Abstract
Tip60 is a key member of the MYST family of histone acetyltransferases and involved in a broad spectrum of cellular pathways and disease conditions. So far, small molecule inhibitors of Tip60 and other members of MYST HATs are rarely reported. To discover new small molecule inhibitors of Tip60 as mechanistic tools for functional study and as chemical leads for therapeutic development, we performed virtual screening using the crystal structure of Esa1 (the yeast homolog of Tip60) on a small molecule library database. Radioactive acetylation assays were carried out to further evaluate the virtual screen hits. Several compounds with new structural scaffolds were identified with micromolar inhibition potency for Tip60 from the biochemical studies. Further, computer modeling and kinetic assays suggest that these molecules target the acetyl-CoA binding site in Tip60. These new inhibitors provide valuable chemical hits to develop further potent inhibitors for the MYST HATs.
Keywords: Tip60, MYST, HAT, Histone, Inhibitor
1. Introduction
The 60-kDa HIV Tat-interacting protein (Tip60), one of the founding members of the MYST (MOZ, Ybf2/Sas3, Sas2, Tip60) family of histone acetyltransferases (HATs), was originally identified as an HIV-1 Tat associating protein by yeast two-hybrid screen [1]. It catalyzes the transfer of acetyl groups from acetyl-CoA to specific lysine residues on the N-terminal tail of nucleosomal core histones. The acetylation of histones results in charge neutralization of lysine residues and decreases the affinity between histones and nucleic acids, and leads to relaxed open chromatin structures [2]. Tip60 is recruited by many key transcription factors to the chromatin template and aberrant Tip60 activity is involved in several types of human diseases such as neurodegeneration and cancer [3–5]. In the pathogenesis of Alzheimer’s disease, the intracellular C-terminal part of β-amyloid precursor protein (APP) associates with the APP-binding protein Fe65 and Tip60, leading to apoptosis and downstream gene activation in parallel with the pathway of the amyloid β-peptide release [6,7]. Research has shown that Tip60 is intimately linked to oncogenesis [8,9]. Oncogenic transcription factors including c-Myc [10], E2F [11], and nuclear factor-κB [12–14] associate with and are coactivated by Tip60. The protein level of Tip60 is substantially high and contributes to the abnormal HAT activity in ODC/Ras tumors [15,16]. In prostate cancer, Tip60 is particularly involved in the progression to the androgen-refractory state [17,18]. Tip60 is upregulated and predominantly resides in the nucleus of hormone resistant prostate cancer cells and tissue specimens from patients who failed endocrine therapy [17]. Importantly, in hormone resistant prostate cancer cells, Tip60 is constitutively recruited to androgen response elements such as prostate-specific antigen (PSA) promoter and enhances PSA expression even in the absence of androgen [17]. Therefore, upregulation of Tip60 may provide a mechanistic explanation why androgen receptor-regulated genes become expressed when the tumor relapses. These observations highlight the significance of Tip60 as a potential pharmacological target in disease therapy.
In light of the significance of HAT inhibitors as potential new therapeutics, quite a few efforts have been invested on developing small molecule inhibitors of HAT enzymes [19]. Most of the developed inhibitors target robust HATs p300/CBP and PCAF/GCN5 [20–24]. On the other hand, inhibitors of the class of MYST HATs are rarely reported. Balasubramanyam et al. [25] reported a naturally occurring HAT inhibitor, anacardic acid, from cashew nut shell liquid. It works as weak nonspecific inhibitors of p300/CBP (CREB-binding protein), and PCAF (p300/CBP-associated factor), and MYST HAT Tip60, and is capable of easily permeating the cells in culture. Curcumin (diferuloylmethane), an ingredient from the plant Curcuma longa rhizome, was reported to inhibit the HAT activity in general [26]. Both curcumin and anacardic acid have been shown to have many non-HAT targets. For example, Shankar and Srivastava [27] showed that curcumin induces apoptosis in prostate cancer cells through activation of multiple signaling pathways, including induction of expression of proapoptotic proteins Bax, Bak, PUMA, Noxa, and Bim, and inhibition of expression of antiapoptotic proteins Bcl-2 and Bcl-XL. Anacardic acid was previously demonstrated as an inhibitor of DNA polymerase β [28]. To develop inhibitors more specifically targeting the MYST family of HATs, our group recently reported substrate-based analog compounds for Tip60 inhibition [29]. Although they present good inhibition activities, the negative charges due to the presence of CoA motif imply that this type of inhibitors may have low pharmacokinetic performance in vivo [30]. To further develop potent inhibitors of MYST HATs with enhanced pharmacological properties, in this work, we have conducted a virtual screening based on the crystal structure of Esa1 (the yeast homolog of Tip60) to search for small molecule inhibitors. In combination with biochemical inhibition studies, several micromolar inhibitors are discovered.
