Abstract
Genetic variation was first shown to be part of the cause of systemic lupus erythematosus (SLE or lupus) in the 1970s with associations in the human leukocyte antigen (HLA) region. Almost four decades later, and with the help of increasingly powerful genetic approaches, more than 25 genes are now known to contribute to the mechanisms that predispose individuals to lupus. Over half of these loci have been discovered in the past two years, underscoring the extraordinary success of recent genome-wide association approaches in SLE. The now well established genetic risk factors include alleles in the MHC region (multiple genes), IRF5, ITGAM, STAT4, BLK, BANK1, PDCD1, PTPN22, TNFSF4, TNFAIP3, SPP1, ATG5, XKR6, PXK, some of the Fcγ receptors, and deficiencies in several complement components, including C1q, C4, and C2. As reviewed here, many of these genes fall into key pathways that are consistent with previous studies implicating immune complexes, host immune signal transduction, and interferon pathways in the pathogenesis of SLE. Other genetic loci have no known function or apparent immunological role and have the potential to reveal novel disease mechanisms. Certainly, as our understanding of the genetic etiology of SLE continues to mature, important new opportunities will emerge for developing more targeted and effective diagnostic and clinical management tools for this complex autoimmune disease.
Introduction
Systemic lupus erythematosus (SLE or lupus) is a complex autoimmune disease with multi-system organ involvement, characterized by autoantibody production and tissue injury. SLE etiology is now at least partially known, involving multiple genetic and environmental factors are involved. A genetic component in lupus is supported by a high sibling risk ratio (λs=8–29) and heritability (>66%), and higher concordance rates between monozygotic twins (>35%) relative to dizygotic twins and other full siblings (2–5%)1–6. The available evidence suggests that the genetic risk for lupus is derived from variation in many (perhaps as many as 100) genes, each of modest effect size (odds ratios generally between 1.15 and 2.0).
Genetic discoveries and emerging technologies are in the process of transforming the basic understanding of human disease. The advent of high throughput genotyping and advances in computing over the past couple of years have led to the rapid discovery of numerous convincingly established genetic associations (Figure 1). Intense efforts are underway to refine the genetic effects at these loci and elucidate the biological mechanisms that lead to SLE. Many more associations in need of careful replication studies have also been identified. Success in this endeavor is determined by four fundamental factors: a germ line genetic basis for the phenotype, rigorous and careful phenotyping sufficient to decrease genetic heterogeneity relative to controls, well-designed and competently executed high-density surveys of the genome, and collaborative efforts permitting the analysis of several thousands of individuals.
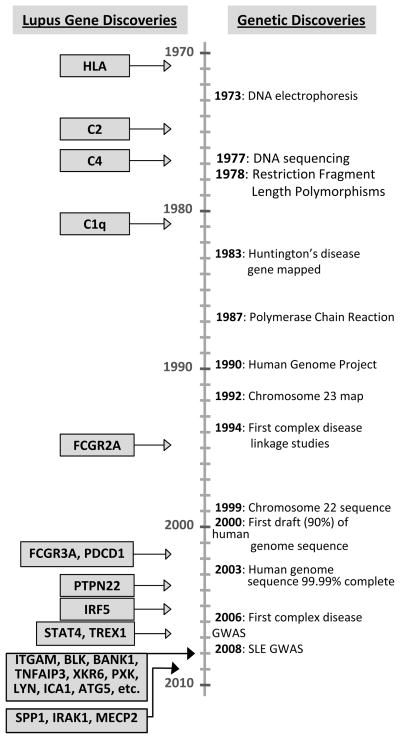
Figure 1.
A timeline of discoveries in human genetics (right) and confirmed genes discovered in lupus (left).
In the past two years, a series of landmark studies have been reported that have revolutionized our understanding of the genetics of lupus7–16. These are composed of insightful candidate gene studies and are anchored by genome-wide association (GWA) studies that include more than 10,000 individuals of European descent genotyped for over 300,000 single nucleotide polymorphisms (SNPs). Before 2007, there were nine established lupus susceptibility genes identified through candidate gene studies or linkage approaches. On the strength of the GWA studies, that number has swelled to more than 25 convincing genetic associations (Table 1, Figures 1 and 2). Many of these genes have important roles in immune functioning while others have no known role in immunity or are located in anonymous stretches of DNA. Non-European derived groups await large scale genetic studies to determine the extent of overlap with risk variants identified in European-derived cohorts and define novel population specific effects. Successful mapping of risk loci for lupus is comparable to inflammatory bowel disease where over 30 loci have now been identified17.
Table 1.
