Abstract
Differential viral recognition by cells bearing Toll-like receptor 4 (TLR4) polymorphisms Asp299Gly and Thr399Ile may influence susceptibility and severity of dengue virus infection. In central Java, Indonesia, we investigated 201 children with dengue hemorrhagic fever (DHF) and 179 healthy controls. Patients and controls were mostly ethnic Javanese. A nearly complete cosegregation of the two mutations was observed. The TLR4 299/399 genotype was found in five patients and four controls. Prevalence of the TLR4 299/399 genotype did not differ significantly between controls and DHF patients or between patients with different severities of DHF. Also, vascular leakage in patients with different TLR4 genotypes did not differ. Thus, the 299/399 TLR4 haplotype has only minor influence on susceptibility and severity of complicated dengue virus infection.
Dengue virus infection usually manifests as non-severe dengue fever, but manifests in some patients as dengue hemorrhagic fever (DHF) with thrombocytopenia and plasma leakage (DHF I and II) and in others with shock (dengue shock syndrome [DSS], DHF III, and IV).1,2 Genetic variation influences the development of DHF and DSS.3,4 Children who have the Toll-like receptor 4 (TLR4) polymorphisms Asp299Gly and Thr399Ile were more susceptible to respiratory syncytial virus (RSV), a flavivirus similar to dengue virus.5–7 Therefore, we assessed in children with DHF to see whether these TLR4 polymorphisms influenced susceptibility to and severity of dengue virus infections.
During 2001–2003 and 2005–2006, we enrolled children 3–14 years of age admitted to the pediatric ward or intensive care unit of Dr. Kariadi Hospital (RSDK) in Semarang, Indonesia, with clinically suspected DHF (either DHF I and II) or DSS into the study.8 Dengue virus infection was confirmed serologically by capture and indirect enzyme-linked immunosorbent assays (Focus Technologies, Cypress, CA) for dengue-specific IgM and IgG. The study was reviewed and approved by the Research Ethics Committee of Dr. Kariadi Hospital. Informed consent was obtained from parents or legal guardians of the patients. Persons with DHF and healthy controls who had no history of DHF/DSS were enrolled from the same geographic area by using consecutive sampling. Children DHF/DSS and controls were of mostly Javanese ethnic descent, as ascertained by the investigator, although no three-generation pedigree was obtained.
Genomic DNA was isolated from venous blood samples stored in EDTA and processed by using a salting-out procedure.9 Detection of TLR4 gene polymorphisms was performed by polymerase chain reaction and restriction fragment length polymorphism assay as described by van der Graaf et al.10 Serum albumin and total protein were measured (Bromocresol–green and Biuret methods; reference values = 3.26 g/dL and ≥ 5.97 g/dL, respectively).
At admission and on day 2, chest radiographs were obtained to calculate the pleural effusion index (PEI = 100 × the maximal width of the effusion divided by the width of the hemithorax). The Fisher exact test was used to analyze the distribution of TLR4 polymorphisms in controls and patients. The Mann-Whitney U test and the Wilcoxon rank tests were used to analyze non-normally distributed data. The paired-sample t-test was used to analyze normally distributed data, and the one-sample t-test was used to compare reference values and normally distributed data. P < 0.05 indicated statistical significance.
A total of 201 children with DHF/DSS (mean age = 7.31 years) and 179 adult controls were included in the study Positive tourniquet test results were found more frequently in persons with DHF without shock (n = 95) than in persons with DSS (n = 38) (P < 0.0001). Platelet counts were significantly lower in persons with DSS than in persons with DHF I/II (P < 0.0001). In 175 patients with available chest radiographs, pleural effusion was more frequent in persons with DSS than in persons with DHF (P = 0.022). The PEI for children with DSS (18.75%, 95% confidence interval [CI] = 9.71–29.05%) was significantly greater (P < 0.0001) than for children with DHF (8.55%, 95% CI = 0.00–20.25%). In persons with DSS, albumin (mean ± SD = 2.9 ± 0.80 g/dL; n = 80) and protein levels (5.23 ± 1.60 g/dL) were decreased and lower than in persons with DHF (3.39 ± 0.56 g/dL; n = 108 and 6.10 g/dL, 95% CI = 5.23–6.70 g/dL, respectively) (P < 0.0001).
The TLR4 299/399 genotype was found in 5 (2.5%) of 201 DHF/DSS patients and in 4 (2.2%) of 179 controls (P = 1.000, by Fisher exact test, P = 1.000). Separation of complicated dengue in DHF and DSS did not influence this finding (P = 0.400, by Fisher exact test).
