Abstract
The regulatory system controlling pheromone-induced plasmid transfer in Enterococcus faecalis is the most thoroughly studied genetic system of this species. Transcription initiation from the target promoter is controlled by a pheromone receptor/repressor protein whose activity is determined by its interaction with two peptide signaling molecules that compete for the same binding site, but have opposing effects on the activity of the receptor protein. For the system to function as a sensitive and robust biological switch, several additional levels of post-transcriptional regulation are also required. Expression of important functions encoded within the enterococcal core genome may also be controlled by multilayered regulatory circuitry. The pheromone system may serve as a useful paradigm to guide comprehensive functional genomic analysis of E. faecalis.
Introduction
Enterococci are significant members of the normal intestinal flora of humans and numerous animal species, including invertebrates such as insects. They have great medical importance because of their high levels of inherent and acquired antibiotic resistance and their ability to cause opportunistic infections [1]. One of the most important features of these organisms is their inherent ability to survive and propagate under a variety of harsh conditions. Systems for sensing and responding to extra cellular signals undoubtedly contribute to their remarkable survival abilities. Here we summarize the current understanding of the sex pheromone-inducible conjugation system of Enterococcus faecalis, a form of intercellular communication where peptide signals inform plasmid-containing cells of the close proximity of potential recipient cells [2]. Recent studies have elucidated the essential nature of multiple levels of regulation for proper function and versatility of the pheromone systems. This review will focus on the pheromone-responsive conjugative plasmid pCF10 [3], and how the experimental tools and knowledge base generated from this research may inform the emerging field of enterococcal functional genomics.
The extracellular phase of pheromone signaling
The sex pheromones are hydrophobic peptides, processed from the signal peptides of secreted lipoproteins [4]. Examination of genome sequences reveals that >50 lipoproteins may be encoded by the E. faecalis chromosome, suggesting that the known pheromones represent a small percentage of the potential signals that are produced. While only a handful of plasmids have been analyzed in detail, it seems likely that most of them encode pheromone-sensing systems that evolved from a common ancestor. For a review of common and unique features of various pheromone plasmids, the reader is referred to [5,6].
The mature pheromone cCF10 is a heptapeptide (LVTLVFV) produced by proteolytic cleavage of the signal peptide segment of the putative secreted lipoprotein CcfA [7]. As is the case for several other pheromones [8], the Eep protease is required for the production of wild-type levels of cCF10 [9•]. Eep-mediated endoproteolytic cleavage occurs within the membrane. The pCF10 pheromone-sensing system (Figure 1b) encompasses two signaling molecules, cCF10 and the plasmid-encoded iCF10 inhibitor peptide (AITLIFI) that competitively inhibits the activity of cCF10 [9•]. The amounts of each peptide produced by pCF10-containing cells are tightly controlled (Figure 1b), such that donor cells secrete a mixture of the two peptides in a molar ratio of ~80:1 in favor of iCF10 [10]. This ratio keeps the conjugation system off, but allows for a sensitive response to increases in relative cCF10 activity. The two peptide system is versatile; a change in the ratio of the two signals can occur either by the addition of exogenous cCF10 (when recipients in close proximity contribute to the cCF10 pool) or by depletion of iCF10 (which occurs when donor cells grow in the bloodstream). The latter phenomenon is critical for the production of the protein product (Asc10) of the pheromone-inducible prgB gene, which increases enterococcal virulence in several models [11• •]. We note that expression of the enterococcal cytolysin toxin is also controlled extracellularly by the balance between antagonistic signaling peptides, which can be altered by interaction with host cells [12•].
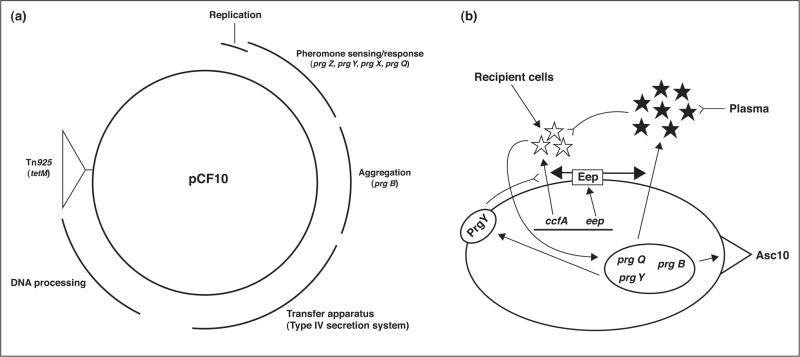
Figure 1.
