Abstract
Most Cryptosporidium infections in humans are caused by C. parvum or C. hominis. However, genotyping techniques have identified infections caused by unusual Cryptosporidium species. Cryptosporidium meleagridis has been identified in ≤ 1% of persons with diarrhea, although prevalence is higher in developing nations. We examined the infectivity of C. meleagridis in healthy adults. Five volunteers were challenged with 105 C. meleagridis oocysts and monitored six weeks for fecal oocysts and clinical manifestations. Four volunteers had diarrhea; three had detectable fecal oocysts; and one infected volunteer remained asymptomatic. Fecal DNA from two volunteers was amplified by using a polymerase chain reaction specific for the Cryptosporidium small subunit ribosomal RNA gene. Nucleotide sequence of these amplicons was diagnostic for C. meleagridis. All infections were self-limited; oocysts were cleared within ≤ 12 days of challenge. These studies establish that healthy adults can be infected and become ill from ingestion of C. meleagridis oocysts.
Introduction
Cryptosporidium spp. are coccidian parasites that cause diarrheal illness in a wide range of animal species. In humans, C. parvum and C. hominis are responsible for most cases of cryptosporidiosis, and C. meleagridis, C. canis, and C. felis make up the remainder.1,2 Cryptosporidium meleagridis appears to be prominent among unusual isolates and in some geographic areas can be as common as C. parvum.3–5
Cryptosporidium meleagridis was originally isolated from a turkey and has subsequently been found in other avian species, including parrots, red-legged partridges, and chickens.6–9 The infection has been experimentally established in immunosuppressed mice,9,10 and C. meleagridis isolated from a human has been successfully passaged in chickens, mice, gnotobiotic piglets, and calves.11 In those animals, the infectivity and virulence of C. meleagridis was similar to that of C. parvum.
Although the first reported human case of C. meleagridis was in a human immunodeficiency virus (HIV)–positive person from Kenya,12 the infection has since been identified in immunocompetent and immunosuppressed humans throughout the world.1–5,7,13–34 Furthermore, the finding of C. meleagridis in humans across geographically distinct regions, such as Peru, Spain, Kenya, Thailand, and Japan, affirmed the potential of the parasite as a public health concern.5,7,23,27 Although widespread geographically, C. meleagridis is frequently attributed to only a small proportion (< 1%) of Cryptosporidium cases from large populations with diarrhea.2,4,14,32 Since 2000, more than 200 C. meleagridis infections have been reported.2–5,12–34 Immune status was known in 135 C. meleagridis infections, and of those, 61.5% were in immunocompromised persons.
Similar to other Cryptosporidium species, the severity of C. meleagridis infection varies with the general health of the host and can range from asymptomatic shedding of oocysts to death associated with severe fluid and weight loss.19,20 Where studied, no differences in illness among adults have been noted between C. meleagridis and C. parvum or C. hominis. However, in HIV-positive children, infections with C. meleagridis were more likely to cause diarrhea than infection with C. parvum or C. hominis.5
Most C. meleagridis cases have been identified in diarrhea studies. However, five asymptomatic cases, all in children, have also been described.13,18 These asymptomatic cases suggest that the actual number of C. meleagridis infections may be higher than reported.
Current literature on C. meleagridis in humans has focused on naturally acquired infections among immunocompetent and immunocompromised persons. Many of these investigations analyzed only those stool samples from patients with diarrhea. Thus, the true prevalence of infection is difficult to assess. Furthermore, the overall prevalence of C. meleagridis may be underestimated because the morphology of oocysts by acid-fast staining is similar to that of C. hominis and C. parvum, and detection by immunofluorescent-based assays is based on the recognition of a shared antigen among the three species. Furthermore, conventional polymerase chain reaction (PCR)–based assays used in many of the studies may be unable to distinguish it from C. parvum.35
Previous human challenge studies have established infectivity and illness outcomes for C. parvum and C. hominis isolates.36–39 The present study was designed 1) to establish the susceptibility of healthy, adult volunteers to C. meleagridis infection after the ingestion of a known dose of oocysts and 2) to describe microbiologic and clinical outcomes of those who became infected. This study represents the first experimental challenge of C. meleagridis in humans.
Materials and Methods
Volunteer selection.
Volunteers 18–50 years of age were solicited at the Texas Medical Center in Houston and screened for general health status as described.40 All studies described herein were reviewed and approved by the Committee for Protection of Human Subjects at The University of Texas Health Science Center at Houston. Informed consent was obtained for initial medical evaluation and blood draw and again before challenge with C. meleagridis oocysts.
Volunteer monitoring and sample collection.
