The DIX domain of Ccd1 was purified and crystallized.
Keywords: Wnt signalling, Ccd1, DIX domain
Abstract
Coiled-coil DIX1 (Ccd1) is a positive regulator that activates the canonical Wnt signalling pathway by inhibiting the degradation of the key signal transducer β-catenin. The C-terminal DIX domain of Ccd1 plays an important role in the regulation of signal transduction through homo-oligomerization and protein complex formation with other DIX domain-containing proteins, i.e. axin and dishevelled proteins. Here, the expression, purification, crystallization and X-ray data collection of the Ccd1 DIX domain are reported. The crystals of the Ccd1 DIX domain belonged to space group P212121, with unit-cell parameters a = 72.9, b = 75.7, c = 125.6 Å. An X-ray diffraction data set was collected at 3.0 Å resolution.
1. Introduction
The Wnt signalling pathway plays an essential role in cell growth, differentiation, polarity formation and neural development (Wodarz & Nusse, 1998 ▶; Peifer & Polakis, 2000 ▶; Moon et al., 2002 ▶; MacDonald et al., 2009 ▶). In the canonical Wnt signalling pathway, two proteins, axin and dishevelled (Dvl), control the stability of the key signal transducer β-catenin (Klingensmith et al., 1994 ▶; Theisen et al., 1994 ▶; Ikeda et al., 1998 ▶). In the absence of Wnt signalling, axin is a negative regulator of Wnt signalling that mediates β-catenin phosphorylation by forming a protein complex with adenomatous polyposis coli (APC), glycogen synthase kinase 3β (GSK3β) and casein kinase 1α (CK1α) termed the β-catenin destruction complex (Hart et al., 1998 ▶; Kishida et al., 1998 ▶; Nakamura et al., 1998 ▶; Dajani et al., 2003 ▶; Xing et al., 2003 ▶). The phosphorylated β-catenin is degraded through the ubiquitin–proteasome system (Aberle et al., 1997 ▶; Orford et al., 1997 ▶). In the presence of Wnt signalling, Dvl suppresses the activity of the destruction complex by binding to axin. As a result, β-catenin remains unphosphorylated and stimulates the transcription of Wnt target genes through interaction with the TCF/LEF transcription factors (Li et al., 1999 ▶; Salic et al., 2000 ▶).
Coiled-coil DIX1 (Ccd1) is a positive regulator of the Wnt signalling pathway (Shiomi et al., 2003 ▶). Ccd1 expression is detected in the neurons of the nervous system and it largely overlaps with the regions of several Wnt genes (Shiomi et al., 2005 ▶; Soma et al., 2006 ▶; Ikeuchi et al., 2009 ▶; Jing et al., 2009 ▶). Expression analysis by RT-PCR and northern blotting of mammalian Ccd1 revealed the generation of multiple isoforms by different promoter usage and alternative splicing (Shiomi et al., 2005 ▶; Wang et al., 2006 ▶). The isoforms of Ccd1 proteins are classified into three subtypes: Ccd1A, Ccd1B and Ccd1C. Ccd1A consists of three domains: an N-terminal calponin-homology (CH) domain, a central coiled-coil region and a C-terminal DIX domain. Ccd1B and Ccd1C lack the N-terminal region containing the CH domain. The N-terminal domains of Ccd1 are required for interaction with target proteins such as MEK kinase 4, γ-tubulin, filamentous actins and disrupted in schizophrenia 1 (DISC1) (Wong et al., 2004 ▶; Wang et al., 2006 ▶; Wu et al., 2009 ▶; Singh et al., 2010 ▶).
All Ccd1 subtypes, as well as axin and Dvl, possess a C-terminal DIX domain. DIX domains play a key role in the regulation of the canonical Wnt signalling pathway through the formation of protein complexes with axin and Dvl (Kishida et al., 1999 ▶; Li et al., 1999 ▶; Shiomi et al., 2003 ▶). DIX domains also confer a unique ability to form cytoplasmic puncta upon overexpression and in a wide variety of mammalian cell lines by self-association (Schwarz-Romond et al., 2005 ▶; Schwarz-Romond, Metcalfe et al., 2007 ▶; Faux et al., 2008 ▶). Recent biochemical and biophysical studies of the Dvl DIX domain in vitro indicated that the DIX domain could polymerize with an estimated K d of 5–20 µM (Schwarz-Romond, Fiedler et al., 2007 ▶). Furthermore, the crystal structure of the axin DIX domain revealed head-to-tail helical filaments formed through an intermolecular parallel β-bridge between the β2 and β4 strands of a ubiquitin-like fold with five β-strands and one α-helix (Schwarz-Romond, Fiedler et al., 2007 ▶). DIX-domain-mediated polymerization could play an important role in amplification of the small signal into a strong response. However, the precise mechanisms by which the DIX domain of Ccd1 forms dynamic polymers and protein complexes with axin and Dvl are unknown. Here, we report the purification, crystallization and preliminary X-ray crystallographic characterization of the Ccd1 DIX domain.
