Abstract
Bacterial exopolysaccharide, succinoglycan, plays an important role in eliciting infection thread formation, which is a key step in the establishment of Sinorhizobium meliloti–alfalfa (Medicago sativa) nitrogen fixing symbiosis. To understand the regulatory mechanisms that control production of succinoglycan, the expression of the key succinoglycan biosynthesis gene, exoY, was analyzed by constructing a set of nested deletions of the exoY promoter region. Two exoY promoters were identified based on the promoter activities and confirmed by direct detection of the transcripts. The expression from both promoters was induced in the exoR95 and exoS96 mutant backgrounds suggesting that both promoters are regulated by the ExoR protein and the ExoS/ChvI two-component signal transduction system. The identification of the exoY promoters provides additional avenue for further analysis of the role of succinoglycan in S. meliloti–alfalfa symbiosis.
Keywords: Symbiosis, Gene expression, Nitrogen fixation, Sinorhizobium meliloti, Succinoglycan biosynthesis, exoR, exoS, Alfalfa, Medicago sativa
1. Introduction
Sinorhizobium meliloti establishes a nitrogen fixing symbiosis with its plant host alfalfa (Medicago sativa) through continuous interactions that elicit plant structure changes, which enable S. meliloti cells to enter and colonize alfalfa root nodules [1-4]. One of the early and critical steps of the symbiosis is the formation of infection threads inside the curled root hairs that are colonized by S. meliloti cells [5-8]. The infection threads are tube-like structures and they are filled with growing S. meliloti cells [7,8]. The infection threads elongate and extend into developing nodule primordium in the root cortex where they release bacterial cells into newly formed plant cells in the middle of root nodules [1,9,10].
The initiation of the formation of infection threads in the curled alfalfa root hairs requires succinoglycan as well as the nodulation factor [7,11]. Succinoglycan is a S. meliloti exopolysaccharide with different degrees of polymerization of a single repeating unit that consists of seven glucoses and one galactose with succinyl, pyruvyl, and acetyl modifications [12,13]. Succinoglycan is produced in small amounts by free-living S. meliloti cells but it is produced in large amounts by S. meliloti exoR95::Tn5 and exoS96::Tn5 mutants [14,15]. The ExoR protein is an unknown regulatory protein [16]. The ExoS is the sensor of the ExoS/ChvI two-component regulatory system [17]. Although it is not clear how ExoR and ExoS/ChvI are related, they both regulate succinoglycan production by regulating the expression of succinoglycan biosynthesis genes, most of which cluster in a 19 kb region of the genome and are organized into several operons [14,18-20]. The ExoR protein and ExoS/ChvI system are also involved in regulating flagella biosynthesis (Cheng, H.-P., manuscript submitted).
While the expression of multiple exo genes, exoA, exoF, exoP, exoQ, and exoT, appear to be regulated [14,18], the expression of the exoY gene appears to be the primary target of regulation [21,22]. The exoY gene is the first gene of the exoYFQ operon and it encodes a galactosyl transferase that carries out the first step of succinoglycan biosynthesis [13,21]. The exoYFQ transcribes divergently from the exoX gene with a 769 bp intergenic region [22]. Transposon insertions in the intergenic region close to the exoY gene suppressed the succinoglycan overproduction caused by exoR95 and exoS96 mutations and brought the levels of succinoglycan production to that of the wild-type [22]. However, transposon insertions in areas further away from the exoY gene did not suppress the succinoglycan overproduction caused by exoR95 and exoS96 mutations [22]. This suggests that the region of the exoY promoter is targeted by the ExoR protein and ExoS/ChvI system in regulating succinoglycan production. In addition to ExoR and ExoS/ChvI, the production of succinoglycan appears to be regulated also by the ExoD [23], ExoX [22], MucR [24], SyrM [25], and SyrA [26] proteins.
To further characterize the regulation of the exoY gene expression and understand the role of succinoglycan in symbiosis, a set of nested deletions of the exoY promoter regions was constructed and fused to the exoY-gfp (green fluorescence protein) fusion. The analyses of fluorescence intensity and mRNA transcripts suggest that there are two different exoY promoters and none of them appear to be controlled directly by the ExoR protein or the ExoS/ChvI two-component regulatory system.
2. Materials and methods
2.1. Strains and growth media
S. meliloti Rm1021 (Strr) was used as the wild-type strain [27], and Rm7095 (exoR95) and Rm7096 (exoS96) were used to determine the effect of the exoR95::Tn5 and exoS96::Tn5 mutations on the expression of the exoY gene. Escherichia coli DH5α was used for plasmid constructions and preparations [17], and E. coli MT616 (pRK600, Cmr) was used as a helper in conjugation [27].
