The crystal structure of Drosphila melanogaster Rab6 bound to the non-hydrolysable GTP analogue GMPPNP has been determined at a resolution of 1.4 Å.
Keywords: Rab6, GTP binding, GMPPNP
Abstract
Rab6 is a small GTPase that belongs to the p21 Ras superfamily. It is involved in vesicle trafficking between the Golgi apparatus and endosomes/ER in eukaryotes. The GDP-bound inactive protein undergoes conformational changes when the nucleotide is exchanged to GTP, allowing Rab6 to interact with a variety of different effector proteins. To further understand how these changes affect downstream protein binding, the crystal structure of Rab6 from Drosophila melanogaster has been solved to 1.4 Å resolution, the highest resolution for a Rab6 structure to date. The crystals belonged to space group C2, with unit-cell parameters a = 116.5, b = 42.71, c = 86.86 Å, α = 90, β = 133.12, γ = 90°. The model was refined to an R factor of 14.5% and an R free of 17.3%.
1. Introduction
Rabs are small GTP-binding proteins that are ubiquitous in eukaryotes and make up a large proportion of the p21 Ras superfamily (Macara et al., 1996 ▶). To date, over 70 rab genes have been identified in the human genome. Rab proteins are involved in a number of pathways to regulate intracellular vesicle trafficking between subcellular compartments of the cell. These include budding, targeting, docking and fusion of vesicles to/with their target membranes (Zerial & McBride, 2001 ▶; Pfeffer, 1994 ▶). Rab activity is regulated via a GTPase cycle. The GDP-bound protein is inactive and is located in the cytosol. The protein is activated by a guanine-exchange factor (GEF), which exchanges the GDP for GTP. The GTP-bound active Rab can then associate with the target membrane and interact with its effector partners to recruit them to their specific subcellular compartments. The GTP is then hydrolysed with the help of a GTPase-activating protein (GAP) and the inactive Rab returns to the cytosol. The binding of GTP is associated with structural changes that allow the protein to interact with its effectors. These changes mostly occur within the conserved switch I and II and interswitch regions of the protein (Dumas et al., 1999 ▶; Ostermeier & Brunger, 1999 ▶). In the inactive form these regions are disordered and they are poorly defined in crystal structures owing to their mobility. When activated, they become ordered and expose a conserved hydrophobic triad of residues on the surface of the protein made up from a Phe and Trp in the interswitch region and a Phe/Tyr in switch II. The arrangement of this hydrophobic patch, along with other residues within the switch regions, is thought to define the specificity of Rab for its different effector partners (Merithew et al., 2001 ▶). To date, only a handful of structures of Rab–effector complexes have been determined. These complexes are characterized by one or two α-helices from the Rab-binding domain (RBD) of the effector protein being in close contact with the switch regions of the Rab protein, emphasizing that it is the GTP-bound Rab which interacts with the effector partners (Ostermeier & Brunger, 1999 ▶; Eathiraj et al., 2005 ▶; Burguete et al., 2008 ▶).
Rab6 has been identified as a specific regulator of retrograde transport between the endosome and the Golgi and between the Golgi and the endoplasmic reticulum (ER) and of the recycling of enzymes through the ER (Martinez & Goud, 1998 ▶). There are several known effector proteins of Rab6, including subunits of the dynein–dynactin complex, Bicaudal D, Rabkinesin-6, GAPCenA, Rab6IP1, Rab6IP2, TMF, Mint3 and GCC185 (Short et al., 2002 ▶; Matanis et al., 2002 ▶; Echard et al., 1998 ▶; Cuif et al., 1999 ▶; Monier et al., 2002 ▶; Fridmann-Sirkis et al., 2004 ▶; Teber et al., 2005 ▶; Burguete et al., 2008 ▶). However, there are only two known structures of Rab6 in complex with an effector protein: Rab6IP1 (Recacha et al., 2009 ▶) and GCC185 (Burguete et al., 2008 ▶). These structures show distinct conformational differences in the switch and hydrophobic triad regions, indicating the importance of these regions in effector specificity. Currently, the only structures of Rab6 in the PDB have come from human or Plasmodium falciparum sources. In order to study the detailed conformational differences in the Rab protein when bound to different effector partners and to identify how each Rab is capable of recognizing a unique subset of partners, it may be necessary to obtain structures from several organisms. Here, we report the structure of Rab6 from Drosophila melanogaster (DmRab6) at 1.4 Å resolution. This is the highest resolution structure of a Rab6 protein to date and the first Drosophila Rab structure.