Genetic organization of pCF10 and the role of extracellular peptide signals in the control of pCF10 conjugation. (a) Map of pCF10, based on DNA sequencing [16] and genetic studies [46]. The curved lines outside the circle denote the segments of the plasmid encoding various biological functions as indicated, with specific genes discussed in the text listed below the respective regions in which they are located. (b) The induction state of pCF10-containing cells is determined by the molar ratios of cCF10 pheromone (white stars; encoded by the chromosomal ccfA gene) to iCF10 inhibitor (black stars; encoded by the prgQ gene of pCF10). Both peptides are processed to their mature heptapeptide form from precursor polypeptides in the membrane by the Eep protease (rectangle in cell membrane). The pCF10-encoded PrgY protein (oval in the membrane) reduces the level of cCF10 produced by its host cell; residual cCF10 escaping PrgY is neutralized by iCF10. Both peptides function by internalization and interaction with PrgX, as described in Figure 2 and in the text, with cCF10 causing increased expression of the prgQ operon leading to production of Asc10, and other proteins involved in conjugation. The peptide ratio can be shifted by recipient cells in close proximity, or in the bloodstream, where iCF10 is sequestered, resulting in self-induction by exogenously produced cCF10. Single arrows indicate a positive effect of one component of the system on another; double-stemmed arrows indicate the polypeptide product of a gene encoded by pCF10 (oval in the cell) or the chromosome (line in the cell); inverted arrows indicate inhibition or destruction of one component by another.
Pheromone import, PrgX and action at the prgQ promoter
In contrast to peptide signaling molecules whose interaction with the surface of the responder cell transduces a signal across the membrane [5], cCF10 binds to PrgZ on the surface of pCF10-carrying cells and is imported into the cytosol [13], where it interacts with PrgX, derepressing the prgQ promoter. Comparison of high-resolution structures of apo-PrgX with those of PrgX::cCF10 and PrgX::iCF10 complexes yielded the model shown in Figure 2 [14•,15• •]. Structural, genetic and biochemical studies indicate that in uninduced cells, PrgX/iCF10 complexes form a repressing PrgX tetramer structure that exhibits enhanced occupancy of two operator sites on pCF10 DNA. The lower affinity site overlaps the prgQ promoter, and reduces transcription when occupied. Binding cCF10 to PrgX displaces iCF10, altering the structure of the PrgX C-terminus. This is predicted to reduce interaction at the tetramer interface, disrupting the DNA loop and ultimately reducing occupancy of the lower affinity DNA binding site that is critical for repression. Quantitative analysis of prgQ transcription under various conditions [16,17] (A. Chatterjee et al., submitted) indicates that repression in vivo is never complete. Moreover, the direct effect of pheromone on enhancement of prgQ transcription initiation is much lower than the level of induction of expression of downstream loci in the prgQ operon, suggesting that post-transcriptional regulation amplifies the pheromone response as described below.
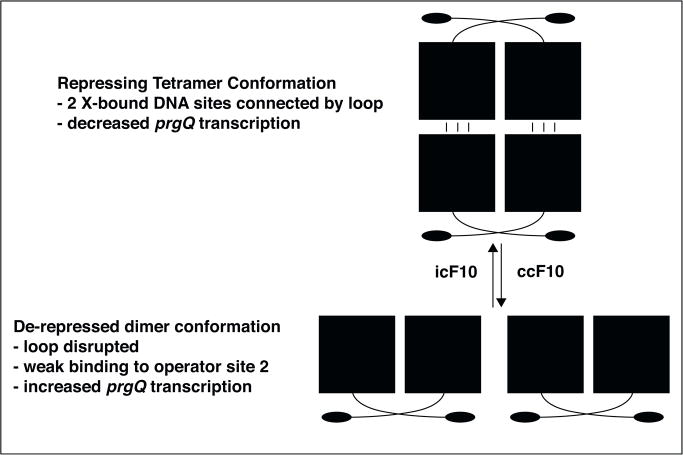
Figure 2.