Luria–Bertani (LB) medium was used for the growth of E. coli strains, and LB supplemented with 2.5 mM MgSO4 and 2.5 mM CaCl2 (LB/MC) was used for all S. meliloti strains [27]. Z-MGS minimal medium [27] was used to prepare cells for exoY promoter activity analyses. Agar (1.5%) was added to make solid media. Antibiotics were used at the indicated concentrations: chloramphenicol, 10 μg ml−1 ; kanamycin, 25 μg ml−1 ; neomycin, 200 μg ml−1 ; and streptomycin 500 μg ml−1.
2.2. Constructing a set of plasmids with nested deletions of the exoY promoter region
To determine the numbers of the exoY promoter, a set of nested deletions in the exoY promoter region was constructed and fused to an exoY-gfp translational fusion in two steps. A DNA fragment containing the entire exoX-exoY 769 bp intergenic region as the exoY promoter region and the first 282 bp of the exoY open reading frame was generated by PCR (polymerase chain reaction) using primers pexoxy588 and pexoy1610r (Table 1). The PCR cycles were performed as 1 min at 95°C, 1 min at 56°C, and 1–2 min at 72°C for 35 cycles. PCR products were purified according to the protocol of QIAquick PCR purification kit (Qiagen Inc., Valencia, CA, USA). This DNA fragment was digested with restriction enzyme HindIII at 37°C for 2 h to generate a HindIII compatible end at the unique HindIII site at the promoter side of the DNA fragment introduced using the PCR primer pexoxy588 (Table 1). A DNA fragment carrying the gfp open reading frame and its transcriptional terminator with an XhoI restriction enzyme compatible end following the transcription terminator was similarly generated using PCR primers pGFP and pGFPr (Table 1), and XhoI digestion at the unique XhoI site introduced through PCR primer pGFPr (Table 1). The DNA fragment containing exoY promoter and the first half of the exoY gene was ligated with the DNA fragment containing the gfp gene at the blunt end of each of the fragments using T4 DNA ligase (New England Biolabs, Beverly, MA, USA) at 4°C overnight. The ligated DNA fragment with exoY promoter region and exoY-gfp fusion was directionally cloned in between the HindIII and XhoI sites on pMB393 [28] to generate plasmid pHC77. The exoY-gfp fusion can not be expressed by any promoters on the vector itself. To generate the nested deletion of the exoY promoter region, a set of oligo DNA primers pexoxy588, and pXY2-15 that anneal to regions of the exoY promoter that are 50 bp apart were individually paired with the primer pGFPr to generate a set of DNA fragments. The set of DNA fragments were similarly digested with HindIII and XhoI restriction enzymes, cloned in between the HindIII and XhoI sites on pMB393, and transformed into E. coli DH5α to generate a set of plasmids, pHC201–pHC215, containing a set of nested deletions of the exoY promoter in front of the exoY-gfp fusion (Table 2). The plasmid pHC201 is the same as the pHC77 and it was reproduced as a control construction. The plasmids pMB393 and pHC201–pHC215 were extracted from same numbers E. coli DH5α cells carrying the plasmids WizardR Plus SV Minipreps DNA purification system kit (Promega Corporation, Madison, WI, USA) following manufacturer’s instruction. The plasmids were digested with HindIII and XhoI restriction enzymes and resolved on agarose gel using electrophoresis to confirm the nested deletion in the exoY promoter region (data not shown). The similarity of the intensity of the DNA bands on agarose gel suggests that copy numbers of the plasmids were not significantly affected by the difference in the regions of the exoY promoter on the plasmids.
Table 1.