2. Materials and methods
2.1. Protein expression
The nucleotide sequence encoding the DIX domain of mouse Ccd1 (residues 388–470) was amplified by PCR using cDNA of the BαL isoform (Gene ID 330938). The amplified fragment was subcloned into a pET49b plasmid (Novagen) using XmaI and XhoI restriction-enzyme sites. The resulting plasmid overexpresses the Ccd1 DIX domain with an N-terminal hexahistidine-tagged glutathione S-transferase (GST) linked by an HRV3C protease site. The construct produces an additional three residues (GPG) at the N-terminus after protease cleavage of the GST tag.
The GST-fused Ccd1 DIX domain was expressed in Escherichia coli BL21 (DE3)-CodonPlus-RILP cells (Stratagene). An overnight culture of cells (15 ml) transformed with pET49b-DIX was inoculated into 1.6 l LB medium containing 50 mg ml−1 kanamycin. The culture was grown at 310 K until the OD600nm reached 0.6. Expression of the recombinant protein was induced by the addition of isopropyl β-d-1-thiogalactopyranoside (IPTG) to the culture to a final concentration of 0.1 mM. Recombinant protein was expressed at 298 K overnight and the cells were harvested by centrifugation at 5000g for 10 min at 277 K.
The Ccd1 DIX domain contains a methionine residue. For multi-wavelength anomalous dispersion (MAD) experiments, selenomethionine (SeMet) substituted protein was expressed using a technique based on the inhibition of methionine biosynthesis (Duyne et al., 1993 ▶; Doublié, 1997 ▶). The amino acids required for the inhibition of methionine biosynthesis were added to the culture when it had grown to mid-log phase. Induction of recombinant protein expression was initiated 30 min after addition of the amino acids. Cultures were grown overnight after IPTG induction.
2.2. Purification
Cells were disrupted by sonication at 277 K. The supernatant was applied onto a glutathione Sepharose 4B column (GE Healthcare) and the column was washed with 50 mM Na HEPES buffer pH 7.5 containing 150 mM NaCl and 1 mM dithiothreitol (DTT). The GST-fusion protein bound to the GST-affinity column was cleaved overnight at 277 K using GST-fused HRV3C protease. The cleaved protein was applied onto a HiTrap Q Sepharose column (GE Healthcare) and eluted using a linear NaCl gradient (0.15–1.0 M). Fractions containing the Ccd1 DIX domain were collected and concentrated. Finally, the Ccd1 DIX domain was purified by gel-filtration chromatography on a Superdex 200 column (GE Healthcare) equilibrated and eluted with 10 mM Na HEPES buffer pH 7.5 containing 150 mM NaCl and 1 mM DTT.
Protein purity was monitored by 17% polyacrylamide gel electrophoresis and staining was carried out with Simply Stain Blue (Invitrogen). Purified samples were validated by N-terminal sequence analysis and matrix-assisted laser desorption/ionization time-of-flight mass spectrometry (MALDI–TOF MS). The presence of three artificial residues, GPG, which remained at the N-terminus of the resultant fragment was confirmed by N-terminal sequence analysis.
2.3. Dynamic light-scattering measurements
Dynamic light-scattering (DLS) measurements were carried out using a Zetasizer µV instrument (Malvern Instruments Ltd). Protein solutions were centrifuged at 20 000g for 10 min to remove debris. All measurements were performed using a 2 µl quartz cuvette filled with 2 µl protein solution at 283 K. Size distributions were calculated from the intensity data acquired at regular time intervals.
2.4. Crystallization and X-ray data collection
The purified Ccd1 DIX domain and SeMet-substituted protein were concentrated to 20 mg ml−1 in a solution consisting of 10 mM Na HEPES pH 7.5, 150 mM NaCl and 1 mM DTT using a Vivaspin 20 (3000 MWCO PES, Vivascience). Initial crystallization screening was performed by the sitting-drop vapour-diffusion method using commercial crystallization solution kits, i.e. Index, Crystal Screen and Crystal Screen 2 (Hampton Research), at 283 K. Crystallization drops were prepared by mixing 1 µl protein solution with 1 µl of each reservoir solution and were equilibrated against 200 µl reservoir solution. The conditions obtained from the screening were optimized using the hanging-drop vapour-diffusion method at 283 K.