Volunteers were challenged and monitored in the University Clinical Research Center (Hermann Memorial Hospital, Houston, TX) as described.40 Briefly, each volunteer received 105 C. meleagridis oocysts instilled in a gelatin capsule. Each person was examined daily for 14 days and three times per week for four weeks. All stool samples were collected for the first 14 days and twice per week thereafter. Stool samples were transported on ice to the laboratory and were diluted 1:4 in buffered formalin for storage at 4°C until assayed. Definitions for diarrhea, illness attack rate, Cryptosporidium infection, cryptosporidiosis, gastrointestinal symptoms, and duration of diarrhea used in this study have been described.37,39
Oocyst isolation and preparation.
Cryptosporidium meleagridis isolate TU1867 was isolated from the stool of a child with diarrhea and identified as C. meleagridis by using the outer cell wall protein (COWP) marker by PCR/restriction fragment length polymorphism (RFLP).14,21 Isolate TU1867 has been established in the gnotobiotic pig model and passaged repeatedly.11 Oocysts were purified, transported to Houston in 2% potassium dichromate, and tested for microbiologic agents as described.40 To further ensure the safety of the inoculum, two tests for the presence of the HIV genome were carried out in the retrovirus laboratory of Dr. Blaine Hollinger (Baylor College of Medicine, Houston, TX). The first test examined the DNA from disrupted C. meleagridis oocysts, which had been prepared in the same way as the inoculum for volunteer studies. For the second test, oocysts were inoculated onto human enterocytes (HCT-8 cells) and cultured for 24–48 hours. Cells were delivered to the Baylor laboratory where DNA was extracted for testing. All tests of the disrupted or cultured parasites were negative for HIV.
Within 48 hours of volunteer challenge, purified C. meleagridis oocysts were examined for excystation rate and adjusted to a concentration of approximately 105 oocysts/10 μL in preparation for volunteer challenge as described.40
Detection of oocysts.
Every fecal sample collected from challenged volunteers over the six-week monitoring period was examined by enzyme immunoassay (EIA) and immunofluorescent assay (IFA) for oocysts. The EIA (SPECTRAFluor Plus; IVD Research Inc., Carlsbad, CA) was carried out and interpreted as per manufacturer's instructions. For the IFA, 5 μL of formalin-fixed stool was added to each well of three wells on a treated microscope slide (SuperStick Slides; Waterborne, Inc., New Orleans, LA) and dried for 1.5 hours at room temperature. Each well then received 50 μL of fluorescein isothiocyanate–conjugated, monoclonal IgM against Cryptosporidium (1:1000; Cellabs Pty. Ltd., Brookvale, New South Wales, Australia) and incubated in a humidified chamber in the dark for 30 minutes. Slides were then gently rinsed with 0.15 M phosphate-buffered saline, pH 7.2, and allowed to air dry. The entire well of the slide was examined by epifluorescence microscopy (20×), and fluorescing oocysts were counted. The mean number of oocysts from three wells was expressed per milliliter after adjusting for the 1:4 formalin dilution. Oocyst counts per stool were estimated as described.41 Oocyst counts from each stool sample were summed to estimate the total oocysts shed during the monitoring period.
To genotype the oocysts excreted by volunteers, DNA was extracted from 200 μL of fecal slurry by using the FastDNA Spin Kit for Soil (MP Biomedicals, Illkirch, France). DNA was recovered in 50 μL of water. A volume of 1 μL of DNA was amplified in a nested PCR protocol specific for a portion of the small-subunit ribosomal RNA (SSU rRNA) as described42 and later amended.35 The primary PCR product was diluted 1:200 or 1:400, and 1 μL of this dilution was used as template in the secondary reaction.
Results
Inoculum.
Cryptosporidium meleagridis oocysts used in the study showed no observable changes in the COWP PCR-RFLP profile as they were passaged through different porcine hosts. In addition, there was no indication of the presence of a subpopulation of oocysts or of contamination with C. parvum laboratory isolates. Two batches of oocysts were used in the study. At the time of challenge, the oocysts were within six weeks of production in the gnotobiotic piglet and had excystation rates of 48.6% and 66%, respectively. Mean ± SD oocyst counts in the inocula were 103,152 ± 10,685 and 100,170 ± 6,108, both within 10.3% or less of the target number (105).
Challenge outcomes.
Five healthy volunteers were enrolled in the study during September–December 2003. Volunteer ages ranged from 22 to 33 years (median = 25 years), and four (80%) of the five volunteers were men. All five volunteers were Caucasian; none had Cryptosporidium-specific serum IgG by enzyme-linked immunosorbent assay before challenge.