Oligo DNA primers used in the construction of nested deletion of the exoY promoter region fused with the exoY-gfp fusion
| Name | Sequence of primers |
|---|---|
| pexoxy588 | 5′-GGCCGGAAGCTTGGCGCTAACCTACCTTCGGGTC-3′ |
| pexoy1610r | 5′-CTCTTCGAGGACCTCGTCGCC-3′ |
| pGFP | 5′-ATGGCTACGAAAGGAGAAGAACTC-3′ |
| pGFPr | 5′-CCGCTCGAGACCCGTCCTGTGGATATCCGG-3′ |
| Primer XY2 | 5′-ATACCCAAGCTTGGGGCCACTATATTAGCGCCC-3′ |
| Primer XY3 | 5′-ATACCCAAGCTTGGGTTGCAGTCGAGCATACATC-3′ |
| Primer XY4 | 5′-ATACCCAAGCTTGGGTCCATTTCGCACAATTCAA-3′ |
| Primer XY5 | 5′-ATACCCAAGCTTGGGCCGGGGCAGTTTGCCGC-3′ |
| Primer XY6 | 5′-ATACCCAAGCTTGGGTCCCCTAAAATTGCCCGG-3′ |
| Primer XY7 | 5′-ATACCCAAGCTTGGGTGGGGCGTGTGGCCGGC-3′ |
| Primer XY8 | 5′-ATACCCAAGCTTGGGTGAGCGGGTAGCCTCAGC-3′ |
| Primer XY9 | 5′-ATACCCAAGCTTGGGAAAAAAGTGAGGGAAAGTTG-3′ |
| Primer XY10 | 5′-ATACCCAAGCTTGGGCGCTGTCCGTCAGTGCAG-3′ |
| Primer XY11 | 5′-ATACCCAAGCTTGGGCGAAATAACTAGCCCGCG-3′ |
| Primer XY12 | 5′-ATACCCAAGCTTGGGGAACTATCCTAACCCCTG-3′ |
| Primer XY13 | 5′-ATACCCAAGCTTGGGTCGCGACTTTCGCCACC-3′ |
| Primer XY14 | 5′-ATACCCAAGCTTGGGGCCATCATTCCGCCTTCA-3′ |
| Primer XY15 | 5′-ATACCCAAGCTTGGGATGAGCCCGCGTCCCAC-3′ |
Table 2.
Plasmids used in the study
| Plasmid | exoY promoter region | Sources |
|---|---|---|
| pMB393 | none | [28] |
| pHC201 (pHC77) | −769 to −1 | this work |
| pHC202 | −701 to −1 | this work |
| pHC203 | −651 to −1 | this work |
| pHC204 | −591 to −1 | this work |
| pHC205 | −551 to −1 | this work |
| pHC206 | −504 to −1 | this work |
| pHC207 | −454 to −1 | this work |
| pHC208 | −404 to −1 | this work |
| pHC209 | −355 to −1 | this work |
| pHC210 | −304 to −1 | this work |
| pHC211 | −254 to −1 | this work |
| pHC212 | −207 to −1 | this work |
| pHC213 | −154 to −1 | this work |
| pHC214 | −104 to −1 | this work |
| pHC215 | −54 to −1 | this work |
2.3. Measuring exoY promoter activities
The level of average GFP fluorescence intensity per cell (‘specific GFP expression’ for short) was determined and used to represent the activities of the exoY promoters since the copy numbers of the plasmids with different parts of the exoY promoter region appear to be similar. To determine the specific GFP expression, S. meliloti cells were collected from LB/MC liquid cultures, washed, resuspended to OD600 0.1 in Z-MGS media, and incubated with shaking at 30°C for two more days. Z-MGS cultures were diluted 1:10, and transferred to wells in a transparent 96-well plate and wells in a black 96-well plate in equal amounts. The cultures in transparent 96-well plates were used to determine the cell density using an absorbance microplate reader (Spectra Max 340PC, Molecular Device, Sunnyvale, CA, USA) and the cultures in black 96-well plates were used to determine the intensity of GFP fluorescence using a fluorescence microplate reader (Spectra Max Germini XS, Molecular Device, Sunnyvale, CA, USA). Both cell density and GFP fluorescence intensity of individual culture were collected and analyzed using a computer program (SpectroSoft, Molecular Device, Sunnyvale, CA, USA). The GFP fluorescence intensity of a culture was normalized to its cell density to generate the average specific GFP expression. The specific GFP expression was used to represent the exoY promoter activities.
