MsDpo4 from M. smegmatis, a member of the Y-family of DNA polymerases, was expressed and purified to homogeneity. Crystals of native MsDpo4 in the apo state were obtained and X-ray diffraction data were collected to a maximum resolution of 2.6 Å.
Keywords: DNA polymerases, MsDpo4, Mycobacterium smegmatis
Abstract
The expression of error-prone DNA polymerases belonging to the Y-family is upregulated in prokaryotes under adverse conditions to facilitate adaptive mutagenesis. However, it has been suggested that representatives of this family in mycobacteria do not participate in adaptive mutagenesis. These studies raise the possibility that mycobacterial representatives might be devoid of biochemical activity. In order to determine whether this possible loss of activity is a consequence of significant changes in the structure of these enzymes, structural studies on a representative enzyme from Mycobacterium smegmatis, MsDpo4, were initiated. The protein crystallized in space group P6122 or P6522. The Matthews coefficient was 4.0 Å3 Da−1 assuming the presence of one molecule in the asymmetric unit; the corresponding solvent content was 69%. X-ray diffraction data were collected to a minimum Bragg spacing of 2.6 Å and crystals of selenomethionine-labelled MsDpo4 have been prepared for ab initio phasing.
1. Introduction
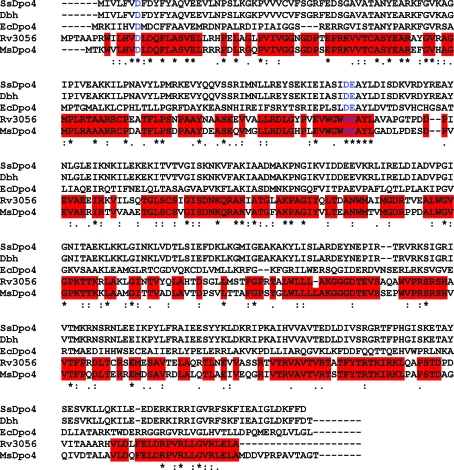
Adaptive mutagenesis involves the synthesis of DNA with low fidelity to increase the frequency at which heritable mutations appear in the genome. This phenomenon provides scope for evolution in the case of adverse conditions and helps to relieve selection pressure imposed by the environment (Galhardo et al., 2007 ▶; Foster, 2007 ▶). Adaptive mutagenesis is held to be responsible for the appearance of point mutations that render resistance to therapeutic agents in the case of pathogenic bacteria (Zhang & Yew, 2009 ▶; Karpinets et al., 2006 ▶). This phenomenon is usually mediated by specialized error-prone DNA polymerases. These enzymes generally belong to the Y-family of DNA polymerases and their expression is upregulated under adverse conditions. The dinB gene in Escherichia coli codes for DNA polymerase IV (EcDpo4), a member of the Y-family, which has been implicated in mutagenesis induced as a response to stress imposed by the environment (Galhardo et al., 2009 ▶). Sequence analysis of the Mycobacterium tuberculosis genome shows that this organism has two possible homologues of DinB, annotated Rv1537 (468 residues) and Rv3056 (346 residues). Rv3056 is of similar length to EcDpo4 (351 residues) and exhibits greater homology to EcDpo4 than Rv1537. Unlike in E. coli, in the case of M. tuberculosis it has been observed that the expression of Rv1537 and Rv3056 is not upregulated under conditions of stress (Brooks et al., 2001 ▶; Kana et al., 2010 ▶). These studies suggest that dinB homologues in mycobacteria code for nonfunctional molecules that are devoid of enzyme activity. The polypeptide sequence of Rv3056 exhibits high homology (71% identity) to a representative member from M. smegmatis named MsDpo4 (356 residues). A comparison of the polypeptide sequence of MsDpo4 with those of EcDpo4, Rv3056 and the structurally well characterized archaeal homologues SsDpo4 (Sulfolobus solfataricus) and Dbh (S. acidocaldarius) shows that although the catalytic residues are conserved there is only a moderate level of sequence identity between the mycobacterial members and EcDpo4/SsDpo4/Dbh (Fig. 1 ▶). In order to ascertain whether the possible loss of activity is a consequence of significant differences in structural architecture compared with known members of this family, we have initiated studies to determine the structure of MsDpo4 in its apo state. In this study, MsDpo4 was cloned, expressed in E. coli and purified by affinity and size-exclusion chromatography. The purified protein crystallized in space group P6122 or P6522 with one molecule per asymmetric unit and a complete data set was collected to a maximum resolution of 2.6 Å. Despite numerous attempts with a number of models, molecular replacement did not yield a satisfactory solution. Hence, crystals of selenomethionine-labelled protein have been prepared for ab initio phasing.