All five volunteers had evidence of infection by either clinical or microbiologic measures or both (Table 1), indicating that the inoculum (105 oocysts) met or exceeded the 100% infectious dose for this parasite. Three (60%) volunteers were positive for fecal oocysts by IFA, and four volunteers had a diarrheal illness, yielding an 80% illness attack rate. Two of the four volunteers with diarrhea had detectable oocysts, and two did not. One volunteer had no unformed stools or symptoms while shedding low, but detectable, numbers of oocysts.
Table 1.
Clinical and microbiologic outcomes of healthy adults challenged with Cryptosporidium meleagridis oocysts*
| Volunteer no. | Diarrhea | Oocyst shedding | ||||||
|---|---|---|---|---|---|---|---|---|
| Onset (dpi) | Duration (hours) | Total no. unformed stools | Total stool weight (g) | GI symptoms | Onset (dpi) | Duration (days) | Total oocysts (log) | |
| 180 | 4 | 61 | 9 | 599.5 | AB pain (day 4); nausea (day 6); gas (days 1–6) | 4 | 4 | 6.41 |
| 181 | NA | NA | NA | NA | Gas (days 5, 7) | 3 | 4 | 6.45 |
| 182 | 7 | 107 | 15 | 2,998.9 | Gas and fecal urgency (days 7–11) | 8 | 3 | 8.65 |
| 183 | 4 | 90 | 4 | 758.5 | Gas (days 2–4, 8) | |||
| 184 | 6 | 50 | 3 | 439.8 | AB cramps and gas (day 6) | |||
dpi = day post-challenge; GI = gastrointestinal; AB = abdominal; NA = not available.
Overall, diarrheal illnesses were characterized by an incubation period of 5.3 days (range = 4–7 days) and passage of approximately eight unformed stools (range = 3–15 stools) in a period of 77 hours (3.2 days; range = 50–105 hours) (Figure 1). Total weight of unformed stools during the diarrheal episode ranged from 0.44 to 3.0 kg (mean = 1.2 kg). In three volunteers, diarrhea was accompanied by additional gastrointestinal symptoms, principally increased gas production.
Figure 1.
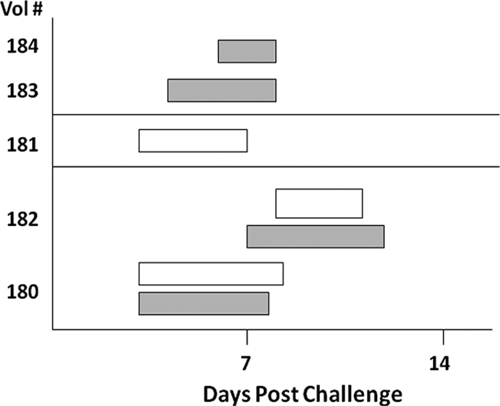
Oocyst shedding (open bars) and diarrhea (shaded bars) in adult volunteers challenged with Cryptosporidium meleagridis oocysts.
Oocyst shedding typically began in conjunction with diarrhea on day 5 post-challenge (range = days 3–8) and lasted for approximately 3.7 days (range = 3–4 days) (Table 1 and Figure 1). Asymptomatic shedding after resolution of diarrhea was noted in only one of the volunteers (no. 181), who shed oocysts for 1.5 additional days. The total number of oocysts shed per diarrheal episode ranged from 2.5 × 106 to 4.5 × 108. All symptoms, including diarrhea, resolved in all volunteers by day 12, and oocyst shedding ceased by day 11 post-challenge.
Volunteer 180.
Volunteer 180 had one episode of diarrhea beginning on day 4 post-challenge that lasted 2.5 days. A total of nine unformed stools were passed: four on day 4 (total weight = 163.8 g), two on day 5 (total weight = 261.3 g), and three on day 6 (weight = 174.5 g). Oocysts were excreted beginning on day 4 and continued one day after resolution of diarrhea and symptoms. No oocysts were detected in 12 subsequent stool samples.
Volunteer 181.
Volunteer 181 did not meet the criteria for a diarrheal illness, but had a microbiologically confirmed infection. Oocysts were excreted for four days beginning on day 3 post-challenge. Five unformed stools were passed during the oocyst shedding period: one on day 3 (weight = 91.9 g), one on day 5 (weight = 77.2 g), two on day 6 (total weight = 136.4 g), and one on day 7 (weight = 77.8 g).
Volunteer 182.