Genes associated with SLE
| Gene | Odds Ratio | Special points |
|---|---|---|
| TREX1 | 25 | Rare, found in 12/417 cases and 2/1712 controls |
| Complement C1q | ~10 | Rare, >90% −/− affected |
| Complement C4A & C4B | 6.5 | Rare, >70% −/− & −/+ affected, CNV |
| Complement C2 | ~5 | <10% −/− affected |
| HLA | 2.36 | Multiple effects |
| TNFAIP3 | 2.28 | Rare haplotype (minor allele frequencies ~0.03–0.07) |
| FCGR3B | 2.21 | CNV |
| ITGAM | 1.62 | H77R |
| FcGR3A | 1.6 | F176V |
| IRF5 | 1.54 | Three functional variants |
| STAT4 | 1.5 | |
| IRAK1 | 1.5 | Chromosome X, adjacent to MECP2 |
| BANK1 | 1.4 | |
| FCGR2A | 1.35 | H131R |
| ICA1 | 1.32 | |
| PTPN22 | 1.3 | N. European; familial lupus |
| CRP | 1.3 | −707 mutation |
| TNFSF4/OX40 | 1.3 | |
| LYN | 1.30 | |
| SCUBE1 | 1.28 | |
| KIAA1542 | 1.28 | rs49663128, SLEGEN GWAS, marker for IRF7? |
| PXK | 1.25 | |
| XKR6 | 1.23 | |
| Chromosome 5q33.3 | 1.23 | rs2431697, SLEGEN GWAS |
| Chromosome 1q25.1 | 1.22 | rs10798269, SLEGEN GWAS |
| BLK/FAM167A(C8orf12) | 1.22 | |
| UBE2L3 | 1.22 | |
| MECP2 | 1.2 | Chromosome X, adjacent to IRAK1 |
| PDCD1 | 1.2 | |
| ATG5 | 1.19 | |
| NMNAT2 | 1.18 | |
| Chromosome 8p21.1 | 1.18 | rs10903340, SLEGEN GWAS |
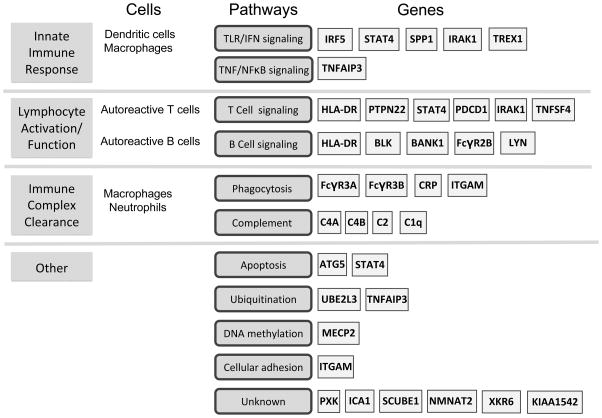
Figure 2.
Examples of immune functions, genetic pathways, and associated genes involved in SLE.
In this review, we summarize the confirmed genetic risk loci identified to date, placed in the context of major SLE disease pathways where possible. We emphasize recent studies that are particularly informative for shaping our view of the emerging genetic landscape of SLE.
Innate immunity genes
Innate immune mechanisms in SLE have received a surge of interest in recent years, which in part have been driven by descriptions of an “interferon (IFN) signature” composed of increased levels of IFN-inducible genes and correlated with more severe disease manifestations18; 19. Most cells produce Type I (IFNα and IFNβ) IFNs during early responses to viral infections, with exceptionally intense production from plasmacytoid dendritic cells (pDCs). Abnormally high levels of INFα have been observed in patients with SLE19. INFα promotes dendritic maturation and proinflammatory cytokine production, leading to diverse effects on immune functions including stimulation of Th1 pathways, promotion of B-cell activation, and regulation of apoptosis.
IRF5 (interferon regulatory factor 5) regulates Type I IFN-responsive genes and has been one of the most consistently associated genes with SLE outside of the Major Histocompatibility Complex (MHC). With an odds ratio 1.5, this transcription factor substantially contributes to SLE risk7; 20–22. In European-derived cases, multiple susceptibility polymorphisms have been identified and include a SNP that creates a novel splice site in exon 1B (rs2004640), a SNP located in the 3′ UTR that creates a polyadenylation site resulting in a shorter transcript with increased stability (rs10954213), and a 30-bp insertion/deletion likely to influence transcription initiation of target genes. Additional studies also support a role for IRF5 in Asian and African-American cohorts22. These studies have led to a model involving toll-like receptor (TLR) signaling, particularly TLR7 and TLR9, thought to be triggered by nucleic acid-containing autoantibody complexes or apoptotic or necrotic cell debris that results in high levels of Type I IFN production by plasmacytoid dendritic cells and is mediated by IRF5. IRF5 levels are increased by INFα and IRF5 induces the production of INFα, generating the possibility that this positive feedback loop is part of a molecular mechanism that produces the clinical abnormalities of SLE19.
Several additional genes with influence on IFN pathway and related innate immune signaling functions have been associated with SLE. IRAK1 (interleukin-1 receptor associated kinase) is a serine/threonine protein kinase located on chromosome X and involved in Toll/interluekin-1 receptor (TIR) family signaling. IRAK1 links several immune receptor complexes to the central adaptor/activator protein, TRAF6, and regulates multiple pathways in both innate and adaptive responses. Recent studies by Jacob et al. offer important insight into IRAK1 function in murine models of SLE and support a possible role for IRAK1 in induction of IFNα/γ, regulation of NFκB in T cells, and TLR activation9; 23. Jacob et al. hypothesized that gene dosage of IRAK1 may contribute to female predominance of SLE by virtue of its location on chromosome X.