Pheromone and inhibitor peptides affect PrgX repression of prgQ transcription by altering the oligomerization state of PrgX in pCF10-containing cells. PrgX monomers contain an N-terminal DNA binding domain (filled oval) and a C-terminal regulatory domain (filled square) connected by a short linker region (curved line). Domain swapping promotes formation of a stable dimer that can occupy one operator binding site on pCF10; the region upstream from prgQ contains two such sites. Interaction of iCF10 with PrgX stabilizes a small-C-terminal loop proposed to function as an interface for protein/protein interactions between pairs of dimers to stabilize a tetramer structure (upper panel) where each dimer occupies one operator site on pCF10, and the two sites are connected by a DNA loop (not shown). Pheromone binding to the same region of PrgX causes a structural change at the extreme C-terminus of PrgX that abolishes the tetramer-stabilizing interaction (lower panel), and indirectly reduces operator site occupancy. This increases transcription initiation at prgQ by RNA polymerase.
Multiple levels of post-transcriptional regulation resulting from genetic organization
There is extensive evidence for additional levels of control of RNAs transcribed from both the prgQ promoter, PQ, and the prgX promoter, PX [18–21]. A picture of complex, reciprocal regulation between these two operons is beginning to develop. The transcriptional start site of prgX mRNA is located downstream of PQ but oriented in the opposite direction [22••]. This makes transcription from these two promoters convergent for approximately 220 bp, generating transcripts that are complementary in this region (Figure 3). Multiple RNA species are produced from each promoter. PX generates both the prgX mRNA and Anti-Q, a small non-coding RNA corresponding to the 5′ 102 nt of the prgX transcript. In the absence of pheromone induction, transcription from PQ generates Qs, a 380 nt RNA that terminates at IRS1, a factor-independent terminator. Qs includes the 66 nt Orf prgQ, which produces the iCF10 inhibitor. When induced, PQ produces increased levels of Qs as well as transcripts that extend past IRS1, into genes encoding the conjugative transfer machinery (Figure 1a) [22••].
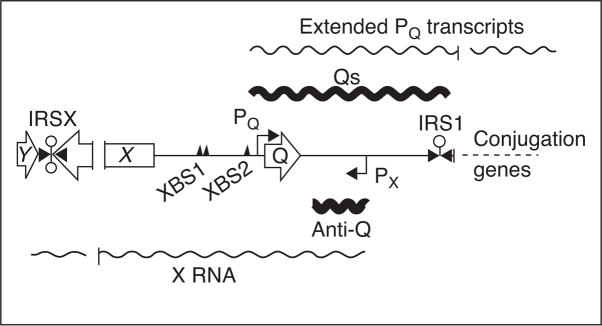
Figure 3.
Genetic organization of prgX and prgQ regions of pCF10 and reciprocal antisense regulation. Map of the prgX and prgQ regions is shown with ORFs for prgY, X, and Q indicated by open arrows. The primary and secondary PrgX binding sites are indicated by triangles labeled XBS1 and XBS2, respectively. The promoters PX and PQ are indicated, along with transcriptional start sites, by bent arrows. Factor-independent terminators are indicated by lollipops. Wavy lines above and below the map indicate RNAs transcribed from PQ and PX, respectively. Transcripts from these two promoters share a 220 bp overlapping complementary region, and published [22••,25• •] and unpublished data C. Johnson & G. Dunny, manuscript in preparation demonstrated that each transcript negatively regulates the corresponding complementary transcript by antisense mechanisms.
Reciprocal regulation of antagonistic operons by multiple mechanisms
In addition to the terminator, IRS1, Qs contains sequences capable of forming an antiterminator that precludes terminator formation in the nascent transcript. Anti-Q is complementary to a portion of the antiterminator and interacts with nascent prgQ transcripts to sequester these sequences, preventing formation of the antiterminator. This allows folding of the terminator helix, which halts transcription at IRS1, attenuating expression of downstream genes [22••]. This mechanism is similar to those that regulate plasmid copy number in other plasmids [23••,24]. As is the case for other systems, Anti-Q and Qs do not form a complete duplex upon interaction. Rather, defined structures within the two RNAs appear to be essential for productive interaction [25••]. Anti-Q can attenuate transcription of sequences downstream of IRS1 in vitro, in the absence of RNA chaperones, suggesting that this activity does not require protein factors beyond RNAP [22••].