2. Materials and methods
2.1. Materials
Small molecule compounds were purchased from ChemBridge Corporation. Peptides were synthesized using Fmoc-based solid phase methodology. Fmoc-protected amino acids and solid phase resins were purchased from NovaBiochem. [14C]-labeled acetyl-CoA was purchased from Perkin Elmer. Tip60 recombinant protein was expressed as previously described [29].
2.2. Virtual screening
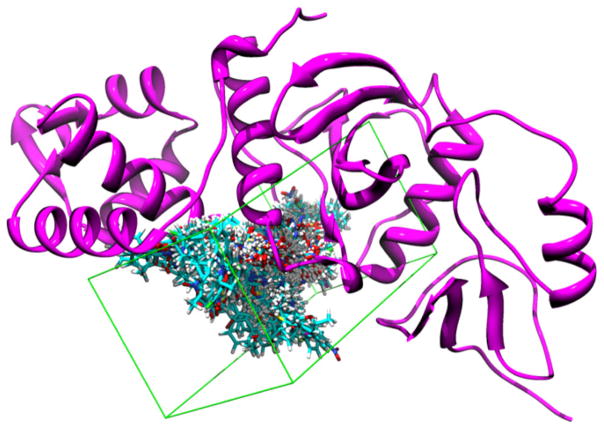
Docking-based virtual screening was conducted by following similar procedures reported earlier [31,32]. Compounds from the ChemBridge database were converted into 3D structures using the CONCORD program [33]. The 3D structures of the compounds had hydrogen atoms added and were assigned AM1-BCC partial charges [34–36]. Esa1 crystal structure (PDB entry: 1FY7) [37] was added hydrogen atoms and then assigned Kollman-all charges with the SYBYL 7.1 program. Residues within a radius of 6 Å around the center of the CoA binding in the Esa1 structure were defined as the active site to construct a grid for the virtual screening. The position and conformation of each compound were minimized by the anchor fragment orientation as well as by the torsion minimization method implemented in the DOCK 6.0 program [38]. Fifty conformations and a maximum of 100 anchor orientations for each compound were generated, and the binding energy of all the docked conformations were minimized by 100 iterations using the standard approach as described in the literature [38]. The docked molecules were ranked based on the sum of the van der Waals contacts and electrostatic energies to obtain the top 1000 compounds. After collecting the top hits, re-analysis of virtual screening results was conducted using drug-like property criteria [39] by the FILTER 2.0.1 software [40]. We then performed consensus scoring evaluation [41] by ChemScore [42,43], PLP [44], ScreenScore [45], Chem-Gauss and ShapeGauss [46] implemented in the FRED 2.2.3 software [40], as well as hydrogen bond and hydrophobic profiles checked by the IDEA 8.8 software [47]. As the final step, a manual binding orientation and conformational analysis was performed to come up with the final 76 hits for biological evaluation (Fig. 1).
Fig. 1.
Docking conformations of the 76 virtual hits (blue sticks) around the binding site (green box) of ESA1 (pink ribbons). (For interpretation of the references to colour in this figure legend, the reader is referred to the web version of this article.)
2.3. Radioactive HAT inhibition assay
Radioisotope-labeled HAT assay was carried out at 30 °C in a reaction volume of 30 μL. The reaction buffer contained 50 mM HEPES (pH 8.0), 0.1 mM EDTA, 50 μg/mL BSA, 10% DMSO, and 1 mM dithiothreitol (DTT). Typically, [14C] acetyl-CoA was used as the acetyl donor and a peptide containing the N-terminal 20-amino acid sequence of histone H4, namely H4-20, was used as the HAT substrate. The reaction was initiated with the HAT enzyme after the other components (acetyl-CoA, H4-20, and inhibitor) were equilibrated at 30 °C for 5 min. Rate measurements were based on initial conditions (generally less than 10% consumption of the limiting substrate). After the reaction, the mixture was loaded onto a Waterman P81 filter paper, dried 30 min, and then washed with 50 mM of sodium bicarbonate (pH 9.0) for two times (10 min each). The paper was air-dried and the amount of radioactivity incorporated into the peptide substrate was quantified by liquid scintillation. For kinetic inhibition pattern analysis of compound a, initial velocities of Tip60 were measured at a range of varied concentrations of AcCoA, a fixed concentration of H4-20 (100 μM), and selected concentrations of inhibitor a, i.e., 0 μM, 80 μM, and 150 μM. The data were displayed in both Michaelis–Menten and Lineweaver–Burk plots. The data points were fitted to the equation which describes noncompetitive inhibition to calculate slope inhibition constant Kis and intercept inhibition constant Kii [29]. In all cases, background acetylation (in the absence of enzyme) was subtracted from the total signals. All the assays were performed at least twice, and duplicates generally agreed within 20%.