2. Materials and methods
2.1. Expression and purification
cDNA of full-length D. melanogaster rab6 was purchased from the Berkeley Drosophila Genome Project (GenBank AY060261). A C-terminally deleted construct containing residues 1–177 was cloned into a modified pET19b vector comprising an N-terminal His10 tag followed by a tobacco etch virus (TEV) protease cleavage site. The plasmid was transformed into competent BL21 (DE3) Star Escherichia coli cells (Invitrogen). Cultures were grown at 310 K to an OD600 of ∼0.6 and protein expression was induced at 291 K for 16 h. Induction was initiated by the addition of isopropyl β-d-1-thiogalactopyranoside (IPTG) to a final concentration of 0.8 mM. Cells were harvested by centrifugation and pellets were resuspended in lysis buffer consisting of 20 mM Tris–HCl, 500 mM NaCl, 20 mM imidazole, 0.1%(v/v) Triton X-100 pH 7.9. Cells were lysed by sonication and the cell debris was removed by centrifugation followed by filtration (0.22 µm). The His-tagged protein was purified by nickel-affinity chromatography (His-Trap HP column, GE Healthcare) and the protein was eluted from the column in 20 mM Tris–HCl, 500 mM NaCl, 400 mM imidazole pH 7.9. The N-terminal His10 tag was cleaved using TEV protease overnight at 277 K after diluting the protein fourfold in 20 mM Tris–HCl pH 7.9. The protein sample was dialysed to remove imidazole and passed through another His-Trap column to remove uncleaved protein and His-tagged TEV protease. Rab6 protein was then subjected to size-exclusion chromatography using a Superdex 75 26/60 column (GE Healthcare) equilibrated with 20 mM HEPES, 200 mM ammonium acetate, 5%(v/v) glycerol pH 7.0. Protein-containing fractions were pooled and concentrated to ∼30 mg ml –1 and 50 µl aliquots were flash-frozen in liquid nitrogen and stored at 197 K.
2.2. Crystallization and data collection
Immediately prior to crystallization screening, equimolar MgCl2 and a nonhydrolysable GTP analogue (GMPPNP; Sigma) were added to the protein. Crystallization trials were carried out at 291 K using the sitting-drop vapour-diffusion method in MRC plates (Molecular Dimensions). The crystallization kits Crystal Screen, Crystal Screen 2, Index, SaltRx, Natrix (Hampton Research) and Wizard 1, Wizard 2 and Wizard 3 (Emerald BioSystems) were used for initial screening. 0.25 µl Rab6/MgCl2/GMPPNP solution was mixed with 0.25 µl well solution and equilibrated over 80 µl well solution. Drops were set up using a Douglas Instruments Oryx crystallization robot. Rod-like crystals grew in 10 mM magnesium acetate, 50 mM MES pH 5.6, 2.5 M ammonium sulfate. To improve the quality of these crystals, several rounds of optimization were carried out, including additive screening. The best crystals were obtained from hanging drops prepared from 1 µl Rab6/MgCl2/GMPPNP solution mixed with 1 µl well solution comprising 2.4 M ammonium sulfate, 0.1 M imidazole pH 8.0, 2.5%(v/v) PEG 400, 2.5%(v/v) DMSO and 3%(w/v) trimethylamine-N-oxide. Crystals were transferred into a drop containing well solution plus 22.5%(v/v) glycerol for approximately 30 s before being cryocooled in liquid nitrogen for storage.