2.4. Detecting transcripts from both exoY promoters
Three special oligo DNA primers were designed to detect the transcripts from the exoY promoters on the plasmid carrying the set of nested deletions of the exoY promoter using RT-PCR (reverse transcription-PCR) (Fig. 2A). Primer 1 (5′-TTTGTATAGTTCATCCATGCC-3′) anneals to the end of the gfp open reading frame on the promoter fusion plasmids and it was used to synthesize the first strand of cDNA based on the RNA transcripts in RT reactions (Fig. 2A). Because the gfp gene is only carried on the plasmid, using a primer that anneals only to the gfp gene, makes it possible to measure the impact of the nested deletions on the expression of the exoY gene from the plasmids in different genetic backgrounds. Primer 2(5′-ATGGCT AGCAAAGGAGAAGA-3′) anneals to a region just downstream of the PexoYdown promoter and primer 3 (5′-TGAGCGGGTAGCCTCAGC-3′) anneals to just downstream of the PexoYup promoter. Total RNA was extracted from the wild-type strain Rm1021 carrying plasmids pHC201 (both promoters), pHC207 (both promoters), pHC208 (PexoYdown promoter), pHC214 (PexoYdown promoter), and pHC215 (no promoter) using RiboPure -Bacteria kit (Ambion Inc. Austin, TX, USA) following manufacturer’s instructions. Briefly, the first strand cDNA synthesis was carried out by mixing MMLV-RT with 2.0 μg total RNA in a final volume of 20 μl. After RT reaction, 2 μl of reaction mixture was used as a template for PCR. Both the minus-RT and the minus-template were used as negative controls for the PCRs to ensure the total RNA used free of DNA. The total RNAs were mixed with primer 1 for RT to generate the cDNA based on the transcript(s). Primers 1 and 2 were added together to the RT reactions for the subsequent PCRs. The cDNA copy of the short transcripts from the PexoYdown promoter can be amplified by primers 1 and 2 in a PCR. The cDNA copy of the long transcript from the PexoYup promoter can be amplified by primers 1 and 3 in a PCR. The RT-PCR products were purified and resolved on agarose gel (Fig. 2B).
-Bacteria kit (Ambion Inc. Austin, TX, USA) following manufacturer’s instructions. Briefly, the first strand cDNA synthesis was carried out by mixing MMLV-RT with 2.0 μg total RNA in a final volume of 20 μl. After RT reaction, 2 μl of reaction mixture was used as a template for PCR. Both the minus-RT and the minus-template were used as negative controls for the PCRs to ensure the total RNA used free of DNA. The total RNAs were mixed with primer 1 for RT to generate the cDNA based on the transcript(s). Primers 1 and 2 were added together to the RT reactions for the subsequent PCRs. The cDNA copy of the short transcripts from the PexoYdown promoter can be amplified by primers 1 and 2 in a PCR. The cDNA copy of the long transcript from the PexoYup promoter can be amplified by primers 1 and 3 in a PCR. The RT-PCR products were purified and resolved on agarose gel (Fig. 2B).
Fig. 2.
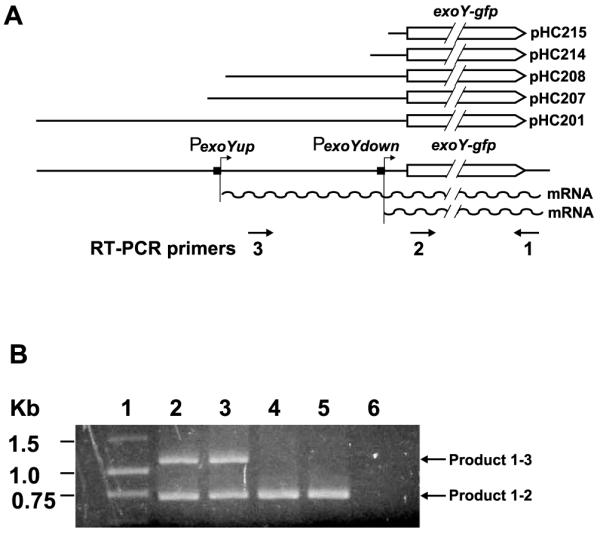
Schematic representations showing the positions of the RT-PCR primers in the exoY promoter region in A and a picture showing the RT-PCR products in B. A: Line graphs showing the exoY promoter region, the positions of proposed exoY promoters, the predicted transcripts, the areas the RT-PCR primer anneals to, and the exoY promoter area in the plasmids used in the study. B: A picture of agarose gel showing DNA size standard (lane 1), RT-PCR products from cells carry plasmid pHC201 (lane 2), pHC207 (lane 3), pHC208 (lane 4), pHC214 (lane 5), and pHC215 (lane 6).