Association with the rs7574865 SNP in the 3rd intron of STAT4 was originally identified in rheumatoid arthritis (RA) patients and SLE through positional cloning24. This variant is thought to contribute to more severe disease including nephritis, early disease onset, and production of anti-dsDNA autoantibodies25. STAT4, or signal transducer and activator of transcription 4, is involved in cellular differentiation, proliferation and apoptosis following cytokine and growth factor signaling26. Like IRAK1, variation in STAT4 likely influences both innate and adaptive functions of immune responses in SLE. The rs7574865 risk variant has recently been shown to confer an increased sensitivity to IFNα signaling in peripheral blood mononuclear cells from SLE patients27. In addition to activation by Type I IFN receptor engagement, STAT4 is also induced by IL-23 and IL-12 (primarily produced by dendritic cells), forms homodimers that translocate to the nucleus, and promotes differentiation of T cells into IFNγ producing Th1 cells. A very recent study by Abelson et al. describes an additional independent genetic association with SLE28. This second variant, rs3821236, is located in intron 16 of STAT4 and may contribute to differential associations across various European-derived populations. They further show an additive effect between STAT4 and IRF5 for increased risk of developing SLE28. The potential repertoire of STAT4 risk variants has also been expanded in African-American, Hispanic-American, Asian and Latin American populations29.
Association with SLE of a risk haplotype spanning the tumor necrosis factor alpha-induced-protein 3 (TNFAIP3) was recently identified through a GWAS study testing association of SLE with over 300,000 SNPs19. An SLE risk haplotype was identified that appears to be of considerable effect size (odds ratio~2.28) present in a minority of patients (minor allele frequencies of ~ 0.03–0.07). Studies in RA, psoriasis and Crohn’s disease suggest TNFAIP3 operates in multiple autoimmune diseases with potentially complex and differential genetic effects30. The TNFAIP3 gene codes for the zinc-finger A20 protein. A20 is a ubiquitin-editing enzyme required for effective termination of NFκB mediated proinflammatory responses downstream of signal transduction through TLRs, IL1R, TNFR, and NOD2. Targets of A20 include TRAF6, RIP1, RIP2, and IKKγ/NEMO.
Association of osteopontin (SPP1) with SLE in males as well as with females with early onset disease has recently been identified31; 32. SPP1 is a multifunctional cytokine that is overexpressed in SLE patients and shown to be critical for IFNα production in murine pDCs. Another gene involved in induction of IFN responses that has been associated with SLE is the 3′ DNA repair exonuclease 1 gene, TREX133. Although rare, genetic association of TREX1 with SLE introduces an additional potential disease mechanism related to innate immunity. TREX1 metabolizes reverse-transcribed single-stranded DNA of endogenous retroelements as a function of cell-intrinsic antiviral surveillance resulting in a potent Type I IFN response33.
MHC
The strongest association in European Americans is in the MHC (major histocompatibility) region at 6p21.3 and extends an amazing 7.2 Mb across a region of unprecedented linkage disequilibrium8; 10. There is no other region of such extraordinary genetic effect in the genome. There are more than 120 genes in the HLA (human leukocyte antigen) Class I, II, and III regions, many of which are important in immune function. While the majority of previous work has focused upon the Class II HLA-DR molecules, recent results raise the serious possibility that important susceptibility genes, perhaps even more important susceptibility genes for SLE, lie in the Class III region of HLA. For example, the gene with the highest association in one study is not in or next to HLA-DR or HLA-DQ, but rather in the MSH5 (mutS homolog 5) gene in Class III10. This gene is important in facilitating DNA re-arrangements that lead to immunoglobulin class switching34. Evidence for an independent Class III effect has been supported by a family-based study of SLE trios centered around a SNP in intron 6 of the SKIV2L (super viralicidic activity 2-like) gene. SKIV2L is thought to be a RNA helicase and is highly expressed in T cells, B cells, and dendritic cells8. Perhaps, MSH5, SKIV2L, or one of its close neighbors is a risk factor for SLE that is independent of the HLA-DR and HLA-DQ loci that have been so frequently studied. Additional independent associations are likely to be defined. Comprehensive evaluation of dense genotype data across this region will be required to fully characterize the genetic contribution of this immunologically rich region of the genome.
Lymphocyte signaling
A hallmark of SLE is autoantibody production, long thought to be an antigen driven process leading to aberrant adaptive immune responses. Multiple genetic risk loci that operate in T and B cell lineages have now been associated with SLE and illustrate the importance of lymphocyte signaling, regulation of activation and proliferation, and careful control of tolerance mechanisms that when disrupted, can lead to autoimmune processes.
Reduced T cell receptor (TCR) signaling is thought to increase risk for autoimmunity. The R620W polymorphism in exon 14 of PTPN22 (protein tyrosine phosphatase 22), which encodes the lymphoid-specific tyrosine phosphatase (LYP) protein, appears to play a role in multiple autoimmune phenotypes (reviewed in Chung et al.35). PTPN22 was originally identified as a candidate gene for Type 1 diabetes and RA, with subsequent demonstration of an association with SLE, autoimmune thyroid disease and other humoral autoimmune disorders. The risk R620W polymorphism is thought to increase phosphatase activity by disruption of the interaction of LYP with CSK and disturb the regulation of the TCR signaling kinases LCK and FYN, as well as ZAP-70 and TCRζ. The 620W risk allele is a gain-of-function variant, with increased catalytic activity compared to the 620R variant and is thought to be a more potent suppressor of T cell receptor signaling. To date, the association with PTPN22 appears to be more robust in familial SLE cases compared to sporadic SLE cases. Interestingly, its effects are strongest in Northern Europeans. In addition, recent work by Orru et al. has identified an association of a loss-of-function variant, R263Q that also affects phosphatase activity of PTPN2236. They demonstrate a protective association of the Q263 allele that appears to be independent of the R620W polymorphism, and is also consistent with alterations in TCR signaling thresholds.