X-ray diffraction data were collected from crystals cryopreserved at 100 K on beamline I03 at Diamond Light Source using 12 700 eV X-rays with an ADSC Quantum 315r CCD detector. The best data were collected to a maximum resolution of 1.4 Å. Data were indexed and integrated using the MOSFLM package (Leslie, 2006 ▶) and scaled using SCALA (Evans, 2006 ▶) from the CCP4 suite (Winn et al., 2011 ▶).
2.3. Structure determination and refinement
The structure was determined by the molecular-replacement method using Phaser (McCoy et al., 2007 ▶) with the structure of human Rab6 (PDB entry 1yzq; Eathiraj et al., 2005 ▶) as the search model. The N-terminal residues 1–7 were truncated from the search model and all ligands were removed prior to Phaser trials. The molecular-replacement solution was subjected to 20 rounds of restrained refinement using REFMAC (Murshudov et al., 2011 ▶) and the initial R factors were determined as R work = 32.86%, R free = 35.90%. The model was subjected to iterative cycles of manual rebuilding in Coot (Emsley & Cowtan, 2004 ▶) and subsequent refinement with REFMAC. No NCS restraints were applied to the model. Waters were added later using the ARP/wARP Solvent program (Lamzin & Wilson, 1993 ▶). Owing to the high resolution of the data, full anisotropic B-factor refinement was used in the final stages.
The model was validated using MolProbity (Chen et al., 2010 ▶) and analysis of the intermolecular interactions was performed using PISA (Krissinel & Henrick, 2007 ▶). All figures were generated with PyMOL. The structure and structure factors were deposited in the Protein Data Bank under PDB code 2y8e. Analysis of this model compared with other known Rab structures was carried out using the SSM pose tool in Coot.
3. Results and discussion
3.1. Expression, crystallization, data collection and structure determination
DmRab6 protein with the hypervariable C-terminal region (residues 178–208; involved in membrane association) removed was expressed with a yield of 15 mg per litre of E. coli culture. Crystals grown using the hanging-drop vapour-diffusion method diffracted to a maximum Bragg spacing of 1.4 Å and belonged to space group C2, with unit-cell parameters a = 116.5, b = 42.71, c = 86.86 Å, α = 90, β = 133.12, γ = 90°. There are two copies of Rab6 per asymmetric unit (referred to as chains A and B) with a solvent content of 35%. Analysis of the interactions between the two protein molecules in the asymmetric unit shows an interface area of 640 Å2 and a free-energy change of −4.8 kJ mol−1. The average surface area of the chains is 8525 Å2; a total of 7.5% of the protein molecule is used in the interface. These data suggest that the protein is monomeric and the interface between the two monomers is nonfunctional and a consequence of crystal packing. This is supported by size-exclusion purification, which shows a single peak corresponding to a protein of 21 kDa, the size of a Rab6 monomer.
The structure was solved by molecular replacement using the structure of human Rab6 (PDB entry 1yzq) as the search model. The initial electron-density map revealed density for GMPPNP (which was absent from the search model) but poor density around a disordered loop incorporating residues 53–66. Subsequent model building and refinement improved this loop, allowing the main chain to be fully built and revealing at least partial density for almost all of the side chains. Overall, residues 10–174 of chain A and 7–173 of chain B were built. The extreme N- and C-termini could not be modelled owing to poor or no visible electron density. The root-mean-square deviation (r.m.s.d.) of the Cα backbone between the two chains is 0.82 Å.