Figure 1.
Sequence alignment of representative DinB homologues. The polypeptide sequences of DinB from S. solfataricus (SsDpo4), S. acidocaldarius (Dbh), E. coli (EcDpo4), M. tuberculosis (Rv3056) and M. smegmatis (MsDpo4) are compared. The active-site residues are displayed in blue and residues that are identical in Rv3056 and MsDpo4 are highlighted by a red background. The asterisks below the sequences highlight identical residues and the colons and dots highlight residues that exhibit high and low similarity, respectively.
2. Methods
2.1. Cloning of MsDpo4
The MsDpo4 (KEGG Database MSMEG_1014) coding sequence was amplified by PCR from genomic DNA of M. smegmatis and was cloned into the pGEX-6P1 (GE Healthcare) expression vector between EcoRI and XhoI flanking sites. Recombinant expression using this vector (pGEX6P1_1014) gives rise to a GST-MsDpo4 fusion protein. The PCR amplification was performed using the forward and reverse oligonucleotide primers 5′-GCT TGC GAA TTC ACC AAA TGG GTG CTC CAC GTC G-3′ and 5′-GCA AGC CTC GAG TTA GGT GCC TGC AGT GAC AGC-3′, respectively.
2.2. Overexpression and purification
Competent E. coli C41 (DE3) cells were transformed with the pGEX6P1_1014 plasmid and the cells were plated onto LB agar containing ampicillin (100 µg ml−1). A starter culture (20 ml) was inoculated with a single colony and grown overnight at 310 K with shaking at 220 rev min−1. This starter culture was used to inoculate 5 l LB medium with ampicillin (100 µg ml−1). When the OD600 reached 0.7, expression of MsDpo4 was induced by the addition of 0.5 mM isopropyl β-d-1-thiogalactopyranoside (IPTG) to the culture and incubation at 291 K for 18 h with shaking at 180 rev min−1. The cells were harvested by centrifugation at 9000g for 30 min at 277 K, and the cell pellet was resuspended in buffer A (25 mM Tris–HCl pH 8.0, 250 mM NaCl, 5% glycerol, 2 mM DTT, 0.01% IGEPAL) and lysed by sonication. After sonication on ice, a clear supernatant was obtained by centrifugation at 18 000 rev min−1 for 45 min at 277 K.
The first step in purifying GST-MsDpo4 involved affinity chromatography utilizing a 5 ml GST-Sepharose column (GE Healthcare) pre-equilibrated with buffer A (lysis buffer). The clear supernatant of the bacterial lysate was applied onto this column and the column was washed with 45 ml buffer A at 277 K. This was followed by washing with 45 ml buffer B (25 mM Tris–HCl pH 8.0, 1000 mM NaCl, 5% glycerol, 2 mM DTT, 0.01% IGEPAL) to remove any nonspecifically bound proteins. After a second wash with 45 ml buffer A, the fusion protein was eluted with 15 mM glutathione in buffer A. To cleave the GST tag, this protein was incubated with PreScission protease (GE Healthcare) overnight at 277 K. MsDpo4 was then purified from the resulting mixture by gel-filtration chromatography using a Superdex 200 column (GE Healthcare). The column was pre-equilibrated with buffer C (25 mM Tris–HCl, 250 mM NaCl, 2 mM DTT pH 8.0). A 1 ml GST-Sepharose column was connected in series to trap the cleaved GST tag and GST-tagged PreScission protease. The purity of the protein was assessed by SDS–PAGE. The purified protein was concentrated using Vivaspin 6 (Argos Technologies) centrifugal concentrators with a molecular-weight cutoff of 10 kDa and the final concentration was estimated using the BCA protein assay (Pierce Biotechnology). The protein was also subjected to mass spectrometry to assess its purity and molecular weight. Prior to recording mass spectra, the final protein preparation was rendered salt-free using C18 ZipTips (Millipore Corporation), with the final elution occurring in a solvent containing 60% formic acid and 0.1% TFA. This salt-free preparation of MsDpo4 was used to record mass spectra on a Q-TOF Ultima machine (Waters Corporation).