2.4. Docking study of the identified hits
For ligand–protein docking, the 3-dimensional structures of hit compounds were constructed in Maestro 9.0.211. The Tip60 PDB file was modified in Maestro 9.0.211 to add hydrogen atoms and remove water molecules and the acetyl-CoA from tip60. AutoDock-Tools4.2 was employed to dock the compounds into Tip60. PDBQT files were generated in AutoDockTools for both of the ligand and the macromolecule for docking. AutoGrid was used to generate a grid box for docking process. AutoDock was then used to dock selected hits individually into Tip60. The Genetic Algorithm with 2500,000 maximum number of evals and 50 generations for picking individuals were used as the docking parameters. Structural analysis with PyMOL was performed following the docking process.
3. Results and discussion
3.1. Virtual screening
Potent small molecule inhibitors are of great value as chemical probes for interrogating cellular functions of biological targets. Tip60 is an important MYST HAT and engaged in multiple cellular pathways [5]. However, there is no report on specific inhibitors of Tip60 so far. We performed a virtual screening with the attempt to search for Tip60 inhibitors from the ChemBridge small molecule collection (about half million compounds) based on the reported crystal structure of Esa1 (the yeast homolog of Tip60) [37,48]. In the virtual screening scheme, the 2D structures of each of these organic molecules are converted into 3D structures and assigned atomic charges with the AM1-BCC protocol [34,35]. Then the molecular docking program, DOCK6.0, is used to perform the initial high-throughput virtual screening of these compounds in binding to Esa1. Residues within a radius of 6 Å around the center of CoA binding in the Esa1 structure were defined as the active site to construct a grid for the virtual screening. After collecting the top hits, a re-analysis of the initial hits, including drug-like properties [39], consensus scoring evaluation [41], absorption, distribution, metabolism, excretion and toxicity (ADMET) prediction [49], hydrogen bond, and hydrophobic profiles and binding orientation examination, was performed. From the virtual screening and analysis, 76 top hits were selected and subjected to experimental tests for their ability to inhibit the HAT activity of Tip60 (Fig. 1).
3.2. Biochemical tests of the virtual screen hits
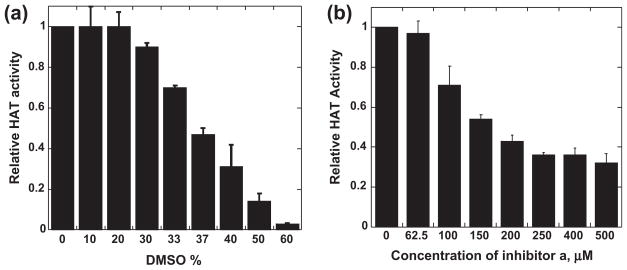
To evaluate the activities of the 76 virtual screen hits for Tip60 inhibition, we carried out histone acetylation with the classic radio-isotope-labeled approach. [14C] acetyl-CoA was used as the acetyl donor and a peptide containing the N-terminal 20 amino acid sequence of histone H4 (H4-20) was used as the Tip60 substrate. The HAT reaction catalyzed by recombinant Tip60 was carried out in the presence and absence of individual virtual screen hits to quantitatively evaluate their inhibition potency. All the compounds were dissolved in DMSO to make a 5-mM stock solution. Because many compounds have poor solubility in the aqueous assay buffer, DMSO had to be present in the acetylation assay buffer. There is a concern that DMSO may affect the enzymatic activity of Tip60. Thus, we first tested the tolerance of Tip60 towards different concentrations of DMSO. As can be seen from Fig. 2a, in the range of 0–20% DMSO (v/v), there is almost no effects observed on Tip60 activity. Even at 30% of DMSO, the retained activity of Tip60 is still 90%. The IC50 of DMSO from this test is 37%. We found that in the presence of 10% DMSO in the assay buffer the individual hit compounds were completely dissolved at 100-μM level. Under this condition, the influence of DMSO on Tip60 activity is negligible.
Fig. 2.
Characterization of chemical inhibition of Tip60 activity. (a) Effect of DMSO on the enzymatic activity of Tip60. (b) Effect of compound a on the enzymatic activity of Tip60. The reactions contain 100 μM of H4-20 and 10 μM of [14C]acetyl-CoA.
