals per pedigree.
Overall, parametric analysis using SIMWALK2 did not support linkage to chromosome 4 (LOD < 1.0) even when allowing for linkage heterogeneity (HLOD < 1.0). Because the results of parametric linkage are dependent on the validity of the model assumptions, and failure to detect linkage can be due to assuming an incorrect model, nonparametric affected-only analysis was also performed.
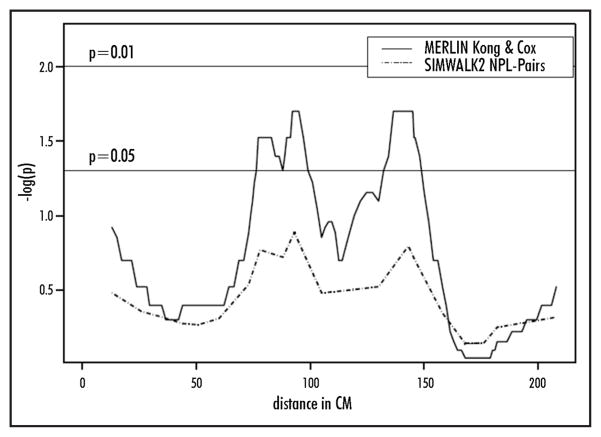
The results of our nonparametric analyses are presented (Table 1). Figure 1 displays the multipoint linkage results. Overall, multipoint nonparametic analysis using SIMWALK2 did not support linkage to chromosome 4q (LOD < 1.0). Two-point linkage analysis using MERLIN provided a Kong and Cox LOD score of 0.84 (p-value 0.02) at marker D4S2394 and a Kong and Cox LOD score of 0.48 (p value 0.07) at marker D4S1625. These markers flank D4S1644, which had a multi-point Kong and Cox LOD of 1.01 and a corresponding p-value of 0.02 near marker D4S1644. This p-value does not reach the conventional threshold level of statistical significance required to confirm that linkage exists in an independent set of families to a region that has previously been shown to harbor linkage. These markers are proximal but adjacent to the linked region reported by Brentnall et al. When examining the family-specific Kong and Cox LOD scores in this region, three families provided LOD scores > 0.40 in this region, but only a single family provided a LOD score > 0.50 (LOD = 0.55 at D4S1644).
Table 1.
Overview of nonparametric linkage analyses results for Chromosome 4q
| Marker | Location (cM) | SIMWALK2 | MERLIN | |
|---|---|---|---|---|
| Nonparametric N PL-pairs p-value | Twopoint Kong & Cox LOD (p-value) | Multipoint Kong & Cox LOD (p-value) | ||
| D4S2394 | 130 | 0.30 | 0.84 (0.02) | 0.44 (0.08) |
| D4S1644 | 143 | 0.16 | 0.11 (0.2) | 1.01 (0.02) |
| D4S1625 | 146 | 0.20 | 0.48 (0.07) | 0.79 (0.03) |
| D4S1629 | 158 | 0.45 | 0.03 (0.3) | 0.04 (0.3) |
| D4S2368 | 168 | 0.72 | −0.47 (0.9) | −0.27 (0.9) |
| D4S2431 | 176 | 0.72 | −0.26 (0.9) | −0.37 (0.9) |
Figure 1.
Results of multipoint non-parametric linkage analyses of chromosome 4
A two-point Kong and Cox LOD score of 1.67 (p = 0.003) was found at D4S2467, which is located on 4p. However, there was no evidence of linkage to this region using either of the multipoint analyses.