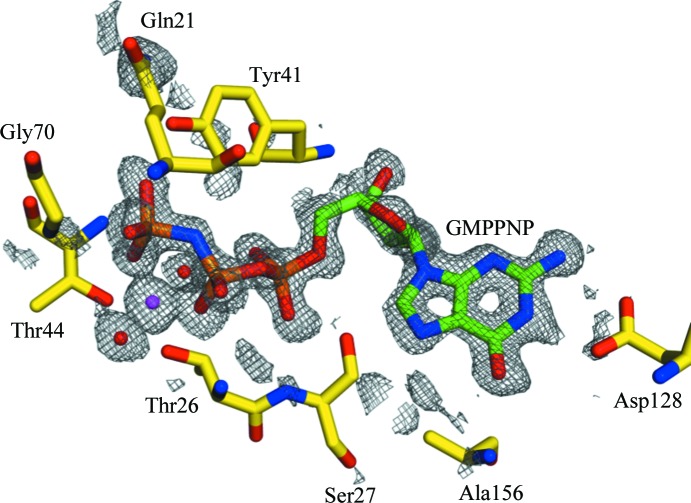
After initial refinement, very clear positive density could be seen for the GMPPNP ligand and an octahedrally coordinated Mg2+ ion in both chains (Fig. 1 ▶). These were built together with a total of 226 waters and three sulfate molecules. The final model has an overall B factor of 11.57; R work and R free were 14.5% and 17.3%, respectively, and the agreement with the expected geometry is excellent. There were two residues in the outlier region of the Ramachandran plot: Lys126 in each chain. This lysine sits directly beneath the ribose ring of the GMPPNP, which may explain the slight strain in the peptide backbone. Electron density around these residues is very well defined. The remaining residues are either in the allowed (99.7%) or favoured (98.5%) regions. The overall MolProbity clash score for the structure was 5.29, putting it in the 89th percentile for structures at this resolution.
Figure 1.
Initial density for the GMPPNP (green/orange) and Mg2+ (magenta) ligands bound in the GTP-binding site of Rab6 (chain A; yellow). The F o − F c map is contoured at 2σ (grey mesh).
Data-collection and refinement statistics are presented in Table 1 ▶ and the model was submitted with PDB code 2y8e.
Table 1. Data-collection and refinement statistics.
Values in parentheses are for the highest resolution shell.
| Data collection | |
| Space group | C2 |
| No. of molecules in asymmetric unit | 2 |
| Unit-cell parameters | |
| a () | 116.52 |
| b () | 42.71 |
| c () | 86.86 |
| () | 90 |
| () | 133.12 |
| () | 90 |
| Wavelength () | 0.9795 |
| Resolution () | 31.71.39 (1.481.39) |
| R merge † (%) | 4.7 (27.9) |
| R p.i.m. ‡ (%) | 2.9 (17.4) |
| No. of unique reflections | 61395 (8718) |
| Completeness (%) | 97.4 (95.3) |
| Multiplicity | 3.5 (3.4) |
| I/(I) | 16.6 (4.4) |
| Refinement | |
| R work § (%) | 14.5 |
| R free ¶ (%) | 17.3 |
| No. of protein atoms | 2721 |
| No. of water molecules | 226 |
| No. of ligand atoms | 81 |
| Average B factor (2) | 11.57 |
| Waters | 28.5 |
| R.m.s.d. from ideal values | |
| Bond lengths () | 0.014 |
| Bond angles () | 1.561 |
| MolProbity scores | |
| All-atom clash score | 5.29 |
| Bad rotamers (%) | 0 |
| Ramachandran favoured (%) | 98.48 |
| Ramachandran outliers (%) | 0.3 |
| PDB code | 2y8e |
R
merge = 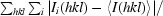
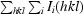 , where Ii(hkl) is the intensity of reflection hkl and
, where Ii(hkl) is the intensity of reflection hkl and 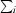 is the sum over all i measurements of reflection hkl.
is the sum over all i measurements of reflection hkl.
R
p.i.m = 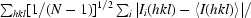
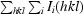 , where N is the number of observations of reflection hkl.
, where N is the number of observations of reflection hkl.
The R factor R
work = 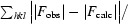
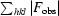 ; F
obs and F
calc are the observed and calculated structure factors, respectively.
; F
obs and F
calc are the observed and calculated structure factors, respectively.
R free isthe R factor calculated over a subset of the data (5.1%) that were excluded from refinement.