Volunteer 182 had diarrheal illness beginning on day 7 post-challenge that lasted 4.5 days. Seventeen unformed stools were passed during the diarrheal illness: one on day 2 (total weight = 141.1 g), one on day 4 (total weight = 188.7 g), five on day 7 (total weight = 847.7 g), five on day 8 (total weight = 955.4 g), one on day 9 (total weight = 218.0 g), one on day 10 (total weight = 154.2 g), and three on day 11 (total weight = 823.4 g). Oocysts were shed for three days during the illness. After day 10, no oocysts were detected in nine subsequent stool samples.
Volunteer 183.
Volunteer 183 had diarrheal illness beginning on day 4 post-challenge that lasted through day 7. Four unformed stools occurred during the illness: one on day 4 (total weight = 248.9 g), two on day 6 (total weight = 354.4 g), and one on day 7 (total weight = 155.2 g). Gas was reported before and during the diarrheal episode with no other gastrointestinal symptoms present. Twenty stool samples were uniformly negative for oocysts.
Volunteer 184.
Volunteer 184 had diarrheal illness on days 6–8 after challenge. Three unformed stools were documented during the illness: two on day 6 (total weight = 305.0 g) and one on day 8 (total weight = 134.7 g). Abdominal cramping and gas were reported on day 6. No oocysts were detected in any of the 17 submitted stool samples.
Fecal DNA from two oocyst-shedding volunteers was amplified by using a nested PCR protocol specific for the SSU rRNA gene. A third sample from volunteer 181 failed to amplify. A PCR product approximating the expected size of 833 basepairs was obtained from volunteers 180 and 182. Sequences of 690 basepairs and 733 basepairs, respectively, were obtained from these PCR products by sequencing both strands. These sequences were 100% identical with the C. meleagridis SSU rRNA sequences found in GenBank, such as accession no. HM116384. A control sample was PCR negative.
Discussion
The C. meleagridis isolate TU1867 used in the study was previously identified by COWP PCR-RFLP and nested PCR of the SSU rRNA gene.11 The resulting 833-basepair fragment from the nested PCR was digested with Ase I, which can distinguish at least nine Cryptosporidium species and C. parvum genotypes. The Ase I RFLP profile of the TU1867 isolate was identical with that reported for C. meleagridis and did not change as the isolate was passaged through different hosts. Also, a second Cryptosporidium population was not detected. The nested PCR products from the original human sample and from the animal host passages were previously cloned and sequenced. These sequences were invariant and were identical with previously published sequences of C. meleagridis.11
In the earlier study, TU1867 was infectious when fed to interferon-γ knockout and immunosuppressed C57BL/6 mice, 2–7-day-old chicks and turkey poults, and colostrum-fed calves.11 Infection of these hosts indicated that C. meleagridis was readily transmissible between mammalian and avian species. However, diarrhea without appreciable signs of dehydration was consistently observed only in infected piglets. In contrast, no clinical signs developed in the infected colostrum-fed calves, even though excretion of oocysts was evident. In comparison, diarrhea has been associated with C. meleagridis infection in immunocompromised and ostensibly healthy humans. However, many of the symptomatic infections reported in immunocompetent persons have been in children and/or in populations that may be undernourished or infected with other pathogens, including chronic parasitoses.
Although these earlier reports of C. meleagridis indicated susceptibility in humans, infectivity and/or illness in healthy adults was less clear and was the focus of the present study. On the basis of previous findings in volunteers who received C. parvum or C. hominis isolates, a C. meleagridis inoculum was chosen that would likely exceed the 100% infectious dose.36–39 This oocyst dose resulted in infection and/or illness in all of the volunteers, thus establishing the susceptibility of healthy adults to C. meleagridis infection. All volunteers had no evidence of previous infection as assessed by the absence of serum antibodies by enzyme-linked immunosorbent assay. It is not known if prior exposure would have provided any protection from subsequent challenge as has been shown with C. parvum.41 Furthermore, because the present study used only one oocyst dose, no estimation of a 50% infectious dose can be made for the TU1867 isolate. It is likely that the 50% infectious dose is far less than the 105 oocysts used.
A self-limited diarrhea was observed in four of the five volunteers in contrast to the paucity of symptoms found earlier in animal studies. In those experiments, only the gnotobiotic piglets had diarrhea, although the neonatal or immunosuppressed animals excreted oocysts. Interestingly, volunteer 182 excreted 100-fold more oocysts than the other volunteers with detectable oocysts. This increased oocyst excretion was associated with longest duration (107 hours) and the greatest severity (approximately 3-kg stool weight and 15 unformed stools).
Volunteers had days of no detectable oocysts between days when large numbers were shed. Also, one of the volunteers shed oocysts detectable by EIA but not by IFA. These results are not particularly surprising because oocyst shedding can be near or below the limit of detection on some days and for one or both assays. This finding may also explain why volunteer 181 was PCR negative. In our experience and in other laboratories, detection limits of the IFA are in the range of 104 oocysts/mL. The absorbance value of the EIA for the EIA-positive/IFA-negative volunteer was nearer the manufacturer's cutoff value than for others volunteers, which suggested a lower intensity of infection.