The tumor necrosis factor superfamily member, TNFSF4 (or OX40L), is expressed on antigen presenting cells. Its receptor, TNFSFR4 or OX40, is expressed on activated T cells and, together, the binding of TNFSF4/TNFSFR4 is a costimulator of T cells37. TNFSF4 is located at 1q25, close to the Fc gamma receptor cluster. A thirteen marker haplotype upstream of TNFSF4 is associated with lupus15 and correlated with increased expression of TNFSF4. Increased levels of TNFSF4 are thought to lead to the increased co-stimulation for CD4+ T cells and further activate the antigen presenting cells that express TNFSF4, thereby destabilizing peripheral tolerance37.
The programmed cell death 1 gene (PD-1 or PDCD1) is an immunoinhibitory receptor that belongs to the B7/CD28 family and leads to the down-regulation of T and B cell activity. PDCD1 was one of the first SLE genes identified through classic linkage and positional cloning approaches in multiplex families. The SLE associated intronic SNP, PD-1.3, disrupts the binding site for the RUNX1 transcription factor, lowering the threshold for resisting immune responses against self38. Although conflicting reports have been published since the original association, a recent meta-analysis supports a role for PDCD1 in SLE39.
Genes involved in B cell receptor signaling contribute to lupus susceptibility. BANK1 (B cell scaffold protein with ankyrin repeats) is a newly described B cell adaptor protein expressed predominantly in B cells. Association of SLE has been identified with three variants13 that are expected to lead to an altered B cell activation threshold to increase lupus risk. LYN (Yamaguchi sarcoma viral oncogene) is a protein tyrosine kinase that physically associates with the B cell receptor, is a binding partner of BANK1, and was shown to be associated with SLE in a recent GWAS13. The B lymphoid tyrosine kinase (BLK) affects functions associated with the development of B cells before the appearance of the B cell receptor (BCR). BLK is a member of the Src family and may influence cell proliferation, differentiation, and B cell tolerance. Interestingly, association of BLK with SLE has been identified in the promoter region which is shared with the C8orf13 (FAM167A) gene. The two genes are transcribed in opposite directions. Risk at this locus is associated with reduced expression of BLK mRNA in transformed B-cell lines and increased expression of C8orf13 transcripts11. The function of C8orf13 is unknown.
Complement and immune complex clearance
The importance of complement components and effective immune complex clearance have long been recognized in SLE. Even with several decades of work, novel members with evidence for association to SLE continue to be identified and expand upon our working knowledge of the relationships to development of SLE and risk for serious manifestations such as nephritis.
Rare variants are likely to play a meaningful role in SLE susceptibility and are at present perhaps best illustrated by associations with complement components. Recent work by Gorlov et al. shows a negative correlation between minor allele frequency and the predicted probability that the variant would adversely affect protein function40. In lupus, the complement component genes, C2, C4A, C4B, and C1q exhibit associations with very strong effect sizes through recessive genetic modes of inheritance of uncommon alleles41–46. Despite their importance, the statistical power of case-control study design using association tests has very low power for detecting recessive effects47. Coupled with the multiple testing problem and difficulty of current genotyping platforms to reliably genotype rare variants, genome-wide association studies will have a difficult time detecting these associations.
Multiple members of the Fcγ receptor family have been associated with SLE and are reviewed in detail elsewhere in this issue48; 49. Briefly, FcγRIIIA and FcγRIIA, the gene products of FCGR3A and FCGR2A respectively, are both activating receptors and members of the immunoglobulin superfamily. FcγRIIIA is expressed on NK cells, macrophages and some monocytes while FcγRIIA is widely expressed on hematopoietic cells and is usually studied on neutrophils. Missense mutations of FcγRIIA (H131R) and FcγRIIIA (F176V) alter their affinity for particular subclasses of IgG50. These changes are thought to influence immune complex processing, leading to alternate fates for immune complexes that encourage autoimmune responses. C-reactive protein (CRP) binds FcγRI, FcγRIIB, and FcγRIIA on the surface of leukocytes. Its binding to FcγRI or FcγRIIA leads to phagocytosis and the release of inflammatory cytokines while binding to FcγRIIB blocks its activating signals51; 52. Heritability of CRP levels has been widely shown and association with SLE is partially accounted for by genetic variation in the promoter52. Other forms of genetic variation such as copy number variations (CNVs) may also play a role in the risk of SLE or lupus nephritis through the Fc gamma receptors, including FcγRIIIB53.
Newly discovered association of SLE with ITGAM was found independently in both GWA and positional cloning studies10–12. ITGAM encodes the α-chain of αMβ2-integrin (also known as Mac-1, CR3, and CD11b/CD18). Functions of ITGAM include immune complex clearance as well as leukocyte activation, adhesion and migration from the bloodstream via interactions with a wide range of ligands including C3bi, fibrinogen, and ICAM-1 (intracellular adhesion molecule-1). The putative causative polymorphism (rs1143679 in exon 3) in ITGAM is not obviously related to either the integrin function or iC3b binding function of this macrophage/monocyte cell surface molecule, but there are many poorly characterized binding properties of ITGAM that are potentially relevant to disease risk12. Strong and robust association of ITGAM was confirmed in a meta-analysis of nine independent cohorts and supported a role for the putative causal allele in European and African-derived populations. Interestingly, this SNP is essentially monomorphic in Asians, providing a clear example of population specific risks for SLE54.