Platelet counts at admission and on day 2 of those who had the 299/399 genotype and wild type/wild type (wt/wt) TLR4 were decreased to values between 53 and 57 × 103 cells/mL, but did not differ significantly. The tourniquet test result was positive for 130 (70.7%) of 184 patients who had wt/wt and for 3 (60%) of 5 patients who had the 299/399 TLR4 polymorphism (P not significant). The presence of petechiae did not differ significantly between patients who had the wt/wt TLR4 polymorphism and patients who had the 299/399 TLR4 polymorphism (33.7% and 40%, respectively). Other hemorrhagic manifestations did not differ significantly between patients with and without the 299/399 TLR4 polymorphism.
Plasma leakage in children with DHF/DSS was assessed at admission and on day 2 by comparing four plasma leakage measurements: hematocrit, albumin levels, protein levels, and PEI, in relation to the TLR4 genotypes and DHF (Figure 1). In patients with the wt/wt DHF/DSS polymorphism, hematocrit, albumin levels, and PEI were statistically different at admission than on day 2 after admission (Figure 1A, B, and D). Patients who had the 299/399 polymorphism showed no differences in hematocrit, protein levels, and PEI (Figure 1A, C, and D) compared with persons who had the wt/wt TLR4 polymorphism. However, on day 2 after admission, albumin levels in persons who had the 299/399 genotype were significantly higher than in persons who had the wt/wt TLR4 genotype (P = 0.013) (Figure 1B). Despite this difference, measured albumin levels were in the reference range. The hospital mortality rate was 11 of 201 children with DHF/DSS. None of these children had the TLR4 299/399 genotype.
Figure 1.
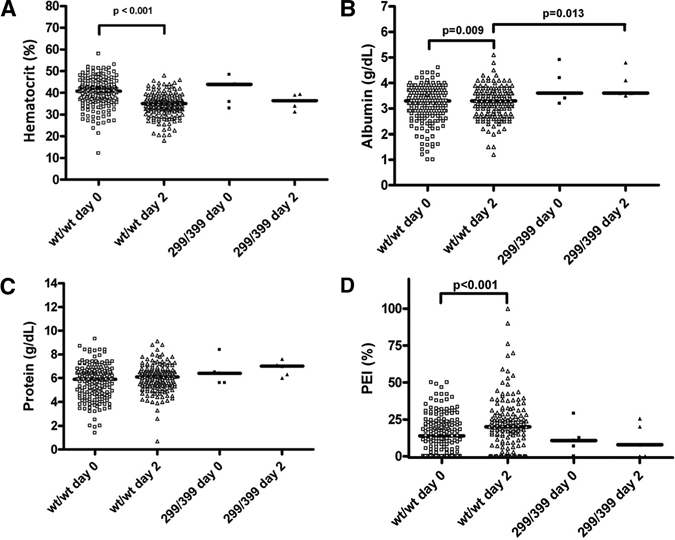
Scatter plot with median showing plasma leakage parameters for dengue virus–infected children, Indonesia. A, Hematocrit was measured on day 0 was measured for wild type/wild type (wt/wt) (n = 196) and 299/399 (n = 5) genotypes and on day 2 in wt/wt (n = 185) and 299/399 (n = 5) genotypes. B, Serum albumin was measured on day 0 in wt/wt (n = 183) and 299/399 genotypes (n = 5) and on day 2 in wt/wt (n = 173) and 299/399 (n = 5) genotypes. C, Protein level was measured on day 0 in wt/wt (n = 184) and 299/399 (n = 5) genotypes and on day 2 in wt/wt (n = 173) and 299/399 (n = 5) genotypes. D, Pleural effusion index (PEI) was measured on day 0 in wt/wt (n = 170) and 299/399 (n = 5) genotypes and on day 2 in wt/wt (n = 151) and 299 of 399 (n = 5) genotypes.
We showed that the distribution of the TLR4 299/399 polymorphisms did not differ between children with DHF/DSS and controls. This finding would have been strengthened if we had been able to exclude previous asymptomatic dengue virus infections by serologic testing. Also, assessment of ancestry genome wide alleles in cases and controls would have provided definite evidence that cases and controls were of similar ethnic decent. Our findings are consistent with those of larger studies that also found no association between these polymorphisms and disease susceptibility.11
Most nonsynonymous TLR4 polymorphisms had a low population frequency (< 1%). However, Asp299Gly and Thr399Ile TLR4 polymorphisms had population frequencies > 5%,12 suggesting that a role of other nonsynonymous TLR4 polymorphisms can be excluded. The 299 TLR4 haplotype alters the conformation of TLR4.13 The observed increase and decrease of TLR4 isoform transcription during dengue virus infection indicate a possible role of TLR4 during the early immune response.14 Dengue virus infection suppressed TLR4 gene expression.15 This finding indicates that instead of the TLR4 299/399 haplotype, reduced TLR4 gene expression caused by dengue virus infection might have a pivotal role in susceptibility and severity of complicated dengue virus infection.