2.3. Crystallization
Crystallization trials were carried out manually at 298 and 277 K using the hanging-drop vapour-diffusion method with 24-well XRL plates (Molecular Dimensions). Equal volumes (1 µl) of protein solution and precipitant solution were mixed and the drops were equilibrated against 500 µl of the same precipitant solution in the reservoir. A number of crystallization conditions were tested using commercially available kits from Emerald BioSystems (Wizard I and Wizard II) and Jena Biosciences (Basic Suite and JCSG screen).
2.4. Preliminary screening and processing of diffraction data
Mounted CryoLoops (Hampton Research) were used to separate and pick up the crystals. Prior to freezing, crystals were treated with different concentrations of cryoprotectants such as glycerol, ethylene glycol and MPD for varying periods. The quality of diffraction was tested using a Bruker Microstar-H X-ray generator with X-ray optics and the frames were recorded on a MAR345 imaging plate at the Molecular Biophysics Unit, Indian Institute of Science. The frames collected were indexed using iMOSFLM (Battye et al., 2011 ▶). X-ray data were collected from a single crystal on the BM14 beamline of the European Synchrotron Radiation Facility (ESRF). The data were integrated and scaled using DENZO and SCALEPACK as part of the HKL-2000 package (Otwinowski & Minor, 1997 ▶).
2.5. Molecular replacement
Potential search models for molecular replacement (MR) were found by comparing the MsDpo4 sequence with those of all structures available in the PDB using the BLAST server (Altschul et al., 1997 ▶). This exercise revealed that SsDpo4 and Dbh were the closest homologues. MsDpo4 exhibits sequence identities of 27% (93 identities in 342 pairs) and 24% (85 identities in 351 pairs) to SsDpo4 and Dbh, respectively. MR was attempted using structures of SsDpo4 in different conformations: the apo state (PDB entry 2rdi; Wong et al., 2008 ▶), a binary protein–DNA complex (PDB entry 2rdj; Wong et al., 2008 ▶) and a ternary protein–DNA–incoming nucleotide complex (PDB entry 1jx4; Ling et al., 2001 ▶). In addition, MR was also attempted with structures of Dbh in the apo state (PDB entry 1im4; Zhou et al., 2001 ▶) and in binary (PDB entry 3bq0; Wilson & Pata, 2008 ▶) and ternary complexes (PDB entry 3bq1; Wilson & Pata, 2008 ▶). The programs Phaser and MOLREP, which are part of the CCP4 suite, were used for MR (McCoy et al., 2007 ▶; Vagin & Teplyakov, 2010 ▶; Winn et al., 2011 ▶).
2.6. Preparation of SeMet-labelled MsDpo4
To prepare selenomethionine-labelled MsDpo4 (Se-MsDpo4), the protein was expressed in the B834 strain of E. coli, which is auxotrophic for methionine, and the cells were grown using an SeMet medium kit (Molecular Dimensions). Fresh competent B834 cells were transformed with the pGEX6P1_1014 plasmid and the cells were plated onto LB agar containing ampicillin (100 µg ml−1). A single colony was used to inoculate a starter culture (20 ml), which was grown overnight at 310 K. 2 l reconstituted SeMet medium (Molecular Dimensions) was supplemented with 50 mg l−1 selenomethionine (Sigma–Aldrich) and this medium was used to wash the pelleted cells from the starter culture thrice before inoculation. When the OD reached 0.7 (∼6 h), expression of MsDpo4 was induced with 0.5 mM IPTG and the flasks were incubated overnight at 291 K with shaking at 180 rev min−1. Se-MsDpo4 was purified using a protocol identical to that used for the native protein and the level of selenium incorporation was assessed by mass spectrometry. Salt-free samples of Se-MsDpo4 prepared using C18 ZipTips (Millipore Corporation) were used to record mass spectra on a Q-TOF Ultima machine (Waters Corporation).
3. Results
3.1. Expression and purification
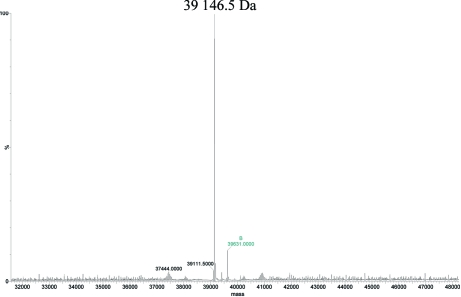
Preliminary expression studies showed that MsDpo4 was optimally expressed in E. coli C41 (DE3) cells, producing significant amounts of soluble protein. The heterologously expressed protein was then purified by affinity and size-exclusion chromatography. The sample was 99% pure as estimated by SDS–PAGE (Fig. 2 ▶). Mass spectrometry gave a molecular weight of 39 146 Da, which is identical to the calculated theoretical molecular weight (Fig. 3 ▶). The calculated molecular weight takes into account seven residues at the N-terminus that appear from the linker to the GST tag and the choice of an EcoRI restriction site during cloning. The final yield from 5 l culture was 22 mg and the protein was concentrated to a final concentration of 1 mM.