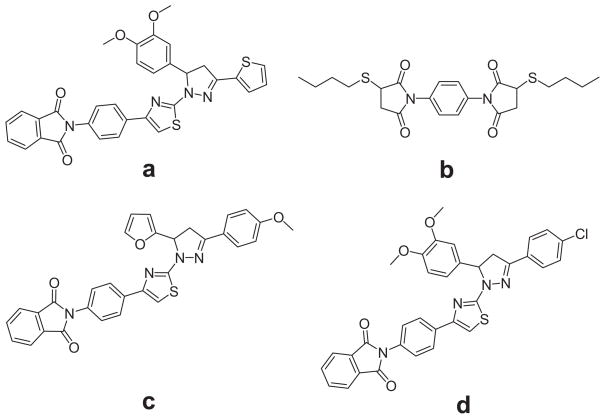
A typical inhibition screen assay was carried out with a reaction mixture containing 0.1 μM of recombinant Tip60, 10 μM of [14C]-labeled acetyl-CoA, 100 μM of H4-20, and 100 μM of individual compounds. The retained fractional activity of Tip60, namely the enzymatic activity in the presence of a hit divided by that in the absence of the hit, was used to quantitatively evaluate the potency of hit compounds. It is notable to mention that, for all the enzymatic assays, the acetylation reaction is maintained under initial condition so that the reaction yields of the limiting substrates are lower than 10%, which is to ensure that the concentrations of acetyl-CoA and the peptide substrate remained constant over the time course of acetylation reaction. Identified hits can be further characterized by its IC50 value. For IC50 measurement, the activity of Tip60 was measured at varied concentrations of inhibitor, and IC50 value is defined as the inhibitor concentration at which 50% of enzyme activity was blocked. A typical activity-concentration titration curve for IC50 measurement was shown in Fig. 2b. From the experimental tests, we found two compounds that possess inhibition activities, i.e. a and b (Fig. 3). Their IC50 values are 149 μM and 181 μM, respectively. In order to look for analogous potent inhibitors, we performed similarity search on the Chem-Bridge database and selected 29 additional compounds that bear structural similarities to compounds a and b. The inhibition activities of these compounds toward Tip60 were also analyzed using the radioactive biochemical assay. From this study, two more inhibitors were found, i.e., c and d (Fig. 3). These two compounds are structural analogs of compound a, confirming that the core structure in the three compounds is a valid structural framework for Tip60 inhibition. For the analogs of compound b, we did not identify additional inhibitors. The inhibition data of the four inhibitors are listed in Table 1.
Fig. 3.
Structures of the identified Tip60 inhibitors.
Table 1.
IC50 of the identified Tip60 inhibitors. Experimental condition: H4-20, 100 μM; [14C]acetyl-CoA, 10 μM; Tip60, 200 nM; and reaction time, 10 min.
| Inhibitors | IC50 (μM) |
|---|---|
| a | 149 ± 36 |
| b | 181 ± 60 |
| c | 400 ± 30 |
| d | 240 ± 15 |
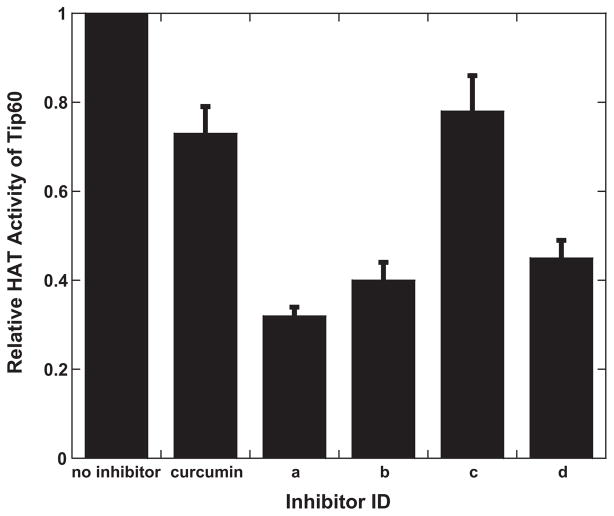
We compared the activities of the four discovered inhibitors with curcumin, a natural product which was reported as a generic HAT inhibitor [26,50]. The data showed that inhibitors a, b and d are more potent than curcumin and compound c has similar activity as curcumin (Fig. 4). Furthermore, we examined whether the inhibition by these new compounds was specific for Tip60. Thus, we tested the inhibition by compound a on several other HAT enzymes, i.e. Esa1, p300, and PCAF. p300 and PCAF belong to different HAT families. Esa1 and Tip60 are both MYST members. H4-20 was used as substrate for Esa1 and p300 as well. For PCAF catalysis, the histone H3 N-terminal 20-aa tail peptide (i.e., H3-20) was used. The data are listed in Table 2 and show that compound a inhibits all the tested HAT enzymes at similar levels, implicating that the inhibition is not specific for MYST HATs. A computational study (see the next) offers structural insight about the nonspecific inhibition by these inhibitors.
Fig. 4.