The innate immune response toward RSV was suppressed in children with mutated TLR4, and children with the TLR4 299/399 polymorphism had a high risk for severe RSV infection.5–7 Because we could not demonstrate such a difference for dengue, the correlation between the TLR4 polymorphism and RSV infection is likely to be virus specific. Interestingly, our screening of TLR4 polymorphisms showed that the distribution of the TLR4 polymorphism has a high frequency of cosegregated 299/399 in all analyzed groups. Previous TLR4 polymorphism reports from Indonesia showed only separate 299 and 399 polymorphisms.16 Therefore, there still could be an effect of the TLR4 299/399 polymorphism in populations with a higher prevalence of this polymorphism.
We found a significant increase of PEI from admission to day 2 in persons who had the wt/wt genotype. We also observed significantly lower albumin levels on day 2 in children with DHF/DSS who had the wt/wt TLR4 polymorphism, but not for other plasma leakage parameters. Therefore, the effect of TLR4 on vascular leakage is negligible. Overall, this finding indicates that in contrast to RSV infections, 299/399 TLR4 polymorphism has a minor role in complicated dengue.
ACKNOWLEDGMENTS
The American Society of Tropical Medicine and Hygiene (ASTMH) assisted with publication expenses.
Footnotes
Financial support: Mihai G. Netea was supported by a Vici Grant from the Netherlands Organization for Scientific Research. Kis Djamiatun was recipient by a PhD Fellowship from Ministry of National Education, Republic of Indonesia.
Authors' addresses: Kis Djamiatun, Parasitology Department, Faculty of Medicine, Diponegoro University, Jl. Sriwijaya 34, Semarang, Indonesia, E-mail: ramus64@yahoo.com. Bart Ferwerda, Radboud University Nijmegen Medical Center, Nijmegen, The Netherlands, E-mail: ferwerda@mail.med.upenn.edu. Mihai G. Netea, André J. A. M. van der Ven, and Will M. V. Dolmans, Department of General Internal Medicine, Radboud University, Nijmegen Medical Center, Nijmegen, The Netherlands, E-mails: m.netea@aig.umcn.nl, a.vanderven@aig.umcn.nl, and dolmansw@xs4all.nl. Sultana M. H. Faradz, Division of Human Genetics, Center for Biomedical Research, Faculty of Medicine, Diponegoro University, Semarang, Indonesia, E-mail: sultana@indosat.net.id.
References
- 1.Halstead SB. Dengue. Lancet. 2007;370:1644–1652. doi: 10.1016/S0140-6736(07)61687-0. [DOI] [PubMed] [Google Scholar]
- 2.Halstead SB. Pathogenesis of dengue: challenges to molecular biology. Science. 1988;239:476–481. doi: 10.1126/science.3277268. [DOI] [PubMed] [Google Scholar]
- 3.Fernandez-Mestre MT, Gendzekhadze K, Rivas-Vetencourt P, Layrisse Z. TNF-alpha-308A allele, a possible severity risk factor of hemorrhagic manifestation in dengue fever patients. Tissue Antigens. 2004;64:469–472. doi: 10.1111/j.1399-0039.2004.00304.x. [DOI] [PubMed] [Google Scholar]
- 4.Sakuntabhai A, Turbpaiboon C, Casademont I, Chuansumrit A, Lowhnoo T, Kajaste-Rudnitski A, Kalayanarooj SM, Tangnararatchakit K, Tangthawornchaikul N, Vasanawathana S, Chaiyaratana W, Yenchitsomanus PT, Suriyaphol P, Avirutnan P, Chokephaibulkit K, Matsuda F, Yoksan S, Jacob Y, Lathrop GM, Malasit P, Despres P, Julier C. A variant in the CD209 promoter is associated with severity of dengue disease. Nat Genet. 2005;37:507–513. doi: 10.1038/ng1550. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Tulic MK, Hurrelbrink RJ, Prele CM, Laing IA, Upham JW, Le Souef P, Sly PD, Holt PG. TLR4 polymorphisms mediate impaired responses to respiratory syncytial virus and lipopolysaccharide. J Immunol. 2007;179:132–140. doi: 10.4049/jimmunol.179.1.132. [DOI] [PubMed] [Google Scholar]