Figure 2.
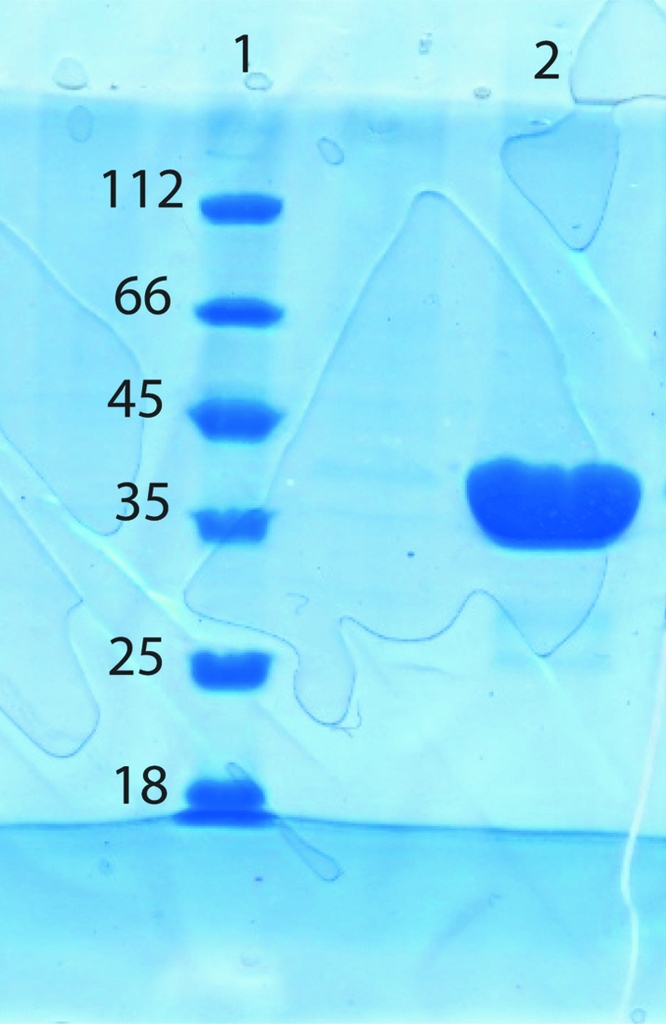
SDS–PAGE analysis of purified MsDpo4. A standard molecular-weight marker (Fermentas) was loaded in lane 1 (labelled in kDa); purified and concentrated MsDpo4 was loaded in lane 2.
Figure 3.
Mass spectrum of MsDpo4. The deconvoluted mass spectrum of MsDpo4 is displayed and indicates a molecular weight of 39 146 Da for the purified MsDpo4.
3.2. Crystallization
Using the hanging-drop method, small needle-shaped crystals were obtained in condition No. 16 (2.5 M sodium chloride, 0.1 M sodium/potassium phosphate pH 6.2) of the Wizard I screen after 3 d at 298 K. The concentration of NaCl and the pH of the buffer were varied to optimize the size of the crystals. Single crystals were finally obtained with 2.4 M sodium chloride pH 6.4 (0.1 M sodium/potassium phosphate). The crystals exhibit an elongated cylindrical morphology and were obtained in 72 h at 298 K (Fig. 4 ▶).
Figure 4.
Crystals of apo MsDpo4. The MsDpo4 crystals exhibit cylindrical morphology with approximate dimensions of 0.1 × 0.03 mm.