The five volunteers that took part in this study were relatively young adults in excellent health. Each received an oocyst dose that would likely be much higher than encountered in a community setting. Nevertheless, the diarrheal illness that they experienced was similar to the illness reported in naturally acquired C. meleagridis infection, which, in turn, was indistinguishable from illness caused by C. parvum or C. hominis. Furthermore, the present study used the same volunteer selection criteria and post-challenge monitoring used in previous volunteer studies of C. parvum or C. hominis. Thus, these earlier highly monitored volunteers may serve as a more apt comparison than in naturally acquired infections. The various microbiologic and clinical measures from the C. meleagridis volunteers were within ranges previously described for C. parvum and C. hominis isolates.36–39
Our study firmly establishes that immunocompetent, healthy persons are susceptible to C. meleagridis infection. Our data further suggest that otherwise healthy persons who have a community-acquired infection will likely experience a mild, self-limited diarrheal disease and will be unlikely to seek medical attention. Furthermore, because current serologic studies of IgG against Cryptosporidium are not specific for the species of Cryptosporidium that elicited the response, the level of exposure to C. meleagridis in the community cannot presently be estimated. The human challenge studies are important for establishing susceptibility to infection and illness from these non-C. parvum species and for contributing to risk assessment and the setting of water quality regulatory standards.
ACKNOWLEDGMENTS
We thank Susan Wu for assistance in preparation of the manuscript and Julia Dilo for expert laboratory support.
Footnotes
Financial support: This study was supported, in part, by the National Center for Environmental Research STAR Program of the Environmental Protection Agency (grant GR828035-01-0 to Cynthia L. Chappell), the National Institutes of Health General Clinical Research Centers (grant RR-02558), and the National Institute of Allergy and Infectious Diseases (grant AI52781 to Giovanni Widmer and grant NO1-AI-25466 to Saul Tzipori).
Authors' addresses: Cynthia L. Chappell, Center for Infectious Diseases, University of Texas School of Public Health, Houston, TX, E-mail: cynthia.l.chappell@uth.tmc.edu. Pablo C. Okhuysen, Division of Infectious Diseases University of Texas Health Medical School, Houston, TX, E-mail: pablo.c.okhuysen@uth.tmc.edu. Rebecca C. Langer-Curry, Bayer CropScience Bioscience, Morrisville, NC, E-mail: becky.langer@bayer.com. Donna E. Akiyoshi, Giovanni Widmer, and Saul Tzipori, Department of Biomedical Sciences, Tufts University, North Grafton, MA, Emails: donna.akiyoshi@tufts.edu, giovanni.widmer@tufts.edu, and saul.tzipori@tufts.edu.
References
- 1.McLauchlin J, Amar C, Pedraza-Diaz S, Nichols GL. Molecular epidemiological analysis of Cryptosporidium spp. in the United Kingdom: results of genotyping Cryptosporidium spp. in 1,705 fecal samples from humans and 105 fecal samples from livestock animals. J Clin Microbiol. 2000;38:3984–3990. doi: 10.1128/jcm.38.11.3984-3990.2000. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Chalmers RM, Elwin K, Thomas AL, Guy EC, Mason B. Long-term Cryptosporidium typing reveals the aetiology and species-specific epidemiology of human cryptosporidiosis in England and Wales, 2000 to 2003. Euro Surveill. 2009;14:pii: 19086. doi: 10.2807/ese.14.02.19086-en. [DOI] [PubMed] [Google Scholar]
- 3.Gatei W, Suputtamongkol Y, Waywa D, Ashford RW, Bailey JW, Greensill J, Beeching NJ, Hart CA. Zoonotic species of Cryptosporidium are as prevalent as the anthroponotic in HIV-infected patients in Thailand. Ann Trop Med Parasitol. 2002;96:797–802. doi: 10.1179/000349802125002202. [DOI] [PubMed] [Google Scholar]