3. Results and discussion
3.1. Locating exoY promoters and determining their activities in free-living cells
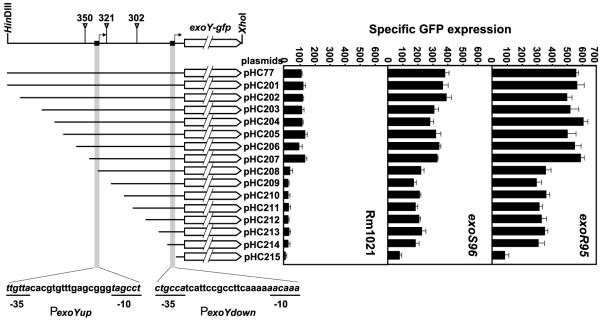
To identify the location of the putative exoY promoters, the set of plasmids (pHC201–pHC215) carrying nested deletions of the exoY promoter region (Table 2) were introduced into the wild-type strain Rm1021 cells by conjugation. The levels of average specific GFP expression were determined for each Rm1021 carrying different plasmids in three independent experiments to determine the levels of the exoY gene expression (Fig. 1).
Fig. 1.
Schematic representation of nested deletions in the exoY promoter region and the effects of the deletions on the expression of the exoY gene in the wild-type, exoR95, and exoS96 mutant backgrounds. A line graph is used to show the exoY and exoX genes, and the positions of three transposon insertions. The positions of the proposed exoY promoters are marked by short solid bars and arrows. The solid lines are used to represent the regions of the exoY promoter carried on each of the plasmids. Two gray vertical bars represent the relative position of the proposed exoY promoters. The sequences of the proposed exoY promoters are shown at the bottom of the bars. The promoter activities of the set of plasmids carrying nested deletions in wild-type Rm1021, exoR95, and exoS96 mutant backgrounds are shown in three different bar graphs.
The level of the specific GFP expression by Rm1021 carrying the plasmid pHC215 with the first 54 bp of the promoter, was the same as Rm1021 carrying the original vector pMB393 (data not shown). This suggests that the exoY-gfp gene fusion was not expressed in Rm1021 (pHC215) cells, which also suggests that the first 54 bp of the exoY promoter region does not contain active exoY promoter. The level of specific GFP expression of the Rm1021 (pHC214) was consistently higher than that of Rm1021 (pHC215), which suggests that the first 104 bp of the exoY promoter region contains a complete exoY promoter that expressed the exoY-gfp fusion. The levels of the specific GFP expression were about the same for the Rm1021 (pHC214) and Rm1021 (pHC213), which suggests that there is no additional sequence required for the exoY promoter on the pHC214 plasmid. Further analysis of the −1 to −153 region of the exoY promoter in the pHC214 using a promoter prediction program (http://www.fruitfly.org/seq_tools/promoter.html), as well as direct comparison of the sequences suggest that the downstream exoY promoter, PexoYdown, is most likely located in the −78 to −104 region of the exoY promoter region (Fig. 1).
The levels of the specific GFP expression by Rm1021 cells carrying the plasmids pHC208–pHC214 were about the same, suggesting there was no additional complete exoY promoter in the −105 to −404 exoY promoter region. The level of the specific GFP expression by Rm1021 (pHC207) was consistently higher than that of Rm1021 (pHC208), suggesting there is another exoY promoter in the −356 to −454 region of the exoY promoter region. Further analysis suggested that this upstream exoY promoter, PexoYup, is most likely located in the −395 to −425 region (Fig. 1). The plasmid pHC208 carries only the −10 region of the PexoYup promoter so it was not expressing exoY gene from the PexoYup promoter.
The levels of the specific GFP expression by Rm1021 cells carrying plasmids pHC207–pHC201 were similar, which suggests that there is no additional exoY promoter in the −426 to −769 region.
All together, these results suggest that the exoY gene can be expressed from two different promoters: an upstream PexoYup promoter and a downstream PexoYdown promoter. The proposed locations of the exoY promoters provide a perfect explanation of the previous transposon insertion analysis of the exoY promoter region [22]. The insertion of Tn5 transposon in the Ω302 and Ω321 mutants (Fig. 1) blocks the exoY expression from the PexoYup promoter but not the PexoYdown promoter in both exoR95 and exoS96 mutant backgrounds so that the colonies appear dim but not dark. This is because the expression of the exoY gene from the PexoYdown promoter was sufficient to support succinoglycan biosynthesis. The Tn5 insertion in the Ω351 mutant was upstream of both exoY promoters, so it can not suppress the overexpression of the exoY gene in neither exoR395 nor exoS396 mutant backgrounds. The exoR395Ω351 and exoS395Ω351 double mutants continue to overproduce succinoglycan.