3.3. Data collection and molecular replacement
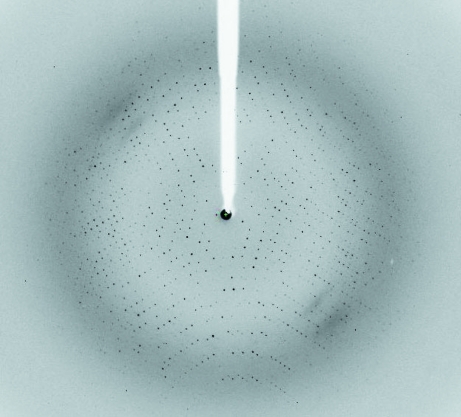
The optimized cryoprotection strategy involved soaking the crystal sequentially for 60 s in reservoir solution supplemented with 5, 10, 15 and 20% glycerol. The crystals diffracted to a resolution of 3.0 Å on a rotating-anode X-ray generator. The crystals belonged to the hexagonal space group P6122 or P6522, with unit-cell parameters a = 124.1, b = 124.1, c = 141.0 Å. A complete X-ray diffraction data set of about sevenfold multiplicity was collected with a minimum Bragg spacing of 2.6 Å. A representative frame recorded during this data collection is shown in Fig. 5 ▶. The volume of the asymmetric unit allows the presence of a monomer, giving a Matthews coefficient (V M) of 4.0 Å3 Da−1 and a solvent content of 69.4% (Matthews, 1968 ▶). Details of the data-collection statistics are reported in Table 1 ▶. Numerous attempts at molecular replacement using a number of models and different programs such as Phaser and MOLREP did not provide any satisfactory solution. DNA polymerases belonging to the Y-family usually exhibit the presence of four domains: fingers, palm, thumb and PAD. The PAD region shows maximal sequence divergence and MR was also attempted by removing the PAD region from the search models, without success.
Figure 5.
Diffraction quality of apo MsDpo4. A representative diffraction image collected from a crystal of apo MsDpo4 on the BM14 beamline of the ESRF is displayed.
Table 1. Data-collection statistics.
Values in parentheses are for the highest resolution shell. Data were collected using one crystal.
| Beamline | BM14, ESRF |
| Wavelength (Å) | 1.0 |
| Space group | P6122/P6522 |
| Unit-cell parameters (Å, °) | a = 124.1, b = 124.1, c = 141.0, α = 90.0, β = 90.0, γ = 120.0 |
| Resolution (Å) | 50–2.6 (2.69–2.60) |
| Reflections measured | 146788 |
| Unique reflections | 20393 |
| Rmerge† | 9.1 (47.4) |
| 〈I/σ(I)〉 | 23.49 (4.5) |
| Completeness (%) | 100 (100) |
| Multiplicity | 7.2 (7.3) |
| Monomers per asymmetric unit | 1 |
| Matthews coefficient (A3 Da−1) | 4.0 |
| Solvent content (%) | 69.4 |
R
merge = 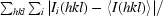
 .
.
3.4. Preparation and crystallization of selenomethionine-labelled protein
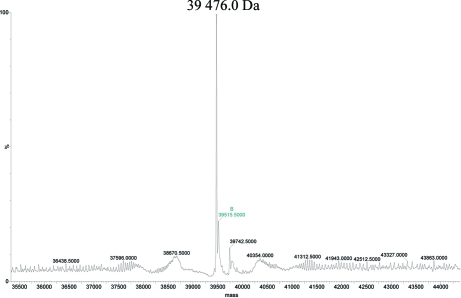
Selenomethionine-labelled MsDpo4 (Se-MsDpo4) was purified using the same protocol as that used for the native protein. Around 3 mg of Se-MsDpo4 was obtained and the protein was concentrated to 22 mg ml−1. The molecular weight of the Se-MsDpo4 protein was estimated by mass spectrometry to be 39 476 Da (Fig. 6 ▶). The difference from the native protein is 330 Da, suggesting that all seven methionines have been substituted by selenomethionine and that the incorporation is 100%. Crystals of Se-MsDpo4 were obtained under identical conditions as used for those of the native protein and will be used for ab initio phasing.
Figure 6.
Mass spectrum of Se-MsDpo4. The deconvoluted mass spectrum of Se-MsDpo4 is displayed and indicates a molecular weight of 39 476 Da for the purified Se-MsDpo4.
4. Discussion
Genetic studies raise the possibility that the representatives of the Y-family of DNA polymerases in mycobacteria may be devoid of enzyme activity. The polypeptide sequences of mycobacterial DinB homologues exhibit only moderate homology to those of EcDpo4 from E. coli and the structurally well characterized archaeal homologues SsDpo4 and Dbh. The possible loss of biochemical activity of mycobacterial DinB homologues could therefore be a consequence of changes in tertiary structure. To test this hypothesis, MsDpo4, a representative DinB homologue from M. smegmatis, was crystallized in its apo state and X-ray diffraction data were collected to a maximum resolution of 2.6 Å. Molecular replacement using structures of SsDpo4 and Dbh in apo, binary (protein–DNA) and ternary (protein–DNA–nucleotide) states did not yield any solutions, suggesting that there may be critical differences in the structure of MsDpo4. Crystals of SeMet-labelled MsDpo4 for ab initio phasing have been prepared and the structure will reveal whether mycobacterial DinB homologues exhibit any unusual structural attributes.