The crystals used for data collection were soaked in reservoir solution containing 25%(v/v) ethylene glycol and flash-cooled at 100 K in an N2-gas stream. X-ray diffraction data were collected from the native crystals of the Ccd1 DIX domain using an ADSC Quantum 315 CCD detector installed on the BL26B2 beamline at SPring-8. Data collection for the native crystals was performed using an angular range of 180°, a step size of 1.0° and an exposure time of 10 s. The camera was fixed at a distance of 200 mm. The data set used for the MAD method was collected on the BL44XU beamline at SPring-8 using a Rayonix MX225HE detector. Optimal peak, edge and remote wavelengths were determined by X-ray fluorescence. Data collection for the SeMet-substituted crystals was performed using an angular range of 360°, a step size of 1.5° and an exposure time of 1.5 s. The camera was fixed at a distance of 300 mm. The diffraction data were processed using HKL-2000 (Otwinowski & Minor, 1997 ▶).
3. Results and discussion
The homogeneity of the purified Ccd1 DIX domain was validated by SDS–PAGE and MALDI–TOF MS. MALDI–TOF MS of the Ccd1 sample resulted in a single peak at 9769.7 Da that corresponded to the calculated value of 9769.9 Da for the recombinant Ccd1 DIX domain. DLS was used to confirm that the purified DIX domain of Ccd1 possessed the property of polymerizing in a reversible fashion. In the DLS measurements the Ccd1 DIX domain oligomer was observed as a single peak in different experiments, although the size distribution of the peaks was not monodisperse (the polydispersity was approximately 30%). However, a concentration-dependent increase in the estimated average hydrodynamic radius of the Ccd1 DIX domain was clearly observed. The size distribution of the Ccd1 DIX domain at two protein concentrations (0.1 and 4.2 mM) indicated average radii of 3.9 ± 1.3 and 5.7 ± 1.7 nm, respectively. The estimated average molecular weights were 33.7 and 214 kDa, respectively. These results suggest that the purified Ccd1 DIX domain polymerizes reversibly in a concentration-dependent manner.
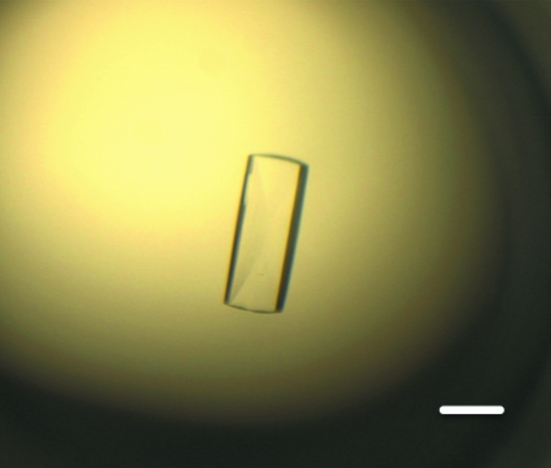
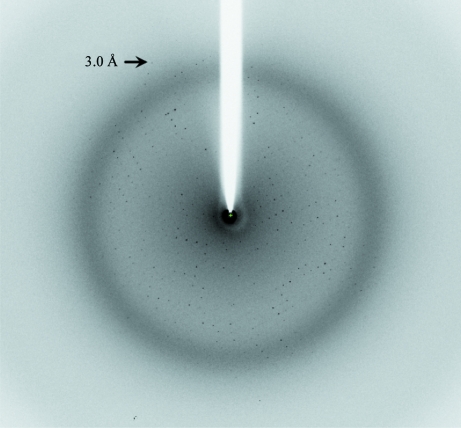
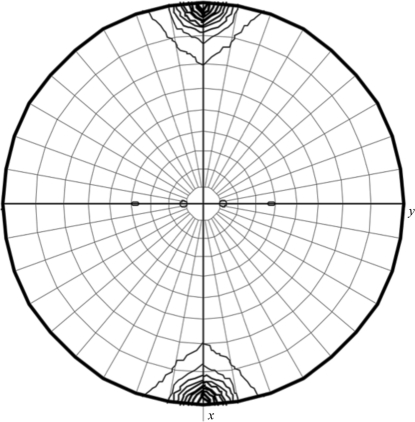
In the initial stages of screening, needle-like crystals were obtained in 3 d using a solution consisting of 6.3%(v/v) PEG 3350, 0.1 M Na HEPES pH 7.8, 80 mM l-proline and 3%(v/v) dimethylsulfoxide (DMSO). Similar needle-like crystals were obtained using the same concentration of glycerol or ethylene glycol in place of DMSO. However, these crystals were unsuitable for crystallographic analysis because their diffraction quality was poor (around 4 Å resolution). Plate-like crystals that diffracted to around 3.1 Å resolution were obtained by additional screening using screening kits containing additive reagents such as glycerol, ethylene glycol or 1,3-propanediol. The best crystals of the Ccd1 DIX domain were obtained using a precipitant solution consisting of 0.1 M Na HEPES pH 7.8, 15%(v/v) ethylene glycol, 3%(v/v) glycerol and 4%(v/v) 