- 4.Cama VA, Ross JM, Crawford S, Kawai V, Chavez-Valdez R, Vargas D, Vivar A, Ticona E, Navincopa M, Williamson J, Ortega Y, Gilman RH, Bern C, Xiao L. Differences in clinical manifestations among Cryptosporidium species and subtypes in HIV-infected persons. J Infect Dis. 2007;196:684–691. doi: 10.1086/519842. [DOI] [PubMed] [Google Scholar]
- 5.Cama VA, Bern C, Roberts J, Cabrera L, Sterling CR, Ortega Y, Gilman RH, Xiao L. Cryptosporidium species and subtypes and clinical manifestations in children, Peru. Emerg Infect Dis. 2008;14:1567–1574. doi: 10.3201/eid1410.071273. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Slavin D. Cryptosporidium meleagridis (sp. nov.) J Comp Pathol. 1955;65:262–266. doi: 10.1016/s0368-1742(55)80025-2. [DOI] [PubMed] [Google Scholar]
- 7.Morgan UM, Xiao L, Limor J, Gelis S, Raidal SR, Fayer R, Lal A, Elliot A, Thompson RC. Cryptosporidium meleagridis in an Indian ring-necked parrot (Psittacula krameri) Aust Vet J. 2000;78:182–183. doi: 10.1111/j.1751-0813.2000.tb10589.x. [DOI] [PubMed] [Google Scholar]
- 8.Pages-Mante A, Pages-Bosch M, Majo-Masferrer N, Gomez-Couso H, Ares-Mazas E. An outbreak of disease associated with cryptosporidia on a red-legged partridge (Alectoris rufa) game farm. Avian Pathol. 2007;36:275–278. doi: 10.1080/03079450701439389. [DOI] [PubMed] [Google Scholar]
- 9.Sreter T, Kovacs G, da Silva AJ, Pieniazek NJ, Szell Z, Dobos-Kovacs M, Marialigeti K, Varga I. Morphologic, host specificity, and molecular characterization of a Hungarian Cryptosporidium meleagridis isolate. Appl Environ Microbiol. 2000;66:735–738. doi: 10.1128/aem.66.2.735-738.2000. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Huang K, Akiyoshi DE, Feng X, Tzipori S. Development of patent infection in immunosuppressed C57Bl/6 mice with a single Cryptosporidium meleagridis oocyst. J Parasitol. 2003;89:620–622. doi: 10.1645/0022-3395(2003)089[0620:DOPIII]2.0.CO;2. [DOI] [PubMed] [Google Scholar]
- 11.Akiyoshi DE, Dilo J, Pearson C, Chapman S, Tumwine J, Tzipori S. Characterization of Cryptosporidium meleagridis of human origin passaged through different host species. Infect Immun. 2003;71:1828–1832. doi: 10.1128/IAI.71.4.1828-1832.2003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Morgan U, Weber R, Xiao L, Sulaiman I, Thompson RC, Ndiritu W, Lal A, Moore A, Deplazes P. Molecular characterization of Cryptosporidium isolates obtained from human immunodeficiency virus-infected individuals living in Switzerland, Kenya, and the United States. J Clin Microbiol. 2000;38:1180–1183. doi: 10.1128/jcm.38.3.1180-1183.2000. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Essid R, Mousli M, Aoun K, Abdelmalek R, Mellouli F, Kanoun F, Derouin F, Bouratbine A. Identification of Cryptosporidium species infecting humans in Tunisia. Am J Trop Med Hyg. 2008;79:702–705. [PubMed] [Google Scholar]
- 14.Pedraza-Díaz S, Amar CF, McLauchlin J, Nichols GL, Cotton KM, Godwin P, Iversen AM, Milne L, Mulla JR, Nye K, Panigrahl H, Venn SR, Wiggins R, Williams M, Youngs ER. Cryptosporidium meleagridis from humans: molecular analysis and description of affected patients. J Infect. 2001;42:243–250. doi: 10.1053/jinf.2001.0839. [DOI] [PubMed] [Google Scholar]
- 15.Cama VA, Bern C, Sulaiman IM, Gilman RH, Ticona E, Vivar A, Kawai V, Vargas D, Zhou L, Xiao L. Cryptosporidium species and genotypes in HIV-positive patients in Lima, Peru. J Eukaryot Microbiol. 2003;50:531–533. doi: 10.1111/j.1550-7408.2003.tb00620.x. [DOI] [PubMed] [Google Scholar]