Inhibition of Tip60 by the identified inhibitors and curcumin at 250 μM. The reaction mixture contains 100 μM of H4-20, 10 μM of [14C]acetyl-CoA, and 200 nM of Tip60. The reaction time is 10 min.
Table 2.
Comparison of the inhibition of Tip60, Esa1, p300, and PCAF by compound a. Tip60 concentration was 200 nM and reaction time was 10 min; Esa1 concentration was 25 nM and reaction time was 5 min; p300 concentration was 5 nM and reaction time was 10 min; PCAF concentration was 5 nM and reaction time was 3.5 min. 10% DMSO was present in Tip60, Esa1 and p300 assay, and 2% DMSO was present in PCAF assay.
| Tip60 | Esa1 | p300 | PCAF | |
|---|---|---|---|---|
| H4-20/H3-20 (μM) | 100 | 100 | 100 | 100 |
| Acetyl-CoA (μM) | 10 | 10 | 10 | 10 |
| IC50 (μM) | 149 | 190 | 150 | >100 |
3.3. Inhibition model of the discovered inhibitors
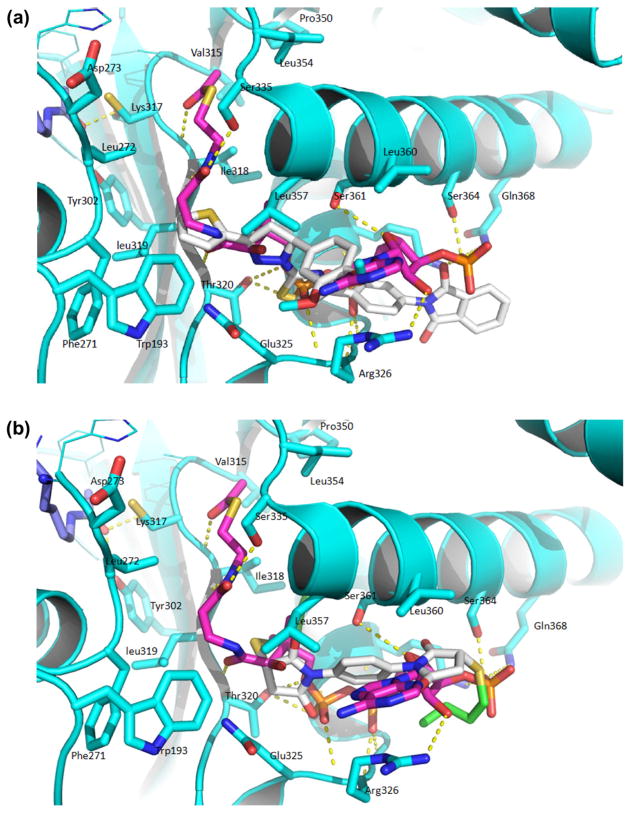
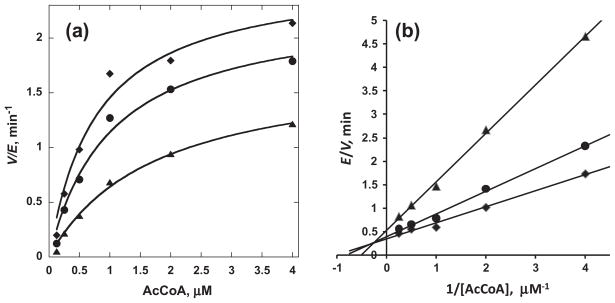
To understand the mechanism by which the discovered small molecule inhibitors block the activity of Tip60, we performed a docking study of compounds a and b with the recently disclosed crystal structure of Tip60 protein (PDB ID 2OU2) (Fig. 5). Briefly, the 3-dimensional structures of compounds a and b were constructed in Maestro 9.0.211. The Tip60 structure file was modified in Maestro9.0.211 to add hydrogen atoms and remove water molecules. Acetyl-CoA ligand was also removed from Tip60 to create a binding pocket. Docking of the compounds into Tip60 was made with Auto- DockTools4.2. Structural analysis with PyMOL was performed following the docking process. As shown in Fig. 5, the results reveal that the docked poses of both compounds a and b superimpose well with the original acetyl-CoA binding ligand in the crystal structure. Thus, the docking result strongly supports that compounds a and b target the acetyl-CoA binding site of Tip60. To confirm the competitive nature of inhibition, we elucidated the relationship between the inhibitors and acetyl-CoA by conducting enzymatic assays. The first experimental evidence comes from the fact that the IC50 values of these inhibitors vary with the concentration of acetyl-CoA. For example, at 10 μM of acetyl-CoA, IC50 of inhibitor a was 149 μM (Table 1), but at 1 μM of acetyl-CoA, it decreased to 87 μM (data not shown). Also, we conducted steady-state kinetic characterization. The initial velocities of Tip60 were measured at several selected concentrations of inhibitor a at a range of concentrations of acetyl-CoA while fixing the concentration of the peptide substrate at 100 μM. The data were plotted in the Michaelis–Menten format with velocity versus concentration of acetyl-CoA as well as in the double reciprocal format with 1/velocity versus 1/(concentration of acetyl-CoA) (Fig. 6). As can be seen from the double-reciprocal plot, a series of straight lines intersected on the left side of the ordinate, a feature characteristic of noncompetitive inhibition. The Michaelis–Menten data points were fitted to the nonlinear noncompetitive inhibition equation [29] and yielded Kii of 460 μM and Kis of 117 μM. Because Kii is 4 folds of higher than Kis, it is concluded that the inhibition of Tip60 by a is essentially competitive with respect to acetyl-CoA. Together with the conclusion from docking study, we suggest that the major binding site of inhibitor a overlaps with the acetyl-CoA binding site in Tip60 although additional weak binding sites may exist. Future work is needed to optimize structures of the chemical hits to obtain inhibitors with improved potency and specificity for Tip60.