1,3-propanediol at 283 K. The crystals grew to maximum dimensions of 0.5 × 0.2 × 0.1 mm after two weeks (Fig. 1 ▶). The crystals diffracted to a resolution of 3.0 Å and belonged to space group P212121, with unit-cell parameters a = 72.9, b = 75.7, c = 125.6 Å (Fig. 2 ▶). We integrated 89 162 reflections to a resolution of 3.0 Å and these were merged to obtain 14 312 unique reflections with an overall R merge of 10.7% and a completeness of 99.0%. The self-rotation function calculated with the MOLREP program (Vagin & Teplyakov, 2010 ▶) shows peaks corresponding to a noncrystallographic sevenfold axis (Fig. 3 ▶). A Matthews coefficient (Matthews, 1968 ▶) of 3.31 Å3 Da−1 was calculated assuming the presence of seven DIX domains with an approximate molecular weight of 10 kDa in an asymmetric unit, which corresponds to 53.1% solvent content by volume.
Figure 1.
Crystal of the Ccd1 DIX domain. The scale bar indicates 200 µm.
Figure 2.
X-ray diffraction image of the native crystal of the Ccd1 DIX domain.
Figure 3.
χ = 51.4° section of the self-rotation function calculated using MOLREP in the resolution range 12–3.5 Å with an integration radius of 30 Å.
Structure determination by molecular replacement using the axin DIX domain (PDB entry 1wsp; Schwarz-Romond, Fiedler et al., 2007 ▶) as a search model (40% sequence identity) was unsuccessful. SeMet-substituted Ccd1 DIX domain was produced using the E. coli expression system in order to determine the structure using the MAD method. The substitution of Met by SeMet was validated by MALDI–TOF MS, which displayed a peak at 9816.6 Da (native protein: 9769.9 Da). SeMet-substituted DIX domain crystals were obtained in two weeks using a precipitant solution consisting of 0.1 M Na HEPES pH 7.0, 6%(v/v) ethylene glycol, 3%(v/v) glycerol, 5 mM EDTA and 4% 1,3-propanediol. Table 1 ▶ summarizes the detailed statistics of the native and MAD data sets.
Table 1. X-ray diffraction data of the Ccd1 DIX domain crystal.
Values in parentheses are for the highest resolution shell.
| SeMet derivative | ||||
|---|---|---|---|---|
| Native | Peak | Edge | Remote | |
| Space group | P212121 | P212121 | P212121 | P212121 |
| Unit-cell parameters (Å) | ||||
| a | 72.9 | 72.9 | 72.9 | 72.9 |
| b | 75.7 | 78.6 | 78.6 | 78.7 |
| c | 125.6 | 125.9 | 125.9 | 126.0 |
| Wavelength (Å) | 1.0000 | 0.9748 | 0.9793 | 0.9641 |
| Resolution (Å) | 50–3.0 (3.11–3.00) | 50–3.2 (3.31–3.20) | 50–3.2 (3.31–3.20) | 50–3.2 (3.31–3.20) |
| Reflections | ||||
| Measured | 89162 | 148041 | 148755 | 148091 |
| Unique | 14312 | 11893 | 11857 | 11848 |
| Multiplicity | 6.2 (4.5) | 12.5 (8.4) | 12.6 (8.6) | 12.5 (8.7) |
| Completeness (%) | 99.0 (91.6) | 95.5 (72.5) | 95.2 (70.1) | 95.0 (68.6) |
| Mean I/σ(I) | 16.3 (2.3) | 15.3 (2.0) | 17.0 (2.2) | 14.5 (2.2) |
| Rmerge† (%) | 10.7 (52.8) | 14.7 (54.7) | 13.8 (51.2) | 14.4 (54.8) |
R
merge = 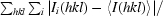
 , where Ii(hkl) is the ith observed intensity of reflection hkl and 〈I(hkl)〉 is the average intensity over symmetry-equivalent measurements.