Transcripts originating at Px are also post-transcriptionally regulated. Pheromone induction leads to a loss of prgX and Anti-Q RNAs [20]. Additionally, experiments using reporter plasmids have demonstrated that transcription from PX is not directly regulated by PrgX. However, when PQ is present in cis, the PrgX protein is required for transcription of prgX [18–20]. Anti-Q levels are modulated in a similar fashion. In the absence of PrgX, a plasmid that contains both PQ and PX produces very little Anti-Q (C Johnson, G Dunny, manuscript in preparation). Inactivating PQ leads to a large increase in Anti-Q expression (C Johnson, G, Dunny, manuscript in preparation). Providing Qs in trans from a compatible plasmid results in intermediate Anti-Q expression. Taken together, these data suggest a model in which transcripts from PX are regulated via multiple mechanisms. Transcription from PQ in cis actively interferes with transcription from PX, and repression of transcription from PQ alleviates this interference. Additionally transcripts from PQ can act in trans to downregulate expression from PX. Recent efforts to apply mathematical modeling and rigorous quantitative analysis to this system suggest that multiple layers of reciprocal regulation between the antagonistic prgQ and prgX operons are required for the system to function as a sensitive and robust biological switch (A Chatterjee et al., submitted). The convergent and overlapping organization of these operons serves to amplify the relatively subtle direct effect of the pheromone signal. It is also worth noting that the gene organization of the enterococcal cytolysin locus also features convergent transcription from opposing promoters [26], suggesting that transcription interference provides an additional layer of regulation in this system as well.
Functional genomics
In bacteria, the analyses of regulatory mechanisms controlling gene expression in mobile elements like phages and plasmids have long provided an essential foundation for chromosomal genetics. Experimental tools and mechanistic insights generated from analysis of the pCF10 pheromone response might also enhance comprehensive analysis of genetic regulatory systems in enterococci (Table 1). The increasing medical significance of enterococci, and the advent of new technologies for genome sequencing and expression analysis, has contributed to a remarkable upsurge in functional genomic studies. Here we consider the extent to which the novel regulatory mechanisms controlling the pCF10 pheromone response might function on a genomic scale. We searched the literature and found that about 50 different enterococcal promoters and/or their cognate transcription factors have been analyzed by various combinations of genetic, biochemical, and in a few cases, structural approaches. Studies of pCF10 and other pheromone plasmids demonstrate that transcription factors very frequently have multiple DNA binding targets (and perhaps RNA targets) and multiple regulatory functions that may not be apparent from focused studies of a single promoter. While PrgX shows very low levels of amino acid sequence relatedness to other transcription factors, recent structural and genetic studies have indicated that PrgX is a member of an emerging superfamily of transcription factors with common structural features [C. Johnson and G. Dunny] that regulate virulence in bacilli [27••] and gram positive cocci [28] in response to extracellular peptide signals. Global effects on host cell physiology mediated by transcription factors targeting multiple operons have already been documented in several cases, such as the CroRS system [29–31] and the Ers system [32–34], and emerging technologies such as RNAseq [35] should enable the definition of the molecular basis for the control of enterococcal physiology on a genomic scale and with high resolution in the future.
Table 1.
| Features of pCF10 regulation applicable to functional genomics | |||
|---|---|---|---|
| Mechanism | pCF10 component(s) | Non-pCF10 example(s) | Considerations |
| Regulation of promoter | PrgX | Many; some positive regulators | Precise mapping of transcription starts, DNA targets. Cofactors and their effects on regulatory proteins |
| Change in mRNA 28 structure to affect termination translation, or RNA processing | Q/Anti-Q RNAs | T box, SMK box, par RNAs, eut riboswitches | Antisense RNA, other RNA-binding ligands |
| Convergent transcription | prgQ/prgX | RIVET clones | Confirmation and demonstration of function; cis/trans effects |
Analysis of the pheromone response has shown that extensive secondary structure and processing of transcripts may produce abundant products whose 5′ ends do not actually represent sites of transcription initiation. Thus it is important to verify putative initiation sites and their corresponding promoters by molecular methods that can distinguish 5′ ends generated by RNA processing from those resulting from true initiation [36], and by careful genetic analysis of the upstream sequences required for proper expression of a gene of interest. Loci encoding specific biological functions frequently are organized as cassettes with the regulated genes and their adjacent cognate regulators are transcribed in the opposite direction. Differentiating between ‘back-to-back’ divergent promoters and convergent promoters where transcription of the regulatory gene(s) actually initiates from the nontemplate strand within the target operon is critical.