Fig. 5.
Docking of inhibitors a and b in the crystal structure of Tip60 (PDB: 2OU2). Both compounds target the acetyl-CoA binding pocket and interact extensively with residues in the pocket.
Fig. 6.
Steady-state kinetic analysis showing that inhibitor a competes with AcCoA. (a) The Michaelis–Menten data of Tip60 activity at different concentrations of acetyl-CoA in the presence of three selected concentrations of compound a (◆0 μM, ◆80 μM, ▲150 μM). (b) The Lineweaver–Burk plot of the Michaelis–Menten data.
4. Conclusion
Tip60 is an important protein target for chromatin remodeling and is involved in multiple cellular processes. Inhibitors of Tip60 have great value as chemical genetics tools for interrogating the function of Tip60 in different pathways and are also potential therapeutic agents. We discovered several micromolar inhibitors of Tip60 from high-throughput virtual screen in combination with experimental inhibition studies. Docking studies and kinetic analyses showed that these new inhibitors target the cofactor-binding site in Tip60. This work provides valuable structural scaffolds for further development of potent inhibitors of Tip60 and the MYST family of HATs.
Acknowledgments
We acknowledge American Heart Association and Georgia Cancer Coalition Distinguish Scholar Award for funding this study. J. Wu is supported by a GSU Molecular Basis of Disease fellowship.
References
- 1.Kamine J, Elangovan B, Subramanian T, Coleman D, Chinnadurai G. Virology. 1996;216:357–366. doi: 10.1006/viro.1996.0071. [DOI] [PubMed] [Google Scholar]
- 2.Roth SY, Denu JM, Allis CD. Annu Rev Biochem. 2001;70:81–120. doi: 10.1146/annurev.biochem.70.1.81. [DOI] [PubMed] [Google Scholar]
- 3.Avvakumov N, Cote J. Subcell Biochem. 2007;41:295–317. [PubMed] [Google Scholar]
- 4.Voss AK, Thomas T. Bioessays. 2009;31:1050–1061. doi: 10.1002/bies.200900051. [DOI] [PubMed] [Google Scholar]
- 5.Sapountzi V, Logan IR, Robson CN. Int J Biochem Cell Biol. 2006;38:1496–1509. doi: 10.1016/j.biocel.2006.03.003. [DOI] [PubMed] [Google Scholar]
- 6.McLoughlin DM, Miller CC. J Neurosci Res. 2008;86:744–754. doi: 10.1002/jnr.21532. [DOI] [PubMed] [Google Scholar]
- 7.Cao X, Sudhof TC. Science. 2001;293:115–120. doi: 10.1126/science.1058783. [DOI] [PubMed] [Google Scholar]
- 8.Squatrito M, Gorrini C, Amati B. Trends Cell Biol. 2006;16:433–442. doi: 10.1016/j.tcb.2006.07.007. [DOI] [PubMed] [Google Scholar]
- 9.Avvakumov N, Cote J. Oncogene. 2007;26:5395–5407. doi: 10.1038/sj.onc.1210608. [DOI] [PubMed] [Google Scholar]
- 10.Patel JH, Du Y, Ard PG, Phillips C, Carella B, Chen CJ, Rakowski C, Chatterjee C, Lieberman PM, Lane WS, Blobel GA, McMahon SB. Mol Cell Biol. 2004;24:10826–10834. doi: 10.1128/MCB.24.24.10826-10834.2004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Taubert S, Gorrini C, Frank SR, Parisi T, Fuchs M, Chan HM, Livingston DM, Amati B. Mol Cell Biol. 2004;24:4546–4556. doi: 10.1128/MCB.24.10.4546-4556.2004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Kim JH, Kim B, Cai L, Choi HJ, Ohgi KA, Tran C, Chen C, Chung CH, Huber O, Rose DW, Sawyers CL, Rosenfeld MG, Baek SH. Nature. 2005;434:921–926. doi: 10.1038/nature03452. [DOI] [PubMed] [Google Scholar]