3.2. Detecting transcription products of the two exoY promoters
Analyses of the nested deletions of the exoY promoter region suggested there were two exoY promoters, which should produce two different transcripts, one long transcript and one short transcript. RT-PCR and three specially designed primers (Fig. 2A) were used to detect the possible transcripts from the exoY promoter as described in details in Section 2. This will allow the simultaneous detection of both long and short exoY-gfp transcripts.
As shown in Fig. 2B, no DNA fragment was detected on the agarose gel when total RNA from Rm1021 (pHC215) was used as the template for RT-PCR. This suggests that there was no transcript produced and that the first 54 bp of the exoY promoter region does not include an active exoY promoter. The fact that no nonspecific DNA fragments were detected also confirmed the specificity of the primers 1, 2, and 3. A 0.7 kb DNA fragment was detected when total RNA from Rm1021 (pHC214) was used as the template. The size of this fragment matches the distance between the sites that primers 1 and 2 anneal to. This suggests that the short transcript of the exoY-gfp gene was produced by Rm1021 (pHC214). This is consistent with the suggestion that the −78 to −104 region contains the PexoYdown promoter. The 0.7 kb DNA fragment was detected again when total RNA from Rm1021 (pHC208) was used as template. This suggests the −1 to −404 region of the exoY promoter contains only the PexoYdown promoter. An additional 1.1 kb DNA fragment was detected when total RNA from either Rm1021 (pHC201) or Rm1021 (pHC207) was used as the template. The size of the 1.1 kb fragment matches the distance between the sites that primers 1 and 3 anneal to. This suggests it was amplified from the long transcript expressed from the PexoYup promoter. These results suggest that the exoY promoter region (−1 to −454) on the plasmid 207 contains another active exoY promoter, which is consistent with the proposed location of the PexoYup promoter.
3.3. Both exoY promoters are induced by the exoR95 and exoS96 mutations
The expression of the exoY gene is upregulated in exoR95 and exoS96 mutant backgrounds [22]. This could be the result of the induction of one or both of the PexoYdown and PexoYup promoters. It is also possible that there are additional exoY promoters that are only active in the exoR95 and exoS96 mutant backgrounds. To examine all these possibilities, the entire set of the plasmids carrying the nested deletions of the exoY promoter was introduced into the S. meliloti strains Rm7095 (exoR95 mutant) and Rm7096 (exoS96 mutant) cells by conjugation. The specific GFP expression was determined for each of strains carrying one of the plasmids (Fig. 1). In the exoR95 mutant background, the specific GFP expression was 80 for Rm7095 (pHC215), 330 for Rm7095 carrying plasmids pHC208–pHC214, and 550 for Rm7095 carrying plasmids pHC77, and pHC201–pHC207. In the exoS96 mutant background, the specific GFP expression was 70 for Rm7096 (pHC215), 200 for Rm7096 carrying plasmids pHC208–pHC214, and 340 for Rm7096 carrying plasmids pHC77, and pHC201–pHC207. The exoY gene was expressed at two levels in both exoR95 and exoS96 mutant background as measured by specific GFP expression, which is similar to that in the wild-type background. The fact that the exoY gene was expressed in two levels by the same plasmids suggests that the exoY gene was expressed from the same two promoters in the exoR95 and exoS96 mutant backgrounds as in the wild-type background. The two levels of the exoY gene expression in the exoR95 and exoS96 mutant backgrounds also suggest that there is no additional exoY promoter that is regulated directly by either ExoR protein or ExoS/ChvI two-component regulatory system.
Further analysis of the levels of the exoY gene expression in different background also suggests that the expression is the highest in the exoR95 mutant background and second highest in the exoS96 mutant background, which is consistent with previous analyses based on lacZ and phoA fusions [14,15]. What is also interesting is that the expressions from PexoYup and PexoYdown promoters were both upregulated in both exoR95 and exoS96 mutant backgrounds. The upregulation of transcription is often the results of the specific interactions between transcriptional regulators and specific DNA sequence elements. Similar DNA sequences were often found around the promoters that are similarly regulated, but no obviously similar DNA sequences or elements were found around PexoYup and PexoYdown promoters. This raises the possibility that other unknown protein factors are directly involved in regulating exoY gene expression, and that neither ExoR protein nor ExoS/ChvI two-component regulatory system directly interact with the exoY promoters. The signals received by either ExoR protein or ExoS/ChvI system are transmitted downstream to other proteins in regulating succinoglycan biosynthesis. This model is consistent with our recent findings that both ExoR protein and the ExoS/ChvI system are involved in regulating flagella biosynthesis in addition to succinoglycan biosynthesis (Cheng, H.-P., manuscript submitted).