Studies of the pheromone system, of plasmid replication [23••,24], and of the par toxin/antitoxin system [37] in enterococci and close relatives suggested that post-transcriptional mechanisms could be as important in gene regulation as promoter control, if not more so. There is growing appreciation of the importance of post-transcriptional gene regulation mediated by antisense RNA interactions, riboswitches, and other RNA-based mechanisms in enterococci on a genomic scale. Competing terminator/antiterminator structures, or structures in which the ribosome binding site of an mRNA is sequestered or free in 5′ untranslated regions have been documented for biosynthetic-resistance, catabolic-resistance, and antibiotic-resistance operons in enterococci and related species [38–40]. The choice between alternative secondary structures is controlled by trans-acting sRNAs in some cases and by small molecule metabolites in other cases. The E. faecalis eut operon encoding the ability to utilize ethanolamine as a nutrient source provides a remarkable new example of multiple forms of post-transcriptional control. This polycistronic locus encodes a two-component regulatory cassette (HK17/RR17 or EutW/EutV; [41••]) that functions by a phosphorelay [42]. However, it appears that the EutV response regulator binds to mRNA and controls transcription termination rather than promoter activity. It is proposed that this system responds to availability of enthanolamine [41••]. Interestingly the operon also contains adenosylcobalamin-responsive riboswitches; the data suggests that these novel features of the mRNAs produced from this locus provide post-transcriptional mechanisms to sense both the availability of the catabolic substrate for the operon and of the adenosylcobalamin cofactor essential for enzymatic activity.
The importance of as yet unidentified antisense RNA regulators in control of essential enterococcal virulence functions is suggested by a recent report that enterococcal RNase J2 is a positive regulator of expression of adhesive pili required for biofilm formation and virulence [43••]. A plausible interpretation of these results is that this RNase degrades an antisense RNA that negatively regulates pilus expression. Further evidence for a global role of sRNAs comes from our genetic screens for promoters activated during biofilm formation [44] and in a subdermal abscess model infection (K Frank et al., submitted for publication). These studies suggest the existence of differentially expressed antisense transcripts from the ‘nontemplate strands’ of protein-encoding genes under specific conditions. The fact that extensive antisense transcription has been observed in the phylo-genetically related Listeria monocytogenes [45] also supports the likely existence of many antisense regulators in enterococci and related genera. Given the rapid pace of progress in analysis of enterococcal gene regulation during the past few years, we suggest that this type of research will provide new approaches to the control and prevention of lethal infections caused by antibiotic-resistant strains.
Acknowledgments
We thank Tim Leonard for help with figures, and our many collaborators for their vital contributions. Our research is supported by grants GM49530 and AI58134 from the National Institutes of Health. CMJ is a recipient of a Dissertation fellowship from the University of Minnesota Graduate School.
References and recommended reading
Papers of particular interest, published within the period of review, have been highlighted as:
• of special interest
•• of outstanding interest
- 1.Gilmore MS, Coburn PS, Nallapareddy SR, Murray BE. Enterococcal virulence. In: Gilmore MS, editor. The Enterococci: Pathogenesis, Molecular Biology and Antibiotic Resistance. American Society for Microbiology Press; 2002. pp. 301–354. [Google Scholar]
- 2.Clewell DB, Dunny GM. Conjugation and genetic exchange in enterococci. In: Gilmore MS, Clewell DB, Courvalin P, Dunny GM, Murray BE, Rice LB, editors. The Enterococci: Pathogenesis, Molecular Biology and Antibiotic Resistance. American Society for Microbiology Press; 2002. pp. 265–300. [Google Scholar]