Chromosome X
Lupus is a disease affecting women ten-fold more than men. This discrepancy in prevalence between genders could be due either to direct effects of sex chromosomes or indirect effects of chromosomes such as those mediated by sex hormones. In a recent study by Scofield et al., support for a direct role was evaluated by assessing the prevalence of Klinefelter’s syndrome in a large cohort of patients with SLE. Prevalence of Klinefelter’s, defined by 47, XXY karyotypes, was approximately 14-fold higher in men with SLE compared to those without SLE. This risk (~1:900) is closer to the female risk of SLE (~1:1400 in European ancestry) instead of the male risk for individuals with normal 46,XY karyotypes,55.
The recent identification of IRAK1 (described above) supports direct effects of chromosome X, possibly through gene dosage effects. In addition to IRAK1, a risk haplotype in the methyl-CpG-binding protein 2 gene (MECP2) has been associated with SLE and suggests a potential role for DNA methylation in the pathogenesis of SLE. Webb et al. reported gene expression profile differences in Epstein-Barr virus (EBV)-transformed B cell lines derived from SLE patients that were identified by comparing cases either with or without the risk haplotype56. Genes that were upregulated had significantly more CpG islands in their promoter regions, including a substantial number of genes regulated by IFNs. As a caveat, the involvement of MECP2 has the potential to also generate a positive feedback loop by accentuating the effect upon binding the methylated cytosines of itself, in the MECP2 gene. As with IRAK1, mapping of MECP2 on chromosome X raises the possibility of a gene dosage effect that may contribute to increased prevalence of SLE among women. The origin of the powerful difference in risk by sex chromosome composition is important, whether or not from here we subsequently establish that lupus is related to the signal transduction function of IRAK1 or the influence of gene expression of MECP2 through gene methylation or some other aspect of associated DNA on chromosome X.
Other loci
The genes with no known relation to immune function (KIAA1542, PXK, XKR6, FAM167A and others yet to be discovered) are potentially the most interesting. The mechanisms by which these mystery genes operate to increase risk will serve to constitute a more complete picture of established lupus pathways (such as IFN signaling), or may constitute novel relationships to lupus autoimmunity. In the case of KIAA1542, the genetic effect observed may be due to close proximity with interferon regulatory factor 7 (IRF7), a gene that also plays a major role in Type I IFN signaling10. Indeed, the associations in anonymous regions of DNA with no obvious connection to a gene product must have mechanisms that influence lupus pathogenesis and autoimmunity in ways that implicate fundamental aspects of biology that are today unknown.
Gene and environment interactions
More complex data analysis approaches using the wealth of genetic information collected to date are only partially explored. As part of the GWA study completed by the International SLEGEN (SLE genetics) consortium, none of the robust associations exhibited evidence of epistasis10. In this study, and using several of the genes mentioned herein, a stepwise multiple logistic regression analysis suggested the variants in the PXK, HLA region, IRF5, KIAA1542 and ITGAM genes act independently. When considered jointly, they are strongly predictive of lupus with significant sensitivity and specificity. Although biased upwards because part of the data were used to identify the associations, the area under the ROC curve, C statistic, was 0.67. In addition, these variants jointly explain ~15% of the lupus sibling risk ratio of 2910. Although refinement of these associations is required and their potential roles in gene-gene or gene-environment interactions need to be further investigated, all of these associations are robust and forged in the fires of further replication and fine mapping.
Conclusions
Genetic variation is the most basic determinant of host contribution to phenotype. The consequences of such variations are now discoverable in such abundance that our capacity to assimilate, much less explain them, is overwhelmed. SLE has long been considered a prototypic autoimmune disease, and from the genetics perspective, has served that title well. The coming years will most certainly continue to reveal additional associations, refinements to our current understanding of those established thus far, and provide new clues to explain the clinical heterogeneity that is a hallmark of SLE. Additional work will include detailed genetic studies in multiple ethnic groups, the development of models of disease that incorporate environmental influences, and studies to determine the overlapping and unique relationships among genetic variants associated with other related autoimmune phenotypes. Every robust, authentic association has a compulsory role in pathogenesis, which promises to profoundly change our capacity to diagnose, predict, and treat human disease.
Acknowledgments
This work has been supported by the NIH (AR043274, AI24717, AR62277, AR42460, AI31584, DE015223, RR015577, RR020143, AR48940, AR049084), the Mary Kirkland Scholarship, the Barrett Scholarship Fund, the Alliance for Lupus Research, and the U.S. Department of Veterans Affairs.