3.2. Overall structure
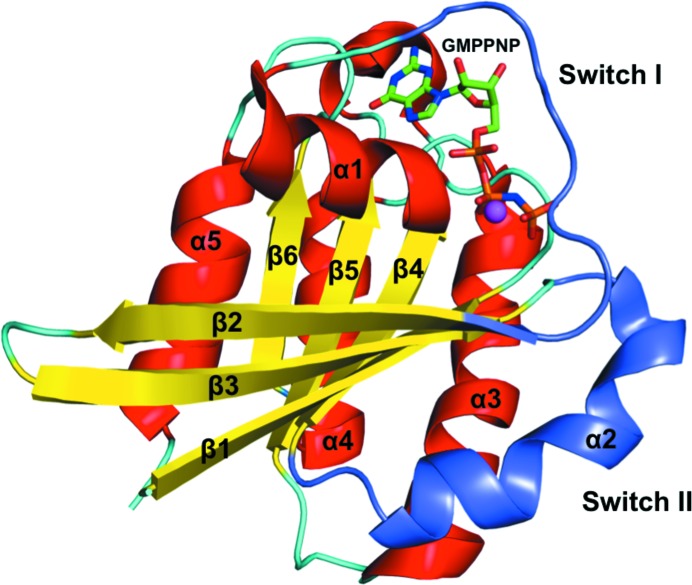
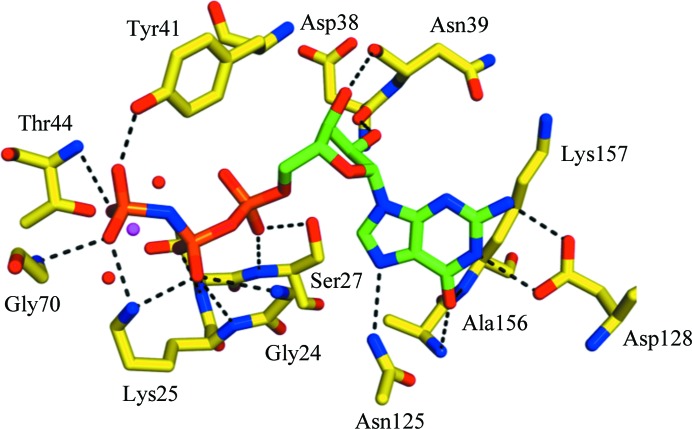
At 1.4 Å resolution, the crystal structure of DmRab6 bound to GMPPNP is the highest resolution structure of any Rab6 protein in the PDB (the previous highest was 1.78 Å; Eathiraj et al., 2005 ▶); it is also the first structure of a Drosophila Rab protein. The structure shows the characteristic fold of the Rab family of GTP-binding proteins: one sheet composed of six β-strands (one antiparallel) surrounded by five α-helices (Fig. 2 ▶). The consensus elements involved in nucleotide binding (Kjeldgaard et al., 1996 ▶; Sprang, 1997 ▶) are well ordered and form hydrogen bonds to the GTP analogue and coordinate the Mg2+ ion. The guanine base is held in position through hydrogen bonds to Asn125, Asp128, Ala156 and Lys157 and the ribose sugar makes contacts with Asp38 and Asn39. The α-phosphate and β-phosphate groups form extensive hydrogen bonds to the P-loop of the Rab6 protein involving Gly24, Lys25, Thr26 and Ser27. The γ-phosphate is then held through bonds to Lys25, Tyr41, Thr44 and Gly70. It is these extra bonds that give the GTP-bound form of the protein a more rigid structure than the GDP-bound form. The complete hydrogen-bonding network of the GMPPNP molecule is presented in Fig. 3 ▶. The Mg2+ ion is classically octahedrally coordinated by 2.1 Å bonds to the oxygen groups of the β- and γ-phosphates of GMPPNP, the hydroxyl groups of Thr26 and Thr44 and two water molecules (Fig. 4 ▶). The switch I and II and interswitch regions known to be important in effector protein binding are well ordered, with the conserved hydrophobic triad (Phe49, Trp66 and Tyr81) exposed on the surface, ready to recognize and interact with available effectors.