- 13.Baek SH, Ohgi KA, Rose DW, Koo EH, Glass CK, Rosenfeld MG. Cell. 2002;110:55–67. doi: 10.1016/s0092-8674(02)00809-7. [DOI] [PubMed] [Google Scholar]
- 14.Inghirami G, Chiarle R, Simmons WJ, Piva R, Schlessinger K, Levy DE. Cell Cycle. 2005;4:1131–1133. doi: 10.4161/cc.4.9.1985. [DOI] [PubMed] [Google Scholar]
- 15.Hobbs CA, Wei G, Defeo K, Paul B, Hayes CS, Gilmour SK. Cancer Res. 2006;66:8116–8122. doi: 10.1158/0008-5472.CAN-06-0359. [DOI] [PubMed] [Google Scholar]
- 16.Wei G, Hobbs CA, Defeo K, Hayes CS, Gilmour SK. Mol Carcinog. 2007;46:611–617. doi: 10.1002/mc.20350. [DOI] [PubMed] [Google Scholar]
- 17.Halkidou K, Gnanapragasam VJ, Mehta PB, Logan IR, Brady ME, Cook S, Leung HY, Neal DE, Robson CN. Oncogene. 2003;22:2466–2477. doi: 10.1038/sj.onc.1206342. [DOI] [PubMed] [Google Scholar]
- 18.Brady ME, Ozanne DM, Gaughan L, Waite I, Cook S, Neal DE, Robson CN. J Biol Chem. 1999;274:17599–17604. doi: 10.1074/jbc.274.25.17599. [DOI] [PubMed] [Google Scholar]
- 19.Zheng YG, Wu J, Chen Z, Goodman M. Med Res Rev. 2008;28:645–687. doi: 10.1002/med.20120. [DOI] [PubMed] [Google Scholar]
- 20.Bowers EM, Yan G, Mukherjee C, Orry A, Wang L, Holbert MA, Crump NT, Hazzalin CA, Liszczak G, Yuan H, Larocca C, Saldanha SA, Abagyan R, Sun Y, Meyers DJ, Marmorstein R, Mahadevan LC, Alani RM, Cole PA. Chem Biol. 2010;17:471–482. doi: 10.1016/j.chembiol.2010.03.006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Chimenti F, Bizzarri B, Maccioni E, Secci D, Bolasco A, Chimenti P, Fioravanti R, Granese A, Carradori S, Tosi F, Ballario P, Vernarecci S, Filetici P. J Med Chem. 2009;52:530–536. doi: 10.1021/jm800885d. [DOI] [PubMed] [Google Scholar]
- 22.Choi K, Park S, Lim BJ, Seong AR, Lee YH, Shiota M, Yokomizo A, Naito S, Na Y, Yoon HG. Biochem J. 2010 doi: 10.1042/BJ20100980. (Ahead of print) [DOI] [PubMed] [Google Scholar]
- 23.Gorsuch S, Bavetsias V, Rowlands MG, Aherne GW, Workman P, Jarman M, McDonald E. Bioorg Med Chem. 2009;17:467–474. doi: 10.1016/j.bmc.2008.11.079. [DOI] [PubMed] [Google Scholar]
- 24.Sbardella G, Castellano S, Vicidomini C, Rotili D, Nebbioso A, Miceli M, Altucci L, Mai A. Bioorg Med Chem Lett. 2008;18:2788–2792. doi: 10.1016/j.bmcl.2008.04.017. [DOI] [PubMed] [Google Scholar]
- 25.Balasubramanyam K, Swaminathan V, Ranganathan A, Kundu TK. J Biol Chem. 2003;278:19134–19140. doi: 10.1074/jbc.M301580200. [DOI] [PubMed] [Google Scholar]
- 26.Balasubramanyam K, Varier RA, Altaf M, Swaminathan V, Siddappa NB, Ranga U, Kundu TK. J Biol Chem. 2004;279:51163–51171. doi: 10.1074/jbc.M409024200. [DOI] [PubMed] [Google Scholar]
- 27.Shankar S, Srivastava RK. Int J Oncol. 2007;30:905–918. [PubMed] [Google Scholar]
- 28.Chen YHZJ, Wang LK, Sucheck SJ, Snow AM, Hecht SM. Chem Commun. 1998:2769–2770. [Google Scholar]