Acknowledgments
The authors would like to thank Professor S. Vijaya for the kind gift of M. smegmatis genomic DNA. The authors thank the Molecular Biophysics Unit, IISc (Indian Institute of Science), Bangalore for access to their X-ray diffraction facility for screening crystals. DTN acknowledges the help rendered by Dr Hassan Belrhali (ESRF) during data collection at the BM-14 beamline at the ESRF. Funding for travel to ESRF and beamtime was provided by the BM-14 project of the Department of Biotechnology (Government of India) and the ESRF. The authors would like to thank the Mass Spectrometry facility at NCBS and Dr Sudarslal for help with mass determination. AS is the recipient of a senior research fellowship from the Council of Scientific and Industrial Research (Government of India). DTN is the recipient of a Ramanujan Fellowship from the Department of Science and Technology (Government of India). The authors thank Vidya Subramanian for help with preparing the figures.
References
- Altschul, S. F., Madden, T. L., Schäffer, A. A., Zhang, J., Zhang, Z., Miller, W. & Lipman, D. J. (1997). Nucleic Acids Res. 25, 3389–3402. [DOI] [PMC free article] [PubMed]
- Battye, T. G. G., Kontogiannis, L., Johnson, O., Powell, H. R. & Leslie, A. G. W. (2011). Acta Cryst. D67, 271–281. [DOI] [PMC free article] [PubMed]
- Brooks, P. C., Movahedzadeh, F. & Davis, E. O. (2001). J. Bacteriol. 183, 4459–4467. [DOI] [PMC free article] [PubMed]
- Foster, P. L. (2007). Crit. Rev. Biochem. Mol. Biol. 42, 373–397. [DOI] [PMC free article] [PubMed]
- Galhardo, R. S., Do, R., Yamada, M., Friedberg, E. C., Hastings, P. J., Nohmi, T. & Rosenberg, S. M. (2009). Genetics, 182, 55–68. [DOI] [PMC free article] [PubMed]
- Galhardo, R. S., Hastings, P. J. & Rosenberg, S. M. (2007). Crit. Rev. Biochem. Mol. Biol. 42, 399–435. [DOI] [PMC free article] [PubMed]
- Kana, B. D., Abrahams, G. L., Sung, N., Warner, D. F., Gordhan, B. G., Machowski, E. E., Tsenova, L., Sacchettini, J. C., Stoker, N. G., Kaplan, G. & Mizrahi, V. (2010). J. Bacteriol. 192, 2220–2227. [DOI] [PMC free article] [PubMed]
- Karpinets, T. V., Greenwood, D. J., Pogribny, I. P. & Samatova, N. F. (2006). Curr. Genomics, 7, 481–496. [DOI] [PMC free article] [PubMed]
- Ling, H., Boudsocq, F., Woodgate, R. & Yang, W. (2001). Cell, 107, 91–102. [DOI] [PubMed]
- Matthews, B. W. (1968). J. Mol. Biol. 33, 491–497. [DOI] [PubMed]
- McCoy, A. J., Grosse-Kunstleve, R. W., Adams, P. D., Winn, M. D., Storoni, L. C. & Read, R. J. (2007). J. Appl. Cryst. 40, 658–674. [DOI] [PMC free article] [PubMed]
- Otwinowski, Z. & Minor, W. (1997). Methods Enzymol. 276, 307–326. [DOI] [PubMed]
- Vagin, A. & Teplyakov, A. (2010). Acta Cryst. D66, 22–25. [DOI] [PubMed]
- Wilson, R. C. & Pata, J. D. (2008). Mol. Cell, 29, 767–779. [DOI] [PubMed]
- Winn, M. D. et al. (2011). Acta Cryst. D67, 235–242.
- Wong, J. H., Fiala, K. A., Suo, Z. & Ling, H. (2008). J. Mol. Biol. 379, 317–330. [DOI] [PubMed]
- Zhang, Y. & Yew, W. W. (2009). Int. J. Tuberc. Lung Dis. 13, 1320–1330. [PubMed]
- Zhou, B.-L., Pata, J. D. & Steitz, T. A. (2001). Mol. Cell, 8, 427–437. [DOI] [PubMed]