Figure 2.
Ribbon diagram of DmRab6 (chain A) bound to GMPPNP. Six β-strands are surrounded by five α-helices. Loop regions are coloured cyan. The GMPPNP is presented as sticks (green/orange) and the Mg2+ ion as a sphere (magenta). The two switch regions are coloured purple.
Figure 3.
The GTP-binding site of DmRab6. Hydrogen bonds between the GMPPNP ligand and DmRab6 side chains (chain A) are shown. Residues that interact with the GMPPNP are labelled. Two water molecules involved in Mg2+ coordination are shown in red.
Figure 4.
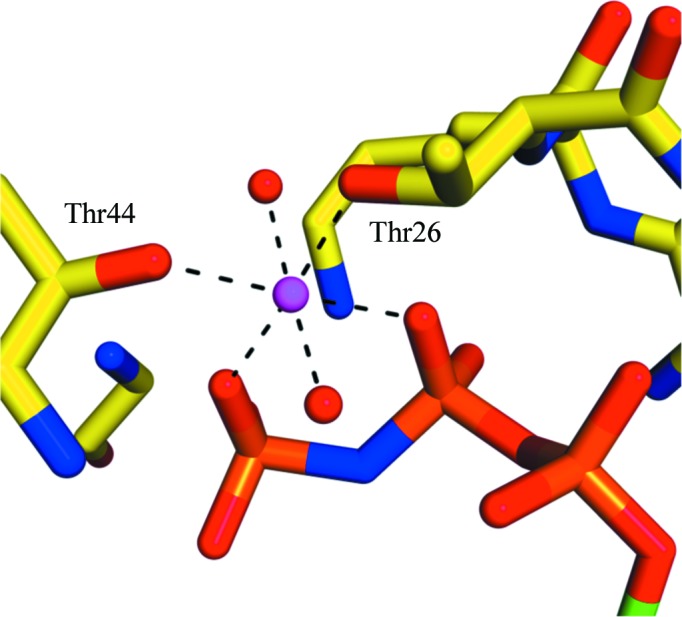
Octahedral Mg2+ coordination. One Mg2+ ion (magenta) is coordinated by O atoms from the β- and γ-phosphates of the GMPPNP (orange), the hydroxyl groups of Thr26 and Thr44 (labelled) and two water molecules (red spheres). The distance of all six coordination bonds is 2.1 Å.
The sequence of DmRab6 is highly homologous to those of Rab6 proteins from other organisms (percentage sequence identities are presented in Table 2 ▶). This is reflected in the structural similarity of DmRab6 to human and P. falciparum Rab6. In particular, the sequence Lys12–Ala88, which incorporates both switch regions, is entirely conserved between D. melanogaster Rab6, mouse Rab6A, human Rab6A (isoform B) and Caenorhabditis elegans Rab6. Furthermore, even in species that show lower sequence identity the hydrophobic triad is completely conserved.
Table 2. Sequence comparison and r.m.s.d. values for known Rab structures with DmRab6.