References
- 1.Deapen D, Escalante A, Weinrib L, Horwitz D, Bachman B, Roy-Burman P, et al. A revised estimate of twin concordance in systemic lupus erythematosus. Arthritis Rheum. 1992;35(3):311–318. doi: 10.1002/art.1780350310. [DOI] [PubMed] [Google Scholar]
- 2.Alarcon-Segovia D, Alarcon-Riquelme ME, Cardiel MH, Caeiro F, Massardo L, Villa AR, et al. Familial aggregation of systemic lupus erythematosus, rheumatoid arthritis, and other autoimmune diseases in 1,177 lupus patients from the GLADEL cohort. Arthritis Rheum. 2005;52 (4):1138–1147. doi: 10.1002/art.20999. [DOI] [PubMed] [Google Scholar]
- 3.Hochberg MC. The application of genetic epidemiology to systemic lupus erythematosus. J Rheumatol. 1987;14(5):867–869. [PubMed] [Google Scholar]
- 4.Lawrence JS, Martins CL, Drake GL. A family survey of lupus erythematosus. 1. Heritability. J Rheumatol. 1987;14(5):913–921. [PubMed] [Google Scholar]
- 5.Sestak AL, Shaver TS, Moser KL, Neas BR, Harley JB. Familial aggregation of lupus and autoimmunity in an unusual multiplex pedigree. J Rheumatol. 1999;26(7):1495–1499. [PubMed] [Google Scholar]
- 6.Block SR. A brief history of twins. Lupus. 2006;15(2):61–64. doi: 10.1191/0961203306lu2263ed. [DOI] [PubMed] [Google Scholar]
- 7.Graham RR, Kyogoku C, Sigurdsson S, Vlasova IA, Davies LR, Baechler EC, et al. Three functional variants of IFN regulatory factor 5 (IRF5) define risk and protective haplotypes for human lupus. Proceedings of the National Academy of Sciences of the United States of America. 2007;104(16):6758–6763. doi: 10.1073/pnas.0701266104. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Fernando MM, Stevens CR, Sabeti PC, Walsh EC, McWhinnie AJ, Shah A, et al. Identification of Two Independent Risk Factors for Lupus within the MHC in United Kingdom Families. PLoS Genet. 2007;3(11):e192. doi: 10.1371/journal.pgen.0030192. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Jacob CO, Reiff A, Armstrong DL, Myones BL, Silverman E, Klein-Gitelman M, et al. Identification of novel susceptibility genes in childhood-onset systemic lupus erythematosus using a uniquely designed candidate gene pathway platform. Arthritis and rheumatism. 2007;56(12):4164–4173. doi: 10.1002/art.23060. [DOI] [PubMed] [Google Scholar]
- 10.Harley JB, Alarcon-Riquelme ME, Criswell LA, Jacob CO, Kimberly RP, Moser KL, et al. Genome-wide association scan in women with systemic lupus erythematosus identifies susceptibility variants in ITGAM, PXK, KIAA1542 and other loci. Nat Genet. 2008;40(2):204–210. doi: 10.1038/ng.81. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Hom G, Graham RR, Modrek B, Taylor KE, Ortmann W, Garnier S, et al. Association of systemic lupus erythematosus with C8orf13-BLK and ITGAM-ITGAX. N Engl J Med. 2008;358(9):900–909. doi: 10.1056/NEJMoa0707865. [DOI] [PubMed] [Google Scholar]
- 12.Nath SK, Han S, Kim-Howard X, Kelly JA, Viswanathan P, Gilkeson GS, et al. A nonsynonymous functional variant in integrin-alpha(M) (encoded by ITGAM) is associated with systemic lupus erythematosus. Nat Genet. 2008;40(2):152–154. doi: 10.1038/ng.71. [DOI] [PubMed] [Google Scholar]
- 13.Kozyrev SV, Abelson AK, Wojcik J, Zaghlool A, Linga Reddy MV, Sanchez E, et al. Functional variants in the B-cellgene BANK1 are associated with systemic lupus erythematosus. Nat Genet. 2008;40(2):211–216. doi: 10.1038/ng.79. [DOI] [PubMed] [Google Scholar]
- 14.Sawalha AH, Webb R, Han S, Kelly JA, Kaufman KM, Kimberly RP, et al. Common variants within MECP2 confer risk of systemic lupus erythematosus. PLoS ONE. 2008;3(3):e1727. doi: 10.1371/journal.pone.0001727. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Graham DS, Graham RR, Manku H, Wong AK, Whittaker JC, Gaffney PM, et al. Polymorphism at the TNF superfamily gene TNFSF4 confers susceptibility to systemic lupus erythematosus. Nat Genet. 2008;40(1):83–89. doi: 10.1038/ng.2007.47. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Graham RR, Cotsapas C, Davies L, Hackett R, Lessard CJ, Leon JM, et al. Genetic variants near TNFAIP3 on 6q23 are associated with systemic lupus erythematosus. Nat Genet. 2008;40(9):1059–1061. doi: 10.1038/ng.200. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Barrett JC, Hansoul S, Nicolae DL, Cho JH, Duerr RH, Rioux JD, et al. Genome-wide association defines more than 30 distinct susceptibility loci for Crohn’s disease. Nat Genet. 2008 doi: 10.1038/NG.175. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Baechler EC, Gregersen PK, Behrens TW. The emerging role of interferon in human systemic lupus erythematosus. Curr Opin Immunol. 2004;16(6):801–807. doi: 10.1016/j.coi.2004.09.014. [DOI] [PubMed] [Google Scholar]
- 19.Crow MK, Kirou KA. Interferon-alpha in systemic lupus erythematosus. Curr Opin Rheumatol. 2004;16(5):541–547. doi: 10.1097/01.bor.0000135453.70424.1b. [DOI] [PubMed] [Google Scholar]