DISCUSSION
Our results suggest that the gene responsible for the familial aggregation of pancreatic cancer in the majority of the families we studied does not reside on chromosome 4q32. Our results are congruent with the recent report of Earl et al14 who could not replicate linkage to this region in a collection of European familial pancreatic cancer kindreds. In our analysis, a single family did provide modest linkage signal (LOD > 0.5) in this region, therefore we cannot rule out the possibility that a disease susceptibility gene is present in this region in a subset of families. The peak LOD score in this family did not overlap at the precise location reported by Eberle but was located centromeric to the original linkage locus.10 However, this finding does not directly preclude the presence of a susceptibility gene in this region because when we examined the haplotypes shared by the affected individuals in the linked family, the haplotype sharing extended beyond marker D4S1629 well into the linkage region reported by Eberle et al.10 Furthermore, linkage signals can extend large distances and the location of the linkage signal can vary around the causal locus. For instance, initial reports of linkage for inflammatory bowel disease to what was later identified as the NOD2 locus varied over a large region.24 In part, this phenomenon reflects the informativeness of individual genetic markers, the statistical power provided by the families and the location of recombination events in relation to the causal locus within a family. Another consideration is that although we used the conventional number of microsatellite markers for a genome wide scan linkage analysis, some additional information could be gained by increasing the density of markers in this region. Thus, the information content (entropy) of our marker set ranged from 0.39–0.68 which limits our power to detect modest to weak linkage signals.
Overall, our observation that this region is not linked to the majority of our familial pancreatic cancer families is not surprising given the unique phenotype of early onset of pancreatic insufficiency, diabetes, and pancreatic cancer in affected members of Family X which showed linkage to chromosome 4q.10 The kindreds in our study did not show early onset pancreatic cancer and did not have pancreatic insufficiency.
Currently, the PACGENE consortium is continuing its efforts to identify the location of susceptibility genes contributing to inherited pancreatic cancer through ongoing linkage analyses. We are currently genotyping 113 families using the Illumina IVb linkage panel, as this platform has been shown to provide better coverage than the STRP microsatellite marker set, and consequently should provide greater power to detect linkage as well as more refined linkage peaks.25 However, the standard microsatellite marker set used in this study does have sufficient power to detect broad regions of strong to modest linkage in these data. Once a susceptibility gene has been discovered, it is anticipated that genetic testing can then be offered to appropriate families. Family members who carry a disease causing mutation can enroll in screening protocols to detect early and potentially curable pancreatic neoplasms.26,27
Acknowledgments
We would like to thank Dr. Daniela Seminara of the National Cancer Institute for her continued advice and support of this work and the patients and families who have participated in this study.
References
- 1.Jemal A, Siegel R, Ward E, Murray T, Xu J, Smigal C, Thun MJ. Cancer statistics 2006. CA Cancer J Clin. 2006;56:106–30. doi: 10.3322/canjclin.56.2.106. [DOI] [PubMed] [Google Scholar]