| PDB entry | Rab | Ligand bound | Sequence identity (%) | R.m.s.d. () |
|---|---|---|---|---|
| 3cwz | Rab6 | GTP | 93.8 | 1.31 |
| 3bbp | Rab6a | GTP | 96.9 | 0.92 |
| 1yzq | Rab6 | GNP | 93.8 | 1.09 |
| 1d5c | Rab6 | GDP | 69.4 | 1.37 |
| 2gil | Rab6a | GTP | 96.9 | 0.94 |
| 2fol | Rab1a | GDP | 37.3 | 1.43 |
| 1z0a | Rab2a | GDP | 38.5 | 1.54 |
| 2gf9 | Rab3d | GDP | 44.0 | 1.61 |
| 1yu9 | Rab4a | GNP | 39.3 | 1.49 |
| 2o52 | Rab4b | GDP | 36.6 | 1.72 |
| 3mjh | Rab5a | GTP | 44.3 | 1.35 |
| 3clv | Rab5a | GDP | 37.8 | 1.65 |
| 1n6h | Rab5a | GNP | 44.4 | 1.40 |
| 2hei | Rab5b | GDP | 42.5 | 1.62 |
| 1huq | Rab5c | GNP | 44.4 | 1.39 |
| 1vg8 | Rab7 | GNP | 39.6 | 1.51 |
| 1vg1 | Rab7 | GDP | 37.1 | 1.59 |
| 2ocb | Rab9b | GNP | 32.9 | 1.71 |
| 1yzl | Rab9 | GNP | 36.7 | 1.56 |
| 3bfk | Rab11a | GDP | 37.1 | 1.51 |
| 1yzk | Rab11 | GNP | 39.8 | 1.74 |
| 1zof | Rab14 | GDP | 35.2 | 1.61 |
| 1x3s | Rab18 | GNP | 40.4 | 1.53 |
| 1yzt | Rab21 | GNP | 37.3 | 1.49 |
| 1yvd | Rab22 | GNP | 39.0 | 1.52 |
| 1z22 | Rab23 | GDP | 37.1 | 1.55 |
| 2oil | Rab25 | GDP | 36.2 | 1.72 |
| 2g6b | Rab26 | GNP | 42.4 | 1.57 |
| 2zet | Rab27b | GTP | 34.2 | 1.35 |
| 2iey | Rab27b | GDP | 28.7 | 1.22 |
| 3e5h | Rab28 | GNP | 36.7 | 1.47 |
| 2hxs | Rab28a | GDP | 36.4 | 1.45 |
| 2ew1 | Rab30 | GNP | 41.9 | 1.52 |
| 2fg5 | Rab31 | GNP | 39.0 | 1.55 |
| 1z06 | Rab33 | GNP | 42.0 | 1.46 |
| 2hup | Rab43 | GDP | 44.3 | 1.56 |
DmRab6 is also very structurally similar to other members of the Rab family. The overall r.m.s.d. of the Cα backbone of DmRab6 to a selection of Rab structures with sequence identities varying between 29% and 97% remains within the range 0.92–1.74 Å (Table 2 ▶). This indicates that very subtle changes in both the sequence and conformation of the Rab protein allow it to distinguish between specific effector partners, highlighting the importance of determining the structures of Rab proteins from a wide variety of organisms.
Supplementary Material
PDB reference: Rab6 GTPase, 2y8e
Acknowledgments
This work was funded by the BBSRC via a studentship (MW) and grant BB/E020070/1 (HTJ and TAE). Data were collected at Diamond Light Source and we would like to thank the beamline staff for ongoing excellent help and support. The authors would like to thank Suzanne Edmunds for her help with the construct cloning and Sandeep Kumar for his help with the purification and crystallization trials.
References
- Burguete, A. S., Fenn, T. D., Brunger, A. T. & Pfeffer, S. R. (2008). Cell, 132, 286–298. [DOI] [PMC free article] [PubMed]
- Chen, V. B., Arendall, W. B., Headd, J. J., Keedy, D. A., Immormino, R. M., Kapral, G. J., Murray, L. W., Richardson, J. S. & Richardson, D. C. (2010). Acta Cryst. D66, 12–21. [DOI] [PMC free article] [PubMed]
- Cuif, M. H., Possmayer, F., Zander, H., Bordes, N., Jollivet, F., Couedel-Courteille, A., Janoueix-Lerosey, I., Langsley, G., Bornens, M. & Goud, B. (1999). EMBO J. 18, 1772–1782. [DOI] [PMC free article] [PubMed]