The finding of two inducible exoY promoters is consistent with our preliminary results that the expression of the exoY gene was upregulated in cells inside infection threads (Cheng, H.-P., unpublished results). The identification of two exoY promoters and the construction of the nested promoter deletion fused to exoY-gfp fusion will facilitate further analyses of the regulation of the succinoglycan biosynthesis gene expression during symbiosis and it will also provide better understanding of the role of succinoglycan and other bacterial exopolysaccharides in microbeplant interactions.
Acknowledgements
This work was supported by grants from NIH (5S06GM08225) and PSC-CUNY (617320030 and 632140032) to H.-P.C.
References
- [1].van Rhijn P, Vanderleyden J. The Rhizobium–plant symbiosis. Microbiol. Rev. 1995;59:124–142. doi: 10.1128/mr.59.1.124-142.1995. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [2].Brewin NJ. The Rhizobium–legume symbiosis: plant morphogenesis in a nodule. Semin. Cell Biol. 1993;4:149–156. doi: 10.1006/scel.1993.1018. [DOI] [PubMed] [Google Scholar]
- [3].Broughton WJ, Perret X. Genealogy of legume–Rhizobium symbioses. Curr. Opin. Plant Biol. 1999;2:305–311. doi: 10.1016/S1369-5266(99)80054-5. [DOI] [PubMed] [Google Scholar]
- [4].Long SR. Rhizobium–legume nodulation: life together in the underground. Cell. 1989;56:203–214. doi: 10.1016/0092-8674(89)90893-3. [DOI] [PubMed] [Google Scholar]
- [5].Kijne JW. In: Biological Nitrogen Fixation. Stacey G, Burris RH, Evans HJ, editors. Chapman&Hall; New York: 1992. pp. 349–398. [Google Scholar]
- [6].Downie JA, Walker SA. Plant responses to nodulation factors. Curr. Opin. Plant Biol. 1999;2:483–489. doi: 10.1016/s1369-5266(99)00018-7. [DOI] [PubMed] [Google Scholar]
- [7].Cheng HP, Walker GC. Succinoglycan is required for initiation and elongation of infection threads during nodulation of alfalfa by Rhizobium meliloti. J. Bacteriol. 1998;180:5183–5191. doi: 10.1128/jb.180.19.5183-5191.1998. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [8].Gage DJ, Margolin W. Hanging by a thread: invasion of legume plants by rhizobia. Curr. Opin. Microbiol. 2000;3:613–617. doi: 10.1016/s1369-5274(00)00149-1. [DOI] [PubMed] [Google Scholar]
- [9].van Brussel AAN, Bakhuizen R, van Spronsen PC, Spaink HP, Tak T, Lugtenberg BJJ, Kijne JW. Induction of pre-infection thread structures in the leguminous host plant by mitogenic lipo-oligosaccharides of Rhizobium. Science. 1992;257:70–72. doi: 10.1126/science.257.5066.70. [DOI] [PubMed] [Google Scholar]
- [10].Timmers AC, Auriac MC, Truchet G. Refined analysis of early symbiotic steps of the Rhizobium–Medicago interaction in relationship with microtubular cytoskeleton rearrangements. Development. 1999;126:3617–3628. doi: 10.1242/dev.126.16.3617. [DOI] [PubMed] [Google Scholar]
- [11].Walker SA, Downie JA. Entry of Rhizobium leguminosarum bv. viciae into root hairs requires minimal nod factor specificity, but subsequent infection thread growth requires nodO or nodE. Mol. Plant-Microbe Interact. 2000;13:754–762. doi: 10.1094/MPMI.2000.13.7.754. [DOI] [PubMed] [Google Scholar]
- [12].Reinhold BB, Chan SY, Reuber TL, Marra A, Walker GC, Reinhold VN. Detailed structural characterization of succinoglycan, the major symbiotically important exopolysaccharide of Rhizobium meliloti strain Rm1021. J. Bacteriol. 1994;176:1997–2002. doi: 10.1128/jb.176.7.1997-2002.1994. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [13].Reuber TL, Walker GC. Biosynthesis of succinoglycan, a symbiotically important exopolysaccharide of Rhizobium meliloti. Cell. 1993;74:269–280. doi: 10.1016/0092-8674(93)90418-p. [DOI] [PubMed] [Google Scholar]