- 2.Lynch HT, Fitzsimmons M, Smyrk TC. Familial pancreatic cancer: Clinicopathological study of 18 nuclear families. Am J Gastroenterol. 1990;85:54–60. [PubMed] [Google Scholar]
- 3.Goggins M, Schutte M, Lu J, Moskaluk CA, Weinstein CL, Petersen GM, Yeo CJ, Jackson CE, Lynch HT, Hruban RH, Kern SE. Germline BRCA2 gene mutations in patients with apparently sporadic pancreatic carcinomas. Cancer Res. 1996;56:5360–4. [PubMed] [Google Scholar]
- 4.Hahn SA, Greenhalf B, Ellis I, Sina-Frey M, Rieder H, Korte B, Gerdes B, Kress R, Ziegler A, Raeburn JA, Campra D, Grutzmann R, Rehder H, Rothmund M, Schmiegel W, Neoptolemos JP, Bartsch DK. BRCA2 germline mutations in familial pancreatic carcinoma. J Natl Cancer Inst. 2003;95:214–21. doi: 10.1093/jnci/95.3.214. [DOI] [PubMed] [Google Scholar]
- 5.Murphy KM, Brune KA, Griffin C, Sollenberger JE, Petersen GM, Bansal R, Hruban RH, Kern SE. Evaluation of candidate genes MAP2K4, MADH4, ACVR1B, and BRCA2 in familial pancreatic cancer: Deleterious BRCA2 mutations in 17% Cancer Res. 2002;62:3789–93. [PubMed] [Google Scholar]
- 6.Goldstein AM, Fraser MC, Struewing JP, Hussussian CJ, Ranade K, Zametkin DP, Fontaine LS, Organic SM, Dracopoli NC, Clark WH., Jr Increased risk of pancreatic cancer in melanoma-prone kindreds with p16INK4 mutations. N Engl J Med. 1995;333:970–4. doi: 10.1056/NEJM199510123331504. [DOI] [PubMed] [Google Scholar]
- 7.Whitcomb DC, Gorry MC, Preston RA, Furey W, Sossenheimer MJ, Ulrich CD, Martin SP, Gates LK, Jr, Amann ST, Toskes PP, Liddle R, McGrath K, Uomo G, Post JC, Ehrlich GD. Hereditary pancreatitis is caused by a mutation in the cationic trypsinogen gene [see comments] Nat Genet. 1996;14:141–5. doi: 10.1038/ng1096-141. [DOI] [PubMed] [Google Scholar]
- 8.Whitcomb DC, Preston RA, Aston CE, Sossenheimer MJ, Barua PS, Zhang Y, Wong-Chong A, White GJ, Wood PG, Gates LK, Jr, Ulrich C, Martin SP, Post JC, Ehrlich GD. A gene for hereditary pancreatitis maps to chromosome 7q35. Gastroenterology. 1996;110:1975–80. doi: 10.1053/gast.1996.v110.pm8964426. [DOI] [PubMed] [Google Scholar]
- 9.Su GH, Hruban RH, Bansal RK, Bova GS, Tang DJ, Shekher MC, Westerman AM, Entius MM, Goggins M, Yeo CJ, Kern SE. Germline and somatic mutations of the STK11/LKB1 Peutz-Jeghers gene in pancreatic and biliary cancers. Am J Pathol. 1999;154:1835–40. doi: 10.1016/S0002-9440(10)65440-5. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Eberle MA, Pfutzer R, Pogue-Geile KL, Bronner MP, Crispin D, Kimmey MB, Duerr RH, Kruglyak L, Whitcomb DC, Brentnall TA. A new susceptibility locus for autosomal dominant pancreatic cancer maps to chromosome 4q32-34. Am J Hum Genet. 2002;70:1044–8. doi: 10.1086/339692. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Meckler KA, Brentnall TA, Haggitt RC, Crispin D, Byrd DR, Kimmey MB, Bronner MP. Familial fibrocystic pancreatic atrophy with endocrine cell hyperplasia and pancreatic carcinoma. Am J Surg Pathol. 2001;25:1047–53. doi: 10.1097/00000478-200108000-00009. [DOI] [PubMed] [Google Scholar]
- 12.Klein AP, Brune KA, Petersen GM, Goggins M, Tersmette AC, Offerhaus GJ, Griffin C, Cameron JL, Yeo CJ, Kern S, Hruban RH. Prospective risk of pancreatic cancer in familial pancreatic cancer kindreds. Cancer Res. 2004;64:2634–8. doi: 10.1158/0008-5472.can-03-3823. [DOI] [PubMed] [Google Scholar]
- 13.Petersen G, de Andrade M, Goggins M, et al. Pancreatic cancer genetic epidemiology consortium Cancer Epidemiology. Biomarkers and Prev. 2006;15:704–10. doi: 10.1158/1055-9965.EPI-05-0734. [DOI] [PubMed] [Google Scholar]
- 14.Earl J, Yan L, Vitone LJ, Risk J, Kemp SJ, McFaul C, Neoptolemos JP, Greenhalf W, Kress R, Sina-Frey M, Hahn SA, Rieder H, Bartsch DK European Registry of Hereditary Pancreatitis and Familial Pancreatic Cancer. German National Case Collection for Familial Pancreatic Cancer. Evaluation of the 4q32-34 locus in European familial pancreatic cancer. Cancer Epidemiol Biomarkers Prev. 2006;15:1948–55. doi: 10.1158/1055-9965.EPI-06-0376. [DOI] [PubMed] [Google Scholar]