, where Ii(hkl) is the ith observed intensity of reflection hkl and 〈I(hkl)〉 is the average intensity over symmetry-equivalent measurements.
Initial experimental phases with an averaged figure of merit (FOM) of 0.27 at 3.2 Å resolution were calculated using autoSHARP (Vonrhein et al., 2007 ▶). Density modification by solvent flipping using SOLOMON improved the averaged FOM to 0.87 at 3.2 Å resolution (Abrahams & Leslie, 1996 ▶). Structural analysis of the Ccd1 DIX domain is now in progress.
Acknowledgments
This work was supported by a Grant-in-Aid for the Global COE program (YH) and Scientific Research on Innovation Areas for ‘Transition Molecular Complex’ (YH) from the Ministry of Education, Culture, Sports, Science and Technology (MEXT) of Japan, JSPS [Category B No. 22370061 (YH), Challenging Exploratory Research No. 22657031 (YH), Creative Scientific Research 18GS0207 (YH) and Young Scientists B No. 22770112 (ST)], JST [CREST (YH)] and JAXA (YH) programs, the Hyogo Science and Technology Association (ST), a Sasakawa Scientific Research Grant from the Japan Science Society (ST) and Leave a Nest Grants Caliper Life Science Award (ST). We acknowledge the staff at beamline BL26B2 and BL44XU, SPring-8, Japan.
References
- Aberle, H., Bauer, A., Stappert, J., Kispert, A. & Kemler, R. (1997). EMBO J. 16, 3797–3804. [DOI] [PMC free article] [PubMed]
- Abrahams, J. P. & Leslie, A. G. W. (1996). Acta Cryst. D52, 30–42. [DOI] [PubMed]
- Dajani, R., Fraser, E., Roe, S. M., Yeo, M., Good, V. M., Thompson, V., Dale, T. C. & Pearl, L. H. (2003). EMBO J. 22, 494–501. [DOI] [PMC free article] [PubMed]
- Doublié, S. (1997). Methods Enzymol. 276, 523–530. [PubMed]
- Faux, M. C., Coates, J. L., Catimel, B., Cody, S., Clayton, A. H., Layton, M. J. & Burgess, A. W. (2008). Oncogene, 27, 5808–5820. [DOI] [PubMed]
- Hart, M. J., de los Santos, R., Albert, I. N., Rubinfeld, B. & Polakis, P. (1998). Curr. Biol. 8, 573–581. [DOI] [PubMed]
- Ikeda, S., Kishida, S., Yamamoto, H., Murai, H., Koyama, S. & Kikuchi, A. (1998). EMBO J. 17, 1371–1384. [DOI] [PMC free article] [PubMed]
- Ikeuchi, Y., Stegmüller, J., Netherton, S., Huynh, M. A., Masu, M., Frank, D., Bonni, S. & Bonni, A. (2009). J. Neurosci. 29, 4312–4321. [DOI] [PMC free article] [PubMed]
- Jing, X.-T., Wu, H.-T., Wu, Y., Ma, X., Liu, S.-H., Wu, Y.-R., Ding, X.-F., Peng, X.-Z., Qiang, B.-Q., Yuan, J.-G., Fan, W.-H. & Fan, M. (2009). Cell. Mol. Neurobiol. 29, 55–67. [DOI] [PMC free article] [PubMed]
- Kishida, S., Yamamoto, H., Hino, S., Ikeda, S., Kishida, M. & Kikuchi, A. (1999). Mol. Cell. Biol. 19, 4414–4422. [DOI] [PMC free article] [PubMed]
- Kishida, S., Yamamoto, H., Ikeda, S., Kishida, M., Sakamoto, I., Koyama, S. & Kikuchi, A. (1998). J. Biol. Chem. 273, 10823–10826. [DOI] [PubMed]
- Klingensmith, J., Nusse, R. & Perrimon, N. (1994). Genes Dev. 8, 118–130. [DOI] [PubMed]