- 20.Cunninghame Graham DS, Manku H, Wagner S, Reid J, Timms K, Gutin A, et al. Association of IRF5 in UK SLE families identifies a variant involved in polyadenylation. Human molecular genetics. 2007;16(6):579–591. doi: 10.1093/hmg/ddl469. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Kozyrev SV, Lewen S, Reddy PM, Pons-Estel B, Witte T, Junker P, et al. Structural insertion/deletion variation in IRF5 is associated with a risk haplotype and defines the precise IRF5 isoforms expressed in systemic lupus erythematosus. Arthritis and rheumatism. 2007;56(4):1234–1241. doi: 10.1002/art.22497. [DOI] [PubMed] [Google Scholar]
- 22.Kelly JA, Kelley JM, Kaufman KM, Kilpatrick J, Bruner GR, Merrill JT, et al. Interferon regulatory factor-5 is genetically associated with systemic lupus erythematosus in African Americans. Genes and immunity. 2008;9(3):187–194. doi: 10.1038/gene.2008.4. [DOI] [PubMed] [Google Scholar]
- 23.Jacob CO, Zhu J, Armstrong DL, Yan M, Han J, Zhou XJ, et al. Identification of IRAK1 as a risk gene with critical role in the pathogenesis of systemic lupus erythematosus. Proceedings of the National Academy of Sciences of the United States of America. 2009 doi: 10.1073/pnas.0901181106. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Remmers EF, Plenge RM, Lee AT, Graham RR, Hom G, Behrens TW, et al. STAT4 and the risk of rheumatoid arthritis and systemic lupus erythematosus. N Engl J Med. 2007;357(10):977–986. doi: 10.1056/NEJMoa073003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Taylor KE, Remmers EF, Lee AT, Ortmann WA, Plenge RM, Tian C, et al. Specificity of the STAT4 genetic association for severe disease manifestations of systemic lupus erythematosus. PLoS Genet. 2008;4(5):e1000084. doi: 10.1371/journal.pgen.1000084. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Levy DE, Darnell JE., Jr Stats: transcriptional control and biological impact. Nat Rev Mol Cell Biol. 2002;3(9):651–662. doi: 10.1038/nrm909. [DOI] [PubMed] [Google Scholar]
- 27.Kariuki SN, Kirou KA, MacDermott EJ, Barillas-Arias L, Crow MK, Niewold TB. Cutting edge: autoimmune disease risk variant of STAT4 confers increased sensitivity to IFN-alpha in lupus patients in vivo. J Immunol. 2009;182(1):34–38. doi: 10.4049/jimmunol.182.1.34. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Abelson AK, Delgado-Vega AM, Kozyrev SV, Sanchez E, Velazquez-Cruz R, Eriksson N, et al. STAT4 Associates with SLE through two independent effects that correlate with gene expression and act additively with IRF5 to increase risk. Annals of the rheumatic diseases. 2008 doi: 10.1136/ard.2008.097642. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Namjou B, Sestak AL, Armstrong DL, Zidovetzki R, Kelly JA, Jacob N, et al. High-density genotyping of STAT4 reveals multiple haplotypic associations with systemic lupus erythematosus in different racial groups. Arthritis and rheumatism. 2009;60(4):1085–1095. doi: 10.1002/art.24387. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Elder JT. Genome-wide association scan yields new insights into the immunopathogenesis of psoriasis. Genes and immunity. 2009 doi: 10.1038/gene.2009.11. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Han S, Guthridge JM, Harley IT, Sestak AL, Kim-Howard X, Kaufman KM, et al. Osteopontin and systemic lupus erythematosus association: a probable gene-gender interaction. PLoS ONE. 2008;3(3):e0001757. doi: 10.1371/journal.pone.0001757. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Kariuki SN, Moore JG, Kirou KA, Crow MK, Utset TO, Niewold TB. Age-and gender-specific modulation of serum osteopontin and interferon-alpha by osteopontin genotype in systemic lupus erythematosus. Genes and immunity. 2009 doi: 10.1038/gene.2009.15. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Stetson DB, Ko JS, Heidmann T, Medzhitov R. Trex1 prevents cell-intrinsic initiation of autoimmunity. Cell. 2008;134(4):587–598. doi: 10.1016/j.cell.2008.06.032. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Sekine H, Ferreira RC, Pan-Hammarstrom Q, Graham RR, Ziemba B, de Vries SS, et al. Role for Msh5 in the regulation of Ig class switch recombination. Proceedings of the National Academy of Sciences of the United States of America. 2007;104(17):7193–7198. doi: 10.1073/pnas.0700815104. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Chung SA, Criswell LA. PTPN22: its role in SLE and autoimmunity. Autoimmunity. 2007;40(8):582–590. doi: 10.1080/08916930701510848. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Orru V, Tsai SJ, Rueda B, Fiorillo E, Stanford SM, Dasgupta J, et al. A loss-of-function variant of PTPN22 is associated with reduced risk of systemic lupus erythematosus. Human molecular genetics. 2009;18(3):569–579. doi: 10.1093/hmg/ddn363. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Stuber E, Strober W. The T cell-B cell interaction via OX40-OX40L is necessary for the T cell-dependent humoral immune response. J Exp Med. 1996;183(3):979–989. doi: 10.1084/jem.183.3.979. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Prokunina L, Castillejo-Lopez C, Oberg F, Gunnarsson I, Berg L, Magnusson V, et al. A regulatory polymorphism in PDCD1 is associated with susceptibility to systemic lupus erythematosus in humans. Nat Genet. 2002;32(4):666–669. doi: 10.1038/ng1020. [DOI] [PubMed] [Google Scholar]