- 15.Fukushima N, Sato N, Ueki T, Rosty C, Walter KM, Wilentz RE, Yeo CJ, Hruban RH, Goggins M. Aberrant methylation of preproenkephalin and p16 genes in pancreatic intraepithelial neoplasia and pancreatic ductal adenocarcinoma. Am J Pathol. 2002;160:1573–81. doi: 10.1016/S0002-9440(10)61104-2. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Matsubayashi H, Canto M, Sato N, Klein A, Abe T, Yamashita K, Yeo CJ, Kalloo A, Hruban R, Goggins M. DNA methylation alterations in the pancreatic juice of patients with suspected pancreatic disease. Cancer Res. 2007 doi: 10.1158/0008-5472.CAN-05-2664. In press. [DOI] [PubMed] [Google Scholar]
- 17.Broman KW, Weber JL. Estimation of pairwise relationships in the presence of genotyping errors. Am J Hum Genet. 1998;63:1563–4. doi: 10.1086/302112. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.O’Connell JR, Weeks DE. PedCheck: A program for identification of genotype incompatibilities in linkage analysis. Am J Hum Genet. 1998;63:259–66. doi: 10.1086/301904. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.SIB-PAIR. A program of elementary genetical analysis. Australia: Queensland Institute of Medical Research; 2002. [Google Scholar]
- 20.Lange EM, Lange K. Powerful allele sharing statistics for nonparametric linkage analysis. Hum Hered. 2004;57:49–58. doi: 10.1159/000077389. [DOI] [PubMed] [Google Scholar]
- 21.Abecasis GR, Cherny SS, Cookson WO, Cardon LR. Merlin-rapid analysis of dense genetic maps using sparse gene flow trees. Nat Genet. 2002;30:97–101. doi: 10.1038/ng786. [DOI] [PubMed] [Google Scholar]
- 22.Kong A, Cox NJ. Allele-sharing models: LOD scores and accurate linkage tests. Am J Hum Genet. 1997;61:1179–88. doi: 10.1086/301592. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Suarez BK, Lin J, Witte JS, Conti DV, Resnick MI, Klein EA, Burmester JK, Vaske DA, Banerjee TK, Catalona WJ. Replication linkage study for prostate cancer susceptibility genes. Prostate. 2000;45:106–14. doi: 10.1002/1097-0045(20001001)45:2<106::aid-pros4>3.0.co;2-h. [DOI] [PubMed] [Google Scholar]
- 24.Hugot JP, Laurent-Puig P, Gower-Rousseau C, Olson JM, Lee JC, Beaugerie L, Naom I, Dupas JL, Van Gossum A, Orholm M, Bonaiti-Pellie C, Weissenbach J, Mathew CG, Lennard-Jones JE, Cortot A, Colombel JF, Thomas G. Mapping of a susceptibility locus for Crohn’s disease on chromosome 16. Nature. 1996;379:821–3. doi: 10.1038/379821a0. [DOI] [PubMed] [Google Scholar]
- 25.Schaid DJ, Guenther JC, Christensen GB, Hebbring S, Rosenow C, Hilker CA, McDonnell SK, Cunningham JM, Slager SL, Blute ML, Thibodeau SN. Comparison of microsatellites versus single-nucleotide polymorphisms in a genome linkage screen for prostate cancer-susceptibility Loci. Am J Hum Genet. 2004;75:948–65. doi: 10.1086/425870. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Canto MI, Goggins M, Hruban RH, Petersen GM, Giardiello FM, Yeo C, Fishman EK, Brune K, Axilbund J, Griffin C, Ali S, Richman J, Jagannath S, Kantsevoy SV, Kalloo AN. Screening for early pancreatic neoplasia in high-risk individuals: A prospective controlled study. Clin Gast Hepatol. 2007 doi: 10.1016/j.cgh.2006.02.005. In press. [DOI] [PubMed] [Google Scholar]
- 27.Canto MI, Goggins M, Hruban RH, Petersen GM, Giardiello FM, Yeo C, Fishman EK, Brune K, Axilbund J, Griffin C, Ali S, Richman J, Jagannath S, Kantsevoy SV, Kalloo AN. Screening for pancreatic neoplasia in high-risk individuals: An EUS-based approach. Clin Gastroenterol Hepatol. 2004;2:606–21. doi: 10.1016/s1542-3565(04)00244-7. [DOI] [PubMed] [Google Scholar]