- Li, L., Yuan, H., Weaver, C. D., Mao, J., Farr, G. H., Sussman, D. J., Jonkers, J., Kimelman, D. & Wu, D. (1999). EMBO J. 18, 4233–4240. [DOI] [PMC free article] [PubMed]
- MacDonald, B. T., Tamai, K. & He, X. (2009). Dev. Cell, 17, 9–26. [DOI] [PMC free article] [PubMed]
- Matthews, B. W. (1968). J. Mol. Biol. 33, 491–497. [DOI] [PubMed]
- Moon, R. T., Bowerman, B., Boutros, M. & Perrimon, N. (2002). Science, 296, 1644–1646. [DOI] [PubMed]
- Nakamura, T., Hamada, F., Ishidate, T., Anai, K., Kawahara, K., Toyoshima, K. & Akiyama, T. (1998). Genes Cells, 3, 395–403. [DOI] [PubMed]
- Orford, K., Crockett, C., Jensen, J. P., Weissman, A. M. & Byers, S. W. (1997). J. Biol. Chem. 272, 24735–24738. [DOI] [PubMed]
- Otwinowski, Z. & Minor, W. (1997). Methods Enzymol. 276, 307–326. [DOI] [PubMed]
- Peifer, M. & Polakis, P. (2000). Science, 287, 1606–1609. [DOI] [PubMed]
- Salic, A., Lee, E., Mayer, L. & Kirschner, M. W. (2000). Mol. Cell, 5, 523–532. [DOI] [PubMed]
- Schwarz-Romond, T., Fiedler, M., Shibata, N., Butler, P. J. G., Kikuchi, A., Higuchi, Y. & Bienz, M. (2007). Nature Struct. Mol. Biol. 14, 484–492. [DOI] [PubMed]
- Schwarz-Romond, T., Merrifield, C., Nichols, B. J. & Bienz, M. (2005). J. Cell Sci. 118, 5269–5277. [DOI] [PubMed]
- Schwarz-Romond, T., Metcalfe, C. & Bienz, M. (2007). J. Cell Sci. 120, 2402–2412. [DOI] [PubMed]
- Shiomi, K., Kanemoto, M., Keino-Masu, K., Yoshida, S., Soma, K. & Masu, M. (2005). Brain Res. Mol. Brain Res. 135, 169–180. [DOI] [PubMed]
- Shiomi, K., Uchida, H., Keino-Masu, K. & Masu, M. (2003). Curr. Biol. 13, 73–77. [DOI] [PubMed]
- Singh, K. K., Ge, X., Mao, Y., Drane, L., Meletis, K., Samuels, B. A. & Tsai, L.-H. (2010). Neuron, 67, 33–48. [DOI] [PMC free article] [PubMed]
- Soma, K., Shiomi, K., Keino-Masu, K. & Masu, M. (2006). Gene Expr. Patterns, 6, 325–330. [DOI] [PubMed]
- Theisen, H., Purcell, J., Bennett, M., Kansagara, D., Syed, A. & Marsh, J. L. (1994). Development, 120, 347–360. [DOI] [PubMed]
- Vagin, A. & Teplyakov, A. (2010). Acta Cryst. D66, 22–25. [DOI] [PubMed]
- Van Duyne, G. D., Standaert, R. F., Karplus, P. A., Schreiber, S. L. & Clardy, J. (1993). J. Mol. Biol. 229, 105–124. [DOI] [PubMed]
- Vonrhein, C., Blanc, E., Roversi, P. & Bricogne, G. (2007). Methods Mol. Biol. 364, 215–230. [DOI] [PubMed]
- Wang, X., Zheng, L., Zeng, Z., Zhou, G., Chien, J., Qian, C., Vasmatzis, G., Shridhar, V., Chen, L. & Liu, W. (2006). Biochem. Biophys. Res. Commun. 347, 22–30. [DOI] [PubMed]
- Wodarz, A. & Nusse, R. (1998). Annu. Rev. Cell Dev. Biol. 14, 59–88. [DOI] [PubMed]
- Wong, C. K., Luo, W., Deng, Y., Zou, H., Ye, Z. & Lin, S.-C. (2004). J. Biol. Chem. 279, 39366–39373. [DOI] [PubMed]
- Wu, Y., Jing, X., Ma, X., Wu, Y., Ding, X., Fan, W. & Fan, M. (2009). Cell Biol. Int. 33, 697–701. [DOI] [PubMed]
- Xing, Y., Clements, W. K., Kimelman, D. & Xu, W. (2003). Genes Dev. 17, 2753–2764. [DOI] [PMC free article] [PubMed]