- 16.Gatei W, Greensill J, Ashford RW, Cuevas LE, Parry CM, Cunliffe NA, Beeching NJ, Hart CA. Molecular analysis of the 18S rRNA gene of Cryptosporidium parasites from patients with or without human immunodeficiency virus infections living in Kenya, Malawi, Brazil, the United Kingdom, and Vietnam. J Clin Microbiol. 2003;41:1458–1462. doi: 10.1128/JCM.41.4.1458-1462.2003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Gatei W, Wamae CN, Mbae C, Waruru A, Mulinge E, Waithera T, Gatika SM, Kamwati SK, Revathi G, Hart CA. Cryptosporidiosis: prevalence, genotype analysis, and symptoms associated with infections in children in Kenya. Am J Trop Med Hyg. 2006;75:78–82. [PubMed] [Google Scholar]
- 18.Gatei W, Das P, Dutta P, Sen A, Cama V, Lal AA, Xiao L. Multilocus sequence typing and genetic structure of Cryptosporidium hominis from children in Kolkata, India. Infect Genet Evol. 2007;7:197–205. doi: 10.1016/j.meegid.2006.08.006. [DOI] [PubMed] [Google Scholar]
- 19.Matos O, Alves M, Xiao L, Cama V, Antunes F. Cryptosporidium felis and C. meleagridis in persons with HIV, Portugal. Emerg Infect Dis. 2004;10:2256–2257. doi: 10.3201/eid1012.031068. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Hung CC, Tsaihong JC, Lee YT, Deng HY, Hsiao WH, Chang SY, Chang SC, Su KE. Prevalence of intestinal infection due to Cryptosporidium species among Taiwanese patients with human immunodeficiency virus infection. J Formos Med Assoc. 2007;106:31–35. doi: 10.1016/S0929-6646(09)60213-8. [DOI] [PubMed] [Google Scholar]
- 21.Tumwine JK, Kekitiinwa A, Nabukeera N, Akiyoshi DE, Rich SM, Widmer G, Feng X, Tzipori S. Cryptosporidium parvum in children with diarrhea in Mulago Hospital, Kampala, Uganda. Am J Trop Med Hyg. 2003;68:710–715. [PubMed] [Google Scholar]
- 22.Tumwine JK, Kekitiinwa A, Bakeera-Kitaka S, Ndeezi G, Downing R, Feng X, Akiyoshi DE, Tzipori S. Cryptosporidiosis and microsporidiosis in Ugandan children with persistent diarrhea with and without concurrent infection with the human immunodeficiency virus. Am J Trop Med Hyg. 2005;73:921–925. [PubMed] [Google Scholar]
- 23.Yagita K, Izumiyama S, Tachibana H, Masuda G, Iseki M, Furuya K, Kameoka Y, Kuroki T, Itagaki T, Endo T. Molecular characterization of Cryptosporidium isolates obtained from human and bovine infections in Japan. Parasitol Res. 2001;87:950–955. doi: 10.1007/s004360100480. [DOI] [PubMed] [Google Scholar]
- 24.Muthusamy D, Rao SS, Ramani S, Monica B, Banerjee I, Abraham OC, Mathai DC, Primrose B, Muliyil J, Wanke CA, Ward HD, Kang G. Multilocus genotyping of Cryptosporidium sp. isolates from human immunodeficiency virus-infected individuals in South India. J Clin Microbiol. 2006;44:632–634. doi: 10.1128/JCM.44.2.632-634.2006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Llorente MT, Clavel A, Goñi MP, Varea M, Seral C, Becerril R, Suarez L, Gómez-Lus R. Genetic characterization of Cryptosporidium species from humans in Spain. Parasitol Int. 2007;56:201–205. doi: 10.1016/j.parint.2007.02.003. [DOI] [PubMed] [Google Scholar]
- 26.Guyot K, Follet-Dumoulin A, Lelièvre E, Sarfati C, Rabodonirina M, Nevez G, Cailliez JC, Camus D, Dei-Cas E. Molecular characterization of Cryptosporidium isolates obtained from humans in France. J Clin Microbiol. 2001;39:3472–3480. doi: 10.1128/JCM.39.10.3472-3480.2001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Tiangtip R, Jongwutiwes S. Molecular analysis of Cryptosporidium species isolated from HIV-infected patients in Thailand. Trop Med Int Health. 2002;7:357–364. doi: 10.1046/j.1365-3156.2002.00855.x. [DOI] [PubMed] [Google Scholar]
- 28.Blanco MA, Iborra A, Vargas A, Nsie E, Mbá L, Fuentes I. Molecular characterization of Cryptosporidium isolates from humans in Equatorial Guinea. Trans R Soc Trop Med Hyg. 2009;103:1282–1284. doi: 10.1016/j.trstmh.2009.02.016. [DOI] [PubMed] [Google Scholar]