- 6.Awomoyi AA, Rallabhandi P, Pollin TI, Lorenz E, Sztein MB, Boukhvalova MS, Hemming VG, Blanco JC, Vogel SN. Association of TLR4 polymorphisms with symptomatic respiratory syncytial virus infection in high-risk infants and young children. J Immunol. 2007;179:3171–3177. doi: 10.4049/jimmunol.179.5.3171. [DOI] [PubMed] [Google Scholar]
- 7.Vogel SN, Awomoyi AA, Rallabhandi P, Medvedev AE. Mutations in TLR4 signaling that lead to increased susceptibility to infection in humans: an overview. J Endotoxin Res. 2005;11:333–339. doi: 10.1179/096805105X58724. [DOI] [PubMed] [Google Scholar]
- 8.World Health Organization . Dengue Haemorrhagic Fever: Diagnosis, Treatment, Prevention and Control. Geneva: World Health Organization; 1997. [Google Scholar]
- 9.Miller SA, Dykes DD, Polesky HF. A simple salting out procedure for extracting DNA from human nucleated cells. Nucleic Acids Res. 1988;16:1215. doi: 10.1093/nar/16.3.1215. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Van der Graaf CA, Netea MG, Morre SA, Den Heijer M, Verweij PE, Van der Meer JW, Kullberg BJ. Toll-like receptor 4 Asp299Gly/Thr399Ile polymorphisms are a risk factor for Candida bloodstream infection. Eur Cytokine Netw. 2006;17:29–34. [PubMed] [Google Scholar]
- 11.Chanock SJ, Manolio T, Boehnke M, Boerwinkle E, Hunter DJ, Thomas G, Hirschhorn JN, Abecasis G, Altshuler D, Bailey-Wilson JE, Brooks LD, Cardon LR, Daly M, Donnelly P, Fraumeni JF, Jr, Freimer NB, Gerhard DS, Gunter C, Guttmacher AE, Guyer MS, Harris EL, Hoh J, Hoover R, Kong CA, Merikangas KR, Morton CC, Palmer LJ, Phimister EG, Rice JP, Roberts J, Rotimi C, Tucker MA, Vogan KJ, Wacholder S, Wijsman EM, Winn DM, Collins FS. Replicating genotype-phenotype associations. Nature. 2007;447:655–660. doi: 10.1038/447655a. [DOI] [PubMed] [Google Scholar]
- 12.Ferwerda B, McCall MB, Verheijen K, Kullberg BJ, van der Ven AJ, Van der Meer JW, Netea MG. Functional consequences of toll-like receptor 4 polymorphisms. Mol Med. 2008;14:346–352. doi: 10.2119/2007-00135.Ferwerda. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Rallabhandi P, Bell J, Boukhvalova MS, Medvedev A, Lorenz E, Arditi M, Hemming VG, Blanco JC, Segal DM, Vogel SN. Analysis of TLR4 polymorphic variants: new insights into TLR4/MD-2/CD14 stoichiometry, structure, and signaling. J Immunol. 2006;177:322–332. doi: 10.4049/jimmunol.177.1.322. [DOI] [PubMed] [Google Scholar]
- 14.de Kruif MD, Setiati TE, Mairuhu AT, Koraka P, Aberson HA, Spek CA, Osterhaus AD, Reitsma PH, Brandjes DP, Soemantri A, van Gorp EC. Differential gene expression changes in children with severe dengue virus infections. PLoS Negl Trop Dis. 2008;2:e215. doi: 10.1371/journal.pntd.0000215. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Modhiran N, Kalayanarooj S, Ubol S. Subversion of innate defenses by the interplay between DENV and pre-existing enhancing antibodies: TLRs signaling collapse. PLoS Negl Trop Dis. 2010;4:e924. doi: 10.1371/journal.pntd.0000924. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Ferwerda B, McCall MB, Alonso S, Giamarellos-Bourboulis EJ, Mouktaroudi M, Izagirre N, Syafruddin D, Kibiki G, Cristea T, Hijmans A, Hamann L, Israel S, ElGhazali G, Troye-Blomberg M, Kumpf O, Maiga B, Dolo A, Doumbo O, Hermsen CC, Stalenhoef AF, van Crevel R, Brunner HG, Oh DY, Schumann RR, de la Rua C, Sauerwein R, Kullberg BJ, van der Ven AJ, van der Meer JW, Netea MG. TLR4 polymorphisms, infectious diseases, and evolutionary pressure during migration of modern humans. Proc Natl Acad Sci USA. 2007;104:16645–16650. doi: 10.1073/pnas.0704828104. [DOI] [PMC free article] [PubMed] [Google Scholar]