- [14].Reuber TL, Long S, Walker GC. Regulation of Rhizobium meliloti exo genes in free-living cells and in planta examined using TnphoA fusions. J. Bacteriol. 1991;173:426–434. doi: 10.1128/jb.173.2.426-434.1991. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [15].Doherty D, Leigh JA, Glazebrook J, Walker GC. Rhizobium meliloti mutants that overproduce the Rhizobium meliloti acidic calcofluor-binding exopolysaccharide. J. Bacteriol. 1988;170:4249–4256. doi: 10.1128/jb.170.9.4249-4256.1988. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [16].Reed JW, Glazebrook J, Walker GC. The exoR gene of Rhizobium meliloti affects RNA levels of other exo genes but lacks homology to known transcriptional regulators. J. Bacteriol. 1991;173:3789–3794. doi: 10.1128/jb.173.12.3789-3794.1991. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [17].Cheng HP, Walker GC. Succinoglycan production by Rhizobium meliloti is regulated through the ExoS-ChvI two-component regulatory system. J. Bacteriol. 1998;180:20–26. doi: 10.1128/jb.180.1.20-26.1998. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [18].Becker A, Kleickmann A, Keller M, Arnold W, Puhler A. Identification and analysis of the Rhizobium meliloti exoAMONP genes involved in exopolysaccharide biosynthesis and mapping of promoters located on the exoHKLAMONP fragment. Mol. Gen. Genet. 1993;241:367–379. doi: 10.1007/BF00284690. [DOI] [PubMed] [Google Scholar]
- [19].Glucksmann MA, Reuber TL, Walker GC. Genes needed for the modification, polymerization, export, and processing of succinoglycan by Rhizobium meliloti: a model for succinoglycan biosynthesis. J. Bacteriol. 1993;175:7045–7055. doi: 10.1128/jb.175.21.7045-7055.1993. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [20].Glucksmann MA, Reuber TL, Walker GC. Family of glycosyl transferases needed for the synthesis of succinoglycan by Rhizobium meliloti. J. Bacteriol. 1993;175:7033–7044. doi: 10.1128/jb.175.21.7033-7044.1993. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [21].Müller P, Weng MWM, Quandt J, Arnold W, Pühler A. Genetic analysis of Rhizobium meliloti exoYFQ operon: ExoY is homologous to sugar transferases and ExoQ represents a transmembrane protein. Mol. Plant-Microbe Interact. 1993;6:55–65. doi: 10.1094/mpmi-6-055. [DOI] [PubMed] [Google Scholar]
- [22].Reed JW, Capage M, Walker GC. Rhizobium meliloti exoG and exoJ mutations affect the exoX-exoY system for modulation of exopolysaccharide production. J. Bacteriol. 1991;173:3776–3788. doi: 10.1128/jb.173.12.3776-3788.1991. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [23].Reed JW, Walker GC. The exoD gene of Rhizobium meliloti encodes a novel function needed for alfalfa nodule invasion. J. Bacteriol. 1991;173:664–677. doi: 10.1128/jb.173.2.664-677.1991. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [24].Bertram-Drogatz PA, Quester I, Becker A, Puhler A. The Sinorhizobium meliloti MucR protein, which is essential for the production of high-molecular-weight succinoglycan exopolysaccharide, binds to short DNA regions upstream of exoH and exoY. Mol. Gen. Genet. 1998;257:433–441. doi: 10.1007/s004380050667. [DOI] [PubMed] [Google Scholar]
- [25].Swanson JA, Mulligan JT, Long SR. Regulation of syrM and nodD3 in Rhizobium meliloti. Genetics. 1993;134:435–444. doi: 10.1093/genetics/134.2.435. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [26].Barnett MJ, Swanson JA, Long SR. Multiple genetic controls on Rhizobium meliloti syrA, a regulator of exopolysaccharide abundance. Genetics. 1998;148:19–32. doi: 10.1093/genetics/148.1.19. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [27].Leigh JA, Signer ER, Walker GC. Exopolysaccharide-deficient mutants of Rhizobium meliloti that form ineffective nodules. Proc. Natl. Acad. Sci. USA. 1985;82:6231–6235. doi: 10.1073/pnas.82.18.6231. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [28].Gage DJ, Bobo T, Long SR. Use of green fluorescent protein to visualize the early events of symbiosis between Rhizobium meliloti and alfalfa (Medicago sativa) J. Bacteriol. 1996;178:7159–7166. doi: 10.1128/jb.178.24.7159-7166.1996. [DOI] [PMC free article] [PubMed] [Google Scholar]