- 29.Wu J, Xie N, Wu Z, Zhang Y, Zheng YG. Bioorg Med Chem. 2009;17:1381–1386. doi: 10.1016/j.bmc.2008.12.014. [DOI] [PubMed] [Google Scholar]
- 30.Cebrat M, Kim CM, Thompson PR, Daugherty M, Cole PA. Bioorg Med Chem. 2003;11:3307–3313. doi: 10.1016/s0968-0896(03)00265-7. [DOI] [PubMed] [Google Scholar]
- 31.Li MY, Huang YJ, Tai PC, Wang BH. Biochem Biophys Res Commun. 2008;368:839–845. doi: 10.1016/j.bbrc.2008.01.135. [DOI] [PubMed] [Google Scholar]
- 32.Li MY, Ni NT, Chou HT, Lu CD, Tai PC, Wang BH. Chem Med Chem. 2008;3:1242–1249. doi: 10.1002/cmdc.200800076. [DOI] [PubMed] [Google Scholar]
- 33.Pearlman RS. Chem Des Autom News. 1987;2:1–7. [Google Scholar]
- 34.Jakalian A, Bush BL, Jack DB, Bayly CI. J Comput Chem. 2000;21:132–146. [Google Scholar]
- 35.Jakalian A, Jack DB, Bayly CI. J Comput Chem. 2002;23:1623–1641. doi: 10.1002/jcc.10128. [DOI] [PubMed] [Google Scholar]
- 36.Tsai KC, Wang SH, Hsiao NW, Li M, Wang B. Bioorg Med Chem Lett. 2008;18:3509–3512. doi: 10.1016/j.bmcl.2008.05.026. [DOI] [PubMed] [Google Scholar]
- 37.Yan Y, Barlev NA, Haley RH, Berger SL, Marmorstein R. Mol Cell. 2000;6:1195–1205. doi: 10.1016/s1097-2765(00)00116-7. [DOI] [PubMed] [Google Scholar]
- 38.Moustakas DT, Lang PT, Pegg S, Pettersen E, Kuntz ID, Brooijmans N, Rizzo RC. J Comput Aided Mol Des. 2006;20:601–619. doi: 10.1007/s10822-006-9060-4. [DOI] [PubMed] [Google Scholar]
- 39.Muegge I. Med Res Rev. 2003;23:302–321. doi: 10.1002/med.10041. [DOI] [PubMed] [Google Scholar]
- 40.FRED, Release version 2.2.3. OpenEye Scientific Software, Inc; Santa Fe, NM: 2007. [Google Scholar]
- 41.Feher M. Drug Discov Today. 2006;11:421–428. doi: 10.1016/j.drudis.2006.03.009. [DOI] [PubMed] [Google Scholar]
- 42.Eldridge MD, Murray CW, Auton TR, Paolini GV, Mee RP. J Comput Aided Mol Des. 1997;11:425–445. doi: 10.1023/a:1007996124545. [DOI] [PubMed] [Google Scholar]
- 43.Murray CW, Auton TR, Eldridge MD. J Comput Aided Mol Des. 1998;12:503–519. doi: 10.1023/a:1008040323669. [DOI] [PubMed] [Google Scholar]
- 44.Verkhivker GM, Bouzida D, Gehlhaar DK, Rejto PA, Arthurs S, Colson AB, Freer ST, Larson V, Luty BA, Marrone T, Rose PW. J Comput Aided Mol Des. 2000;14:731–751. doi: 10.1023/a:1008158231558. [DOI] [PubMed] [Google Scholar]
- 45.Stahl M, Rarey M. J Med Chem. 2001;44:1035–1042. doi: 10.1021/jm0003992. [DOI] [PubMed] [Google Scholar]
- 46.McGann MR, Almond HR, Nicholls A, Grant JA, Brown FK. Biopolymers. 2003;68:76–90. doi: 10.1002/bip.10207. [DOI] [PubMed] [Google Scholar]
- 47.IDEA, Version 8.8. Breadth Technology; Taipei, Taiwan: 2007. [Google Scholar]
- 48.Yan Y, Harper S, Speicher DW, Marmorstein R. Nat Struct Biol. 2002;9:862–869. doi: 10.1038/nsb849. [DOI] [PubMed] [Google Scholar]
- 49.van de Waterbeemd H, Gifford E. Nat Rev Drug Discov. 2003;2:192–204. doi: 10.1038/nrd1032. [DOI] [PubMed] [Google Scholar]
- 50.Marcu MG, Jung YJ, Lee S, Chung EJ, Lee MJ, Trepel J, Neckers L. Med Chem. 2006;2:169–174. doi: 10.2174/157340606776056133. [DOI] [PubMed] [Google Scholar]