- 29.Al-Brikan FA, Salem HS, Beeching N, Hilal N. Multilocus genetic analysis of Cryptosporidium isolates from Saudi Arabia. J Egypt Soc Parasitol. 2008;38:645–658. [PubMed] [Google Scholar]
- 30.Araújo AJ, Kanamura HY, Almeida ME, Gomes AH, Pinto TH, Da Silva AJ. Genotypic identification of Cryptosporidium spp. isolated from HIV-infected patients and immunocompetent children of São Paulo, Brazil. Rev Inst Med Trop Sao Paulo. 2008;50:139–143. doi: 10.1590/s0036-46652008005000003. [DOI] [PubMed] [Google Scholar]
- 31.Bajer A, Bednarska M, Cacciò SM, Wolska-Kusśnierz B, Heropolitanska-Pliszka E, Bernatowska E, Wielopolska M, Paziewska A, Welc-Faleciak R, Sinński E. Genotyping of Cryptosporidium isolates from human clinical cases in Poland. Parasitol Res. 2008;103:37–42. doi: 10.1007/s00436-008-0924-5. [DOI] [PubMed] [Google Scholar]
- 32.Leoni F, Amar C, Nichols G, Pedraza-Díaz S, McLauchlin J. Genetic analysis of Cryptosporidium from 2414 humans with diarrhoea in England between 1985 and 2000. J Med Microbiol. 2006;55:703–707. doi: 10.1099/jmm.0.46251-0. [DOI] [PubMed] [Google Scholar]
- 33.Enemark HL, Ahrens P, Juel CD, Petersen E, Petersen RF, Andersen JS, Lind P, Thamsborg SM. Molecular characterization of Danish Cryptosporidium parvum isolates. Parasitology. 2002;125:331–341. doi: 10.1017/s0031182002002226. [DOI] [PubMed] [Google Scholar]
- 34.Xiao L, Bern C, Limor J, Sulaiman I, Roberts J, Checkley W, Cabre L, Gilman RH, Lal AA. Identification of 5 types of Cryptosporidium parasites in children in Lima, Peru. J Infect Dis. 2001;183:492–497. doi: 10.1086/318090. [DOI] [PubMed] [Google Scholar]
- 35.Champliaud D, Gobet P, Naciri M, Vagner O, Lopez J, Buisson JC, Varga I, Harly G, Mancassola R, Bonnin A. Failure to differentiate Cryptosporidium parvum from C. meleagridis based on PCR amplification of eight DNA sequences. Appl Environ Microbiol. 1998;64:1454–1458. doi: 10.1128/aem.64.4.1454-1458.1998. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.DuPont HL, Chappell CL, Sterling CR, Okhuysen PC, Rose JB, Jakubowski W. The infectivity of Cryptosporidium parvum in healthy volunteers. N Engl J Med. 1995;332:855–859. doi: 10.1056/NEJM199503303321304. [DOI] [PubMed] [Google Scholar]
- 37.Okhuysen PC, Chappell CL, Crabb JH, Sterling CR, DuPont HL. Virulence of three distinct Cryptosporidium parvum isolates for healthy adults. J Infect Dis. 1999;180:1275–1281. doi: 10.1086/315033. [DOI] [PubMed] [Google Scholar]
- 38.Okhuysen PC, Rich SM, Chappell CL, Grimes KA, Widmer G, Feng X, Tzipori S. Infectivity of a Cryptosporidium parvum isolate of cervine origin for healthy adults and interferon-γ knockout mice. J Infect Dis. 2002;185:1320–1325. doi: 10.1086/340132. [DOI] [PubMed] [Google Scholar]
- 39.Chappell CL, Okhuysen PC, Langer-Curry R, Widmer G, Akiyoshi DE, Tanriverdi S, Tzipori S. Cryptosporidium hominis: experimental challenge of healthy adults. Am J Trop Med Hyg. 2006;75:851–857. [PubMed] [Google Scholar]
- 40.Chappell CL, Okhuysen PC, White AC., Jr . In: Cryptosporidium: From Molecules to Disease. Thompson R, Armson A, Ryan U, editors. Bridgewater, NJ: Elsevier Science; 2004. pp. 19–50. (Cryptosporidium parvum: infectivity, pathogenesis and the host-parasite relationship). [Google Scholar]
- 41.Chappell CL, Okhuysen PC, Sterling CR, Wang C, Jakubowski W, Dupont HL. Infectivity of Cryptosporidium parvum in healthy adults with pre-existing anti-C. parvum serum immunoglobulin G. Am J Trop Med Hyg. 1999;60:157–164. doi: 10.4269/ajtmh.1999.60.157. [DOI] [PubMed] [Google Scholar]
- 42.Xiao L, Morgan UM, Limor J, Escalante A, Arrowood M, Shulaw W, Thompson RC, Fayer R, Lal AA. Genetic diversity within Cryptosporidium parvum and related Cryptosporidium species. Appl Environ Microbiol. 1999;65:3386–3391. doi: 10.1128/aem.65.8.3386-3391.1999. [DOI] [PMC free article] [PubMed] [Google Scholar]