- Dumas, J. J., Zhu, Z., Connolly, J. L. & Lambright, D. G. (1999). Structure, 7, 413–423. [DOI] [PubMed]
- Eathiraj, S., Pan, X., Ritacco, C. & Lambright, D. G. (2005). Nature (London), 436, 415–419. [DOI] [PMC free article] [PubMed]
- Echard, A., Jollivet, F., Martinez, O., Lacapère, J. J., Rousselet, A., Janoueix-Lerosey, I. & Goud, B. (1998). Science, 279, 580–585. [DOI] [PubMed]
- Emsley, P. & Cowtan, K. (2004). Acta Cryst. D60, 2126–2132. [DOI] [PubMed]
- Evans, P. (2006). Acta Cryst. D62, 72–82. [DOI] [PubMed]
- Fridmann-Sirkis, Y., Siniossoglou, S. & Pelham, H. R. (2004). BMC Cell Biol. 5, 18. [DOI] [PMC free article] [PubMed]
- Kjeldgaard, M., Nyborg, J. & Clark, B. F. (1996). FASEB J. 10, 1347–1368. [PubMed]
- Krissinel, E. & Henrick, K. (2007). J. Mol. Biol. 372, 774–797. [DOI] [PubMed]
- Lamzin, V. S. & Wilson, K. S. (1993). Acta Cryst. D49, 129–147. [DOI] [PubMed]
- Leslie, A. G. W. (2006). Acta Cryst. D62, 48–57. [DOI] [PubMed]
- Macara, I. G., Lounsbury, K. M., Richards, S. A., McKiernan, C. & Bar-Sagi, D. (1996). FASEB J. 10, 625–630. [DOI] [PubMed]
- Martinez, O. & Goud, B. (1998). Biochim. Biophys. Acta, 1404, 101–112. [DOI] [PubMed]
- Matanis, T., Akhmanova, A., Wulf, P., Del Nery, E., Weide, T., Stepanova, T., Galjart, N., Grosveld, F., Goud, B., De Zeeuw, C. I., Barnekow, A. & Hoogenraad, C. C. (2002). Nature Cell Biol. 4, 986–992. [DOI] [PubMed]
- McCoy, A. J., Grosse-Kunstleve, R. W., Adams, P. D., Winn, M. D., Storoni, L. C. & Read, R. J. (2007). J. Appl. Cryst. 40, 658–674. [DOI] [PMC free article] [PubMed]
- Merithew, E., Hatherly, S., Dumas, J. J., Lawe, D. C., Heller-Harrison, R. & Lambright, D. G. (2001). J. Biol. Chem. 276, 13982–13988. [DOI] [PubMed]
- Monier, S., Jollivet, F., Janoueix-Lerosey, I., Johannes, L. & Goud, B. (2002). Traffic, 3, 289–297. [DOI] [PubMed]
- Murshudov, G. N., Skubák, P., Lebedev, A. A., Pannu, N. S., Steiner, R. A., Nicholls, R. A., Winn, M. D., Long, F. & Vagin, A. A. (2011). Acta Cryst. D67, 355–367. [DOI] [PMC free article] [PubMed]
- Ostermeier, C. & Brunger, A. T. (1999). Cell, 96, 363–374. [DOI] [PubMed]
- Pfeffer, S. R. (1994). Curr. Opin. Cell Biol. 6, 522–526. [DOI] [PubMed]
- Recacha, R., Boulet, A., Jollivet, F., Monier, S., Houdusse, A., Goud, B. & Khan, A. R. (2009). Structure, 17, 21–30. [DOI] [PubMed]
- Short, B., Preisinger, C., Schaletzky, J., Kopajtich, R. & Barr, F. A. (2002). Curr. Biol. 12, 1792–1795. [DOI] [PubMed]
- Sprang, S. R. (1997). Annu. Rev. Biochem. 66, 639–678. [DOI] [PubMed]
- Teber, I., Nagano, F., Kremerskothen, J., Bilbilis, K., Goud, B. & Barnekow, A. (2005). Biol. Chem. 386, 671–677. [DOI] [PubMed]
- Winn, M. D. et al. (1994). Acta Cryst. D67, 235–242.
- Zerial, M. & McBride, H. (2001). Nature Rev. Mol. Cell Biol. 2, 107–117. [DOI] [PubMed]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
PDB reference: Rab6 GTPase, 2y8e