- 39.Lee YH, Woo JH, Choi SJ, Ji JD, Song GG. Association of programmed cell death 1 polymorphisms and systemic lupus erythematosus: a meta-analysis. Lupus. 2009;18(1):9–15. doi: 10.1177/0961203308093923. [DOI] [PubMed] [Google Scholar]
- 40.Gorlov IP, Gorlova OY, Sunyaev SR, Spitz MR, Amos CI. Shifting paradigm of association studies: value of rare single-nucleotide polymorphisms. American journal of human genetics. 2008;82(1):100–112. doi: 10.1016/j.ajhg.2007.09.006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Walport MJ. The Roche Rheumatology Prize Lecture. Complement deficiency and disease. Br J Rheumatol. 1993;32(4):269–273. doi: 10.1093/rheumatology/32.4.269. [DOI] [PubMed] [Google Scholar]
- 42.Sullivan KE, Petri MA, Schmeckpeper BJ, McLean RH, Winkelstein JA. Prevalence of a mutation causing C2 deficiency in systemic lupus erythematosus. J Rheumatol. 1994;21(6):1128–1133. [PubMed] [Google Scholar]
- 43.Fielder AH, Walport MJ, Batchelor JR, Rynes RI, Black CM, Dodi IA, et al. Family study of the major histocompatibility complex in patients with systemic lupus erythematosus: importance of null alleles of C4A and C4B in determining disease susceptibility. Br Med J (Clin Res Ed) 1983;286(6363):425–428. doi: 10.1136/bmj.286.6363.425. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Reveille JD, Arnett FC, Wilson RW, Bias WB, McLean RH. Null alleles of the fourth component of complement and HLA haplotypes in familial systemic lupus erythematosus. Immunogenetics. 1985;21(4):299–311. doi: 10.1007/BF00430796. [DOI] [PubMed] [Google Scholar]
- 45.Morgan BP, Walport MJ. Complement deficiency and disease. Immunol Today. 1991;12(9):301–306. doi: 10.1016/0167-5699(91)90003-C. [DOI] [PubMed] [Google Scholar]
- 46.Botto M, Walport MJ. C1q, autoimmunity and apoptosis. Immunobiology. 2002;205(4–5):395–406. doi: 10.1078/0171-2985-00141. [DOI] [PubMed] [Google Scholar]
- 47.Harley JB. IL-7Ralpha and multiple sclerosis risk. Nat Genet. 2007;39(9):1053–1054. doi: 10.1038/ng0907-1053. [DOI] [PubMed] [Google Scholar]
- 48.Edberg JC, Langefeld CD, Wu J, Moser KL, Kaufman KM, Kelly J, et al. Genetic linkage and association of Fcgamma receptor IIIA (CD16A) on chromosome 1q23 with human systemic lupus erythematosus. Arthritis Rheum. 2002;46(8):2132–2140. doi: 10.1002/art.10438. [DOI] [PubMed] [Google Scholar]
- 49.Salmon JE, Millard S, Schachter LA, Arnett FC, Ginzler EM, Gourley MF, et al. Fc gamma RIIA alleles are heritable risk factors for lupus nephritis in African Americans. J Clin Invest. 1996;97(5):1348–1354. doi: 10.1172/JCI118552. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.van der Pol W, van de Winkel JG. IgG receptor polymorphisms: risk factors for disease. Immunogenetics. 1998;48(3):222–232. doi: 10.1007/s002510050426. [DOI] [PubMed] [Google Scholar]
- 51.Marnell L, Mold C, Du Clos TW. C-reactive protein: ligands, receptors and role in inflammation. Clin Immunol. 2005;117(2):104–111. doi: 10.1016/j.clim.2005.08.004. [DOI] [PubMed] [Google Scholar]
- 52.Edberg JC, Wu J, Langefeld CD, Brown EE, Marion MC, McGwin G, Jr, et al. Genetic variation in the CRP promoter: association with systemic lupus erythematosus. Hum Mol Genet. 2008;17(8):1147–1155. doi: 10.1093/hmg/ddn004. [DOI] [PubMed] [Google Scholar]
- 53.Fanciulli M, Norsworthy PJ, Petretto E, Dong R, Harper L, Kamesh L, et al. FCGR3B copy number variation is associated with susceptibility to systemic, but not organ-specific, autoimmunity. Nat Genet. 2007;39(6):721–723. doi: 10.1038/ng2046. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Han S, Kim-Howard X, Deshmukh H, Kamatani Y, Viswanathan P, Guthridge JM, et al. Evaluation of imputation-based association in and around the integrin-alpha-M (ITGAM) gene and replication of robust association between a non-synonymous functional variant within ITGAM and systemic lupus erythematosus (SLE) Hum Mol Genet. 2009;18(6):1171–1180. doi: 10.1093/hmg/ddp007. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55.Scofield RH, Bruner GR, Namjou B, Kimberly RP, Ramsey-Goldman R, Petri M, et al. Klinefelter’s syndrome (47, XXY) in male systemic lupus erythematosus patients: support for the notion of a gene-dose effect from the X chromosome. Arthritis Rheum. 2008;58(8):2511–2517. doi: 10.1002/art.23701. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56.Webb R, Wren JD, Jeffries M, Kelly JA, Kaufman KM, Tang Y, et al. Variants within MECP2, a key transcription regulator, are associated with increased susceptibility to lupus and differential gene expression in patients with systemic lupus erythematosus. Arthritis Rheum. 2009;60(4):1076–1084. doi: 10.1002/art.24360. [DOI] [PMC free article] [PubMed] [Google Scholar]