- 3.Haemig HAH, Dunny GM. New insights into pheromone control and response in Enterococcus faecalis pCF10. In: Bassler B, Winans S, editors. Chemical Communication Among Bacteria. American Society for Microbiology Press; 2008. pp. 31–49. [Google Scholar]
- 4.Clewell DB, An FY, Flannagan SE, Antiporta M, Dunny GM. Enterococcal sex pheromone precursors are part of signal sequences for surface lipoproteins. Mol Microbiol. 2000;35:246–248. doi: 10.1046/j.1365-2958.2000.01687.x. [DOI] [PubMed] [Google Scholar]
- 5.Chandler JR, Dunny GM. Enterococcal peptide sex pheromones: synthesis and control of biological activity. Peptides. 2004;25:1377–1388. doi: 10.1016/j.peptides.2003.10.020. [DOI] [PubMed] [Google Scholar]
- 6.Hirt H, Wirth R, Muscholl A. Comparative analysis of 18 sex pheromone plasmids from Enterococcus faecalis: detection of a new insertion element on pPD1 and hypotheses on the evolution of this plasmid family. Mol Gen Genet. 1996;252:640–647. doi: 10.1007/BF02173969. [DOI] [PubMed] [Google Scholar]
- 7.Antiporta MH, Dunny GM. ccfA, the genetic determinant for the cCF10 peptide pheromone in Enterococcus faecalis OG1RF. J Bacteriol. 2002;184:1155–1162. doi: 10.1128/jb.184.4.1155-1162.2002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.An FY, Sulavik MC, Clewell DB. Identification and characterization of a determinant (eep) on the Enterococcus faecalis chromosome that is involved in production of the peptide sex pheromone cAD1. J Bacteriol. 1999;181:5915–5921. doi: 10.1128/jb.181.19.5915-5921.1999. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9•.Chandler JR, Dunny GM. Characterization of the sequence specificity determinants required for processing and control of sex pheromone by the intramembrane protease Eep and the plasmid-encoded protein PrgY. J Bacteriol. 2008;190:1172–1183. doi: 10.1128/JB.01327-07. This paper presents genetic evidence that the peptide sequence recognition specificity for PrgY lies within mature cCF10, but within cCF10 precursors in the case of Eep. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Hirt H, Schlievert PM, Dunny GM. In vivo induction of virulence and antibiotic resistance transfer in Enterococcus faecalis mediated by the sex pheromone-sensing system of pCF10. Infect Immun. 2002;70:716–723. doi: 10.1128/iai.70.2.716-723.2002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11••.Chandler JR, Hirt H, Dunny GM. A paracrine peptide sex pheromone also acts as an autocrine signal to induce plasmid transfer and virulence factor expression in vivo. Proc Natl Acad Sci U S A. 2005;102:15617–15622. doi: 10.1073/pnas.0505545102. This paper showed that induction of the pheromone system in the bloodstream was an autocrine process resulting from iCF10 sequestration, and endogenous cCF10. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12•.Coburn PS, Pillar CM, Jett BD, Haas W, Gilmore MS. Enterococcus faecalis senses target cells and in response expresses cytolysin. Science. 2004;306:2270–2272. doi: 10.1126/science.1103996. This paper shows that sequestration of a inhibitory peptide by the mammalian host induces toxin production in another system controlled by two signaling peptides. [DOI] [PubMed] [Google Scholar]
- 13.Leonard BAB, Podbielski A, Hedberg PJ, Dunny GM. Enterococcus faecalis pheromone binding protein, PrgZ, recruits a chromosomal oligopeptide permease system to import sex pheromone cCF10 for induction of conjugation. Proc Natl Acad Sci U S A. 1996;93:260–264. doi: 10.1073/pnas.93.1.260. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14•.Kozlowicz BK, Shi K, Gu ZY, Ohlendorf DH, Earhart CA, Dunny GM. Molecular basis for control of conjugation by bacterial pheromone and inhibitor peptides. Mol Microbiol. 2006;62:958–969. doi: 10.1111/j.1365-2958.2006.05434.x. This paper shows that iCF10 acts as a co-repressor with PrgX by binding to the same site as cCF10, but causing a different structural effect. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15•.Shi K, Brown CK, Gu ZY, Kozlowicz BK, Dunny GM, Ohlendorf DH, Earhart CA. Structure of peptide sex pheromone receptor PrgX and PrgX/pheromone complexes and regulation of conjugation in Enterococcus faecalis. Proc Natl Acad Sci U S A. 2005;102:18596–18601. doi: 10.1073/pnas.0506163102. This paper reported the structure of PrgX and PrgX/cCF10 complexes leading to the model shown in Figure 2. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Hirt H, Manias DA, Bryan EM, Klein JR, Marklund JK, Staddon JH, Paustian ML, Kapur V, Dunny GM. Characterization of the pheromone response of the Enterococcus faecalis conjugative plasmid pCF10: complete sequence and comparative analysis of the transcriptional and phenotypic responses of pCF10-containing cells to pheromone induction. J Bacteriol. 2005;187:1044–1054. doi: 10.1128/JB.187.3.1044-1054.2005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Bensing BA, Manias DA, Dunny GM. Pheromone cCF10 and plasmid pCF10-encoded regulatory molecules act post-transcriptionally to activate expression of downstream conjugation functions. Mol Microbiol. 1997;24:295–308. doi: 10.1046/j.1365-2958.1997.3301710.x. [DOI] [PubMed] [Google Scholar]
- 18.Bae T, Clerc-Bardin S, Dunny GM. Analysis of expression of prgX, a key negative regulator of the transfer of the Enterococcus faecalis pheromone-inducible plasmid pCF10. J Mol Biol. 2000;297:861–875. doi: 10.1006/jmbi.2000.3628. [DOI] [PubMed] [Google Scholar]
- 19.Bae T, Kozlowicz BK, Dunny GM. Characterization of cis-acting prgQ mutants: evidence for two distinct repression mechanisms by Qa RNA and PrgX protein in pheromone-inducible enterococcal plasmid pCF10. Mol Microbiol. 2004;51:271–281. doi: 10.1046/j.1365-2958.2003.03832.x. [DOI] [PubMed] [Google Scholar]
- 20.Kozlowicz BK, Bae T, Dunny GM. Enterococcus faecalis pheromone-responsive protein PrgX: genetic separation of positive autoregulatory functions from those involved in negative regulation of conjugative plasmid transfer. Mol Microbiol. 2004;54:520–532. doi: 10.1111/j.1365-2958.2004.04286.x. [DOI] [PubMed] [Google Scholar]
- 21.Bae T, Kozlowicz B, Dunny GM. Two targets in pCF10 DNA for PrgX binding: their role in production of Qa and prgX mRNA and in regulation of pheromone-inducible conjugation. J Mol Biol. 2002;315:995–1007. doi: 10.1006/jmbi.2001.5294. [DOI] [PubMed] [Google Scholar]
- 22••.Johnson CM, Manias DA, Haemig HA, Shokeen S, Weaver KE, Henkin TM, Dunny GM. Direct evidence for control of the pheromone-inducible prgQ operon of Enterococcus faecalis plasmid pCF10 by a countertranscript-driven attenuation mechanism. J Bacteriol. 2009;192:1634–1642. doi: 10.1128/JB.01525-09. This paper provided compelling genetic and biochemical evidence for regulation of prgQ transcription in trans by the Anti-Q countertranscript, confirming predictions from previous studies. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23••.Novick RP, Iordanescu S, Projan SJ, Kornblum J, Edelman I. pT181 plasmid replication is regulated by a countertranscript-driven transcriptional attenuator. Cell. 1989;59:395–404. doi: 10.1016/0092-8674(89)90300-0. A classic paper representing the first report of the countertranscript-mediated termination mechanism used in multiple regulatory systems in gram positive bacteria. [DOI] [PubMed] [Google Scholar]
- 24.Le Chatelier E, Ehrlich SD, Janniere L. Countertranscript-driven attenuation system of the pAMbeta1 repE gene. Mol Microbiol. 1996;20:1099–1112. doi: 10.1111/j.1365-2958.1996.tb02550.x. [DOI] [PubMed] [Google Scholar]
- 25••.Shokeen S, Johnson CM, Greenfield TJ, Manias DA, Dunny GM, Weaver KE. Structural analysis of the Anti-Q-Qs interaction: RNA-mediated regulation of E. faecalis plasmid pCF10 conjugation. Plasmid. 2010;64:26–35. doi: 10.1016/j.plasmid.2010.03.002. Detailed analysis of the cirtical secondary structures and base-pariing interactions mediating Q/Anti-Q interactions. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Haas W, Shepard BD, Gilmore MS. Two-component regulator of Enterococcus faecalis cytolysin responds to quorum-sensing autoinduction. Nature. 2002;415:84–87. doi: 10.1038/415084a. [DOI] [PubMed] [Google Scholar]
- 27••.Declerck N, Bouillaut L, Chaix D, Rugani N, Slamti L, Hoh F, Lereclus D, Arold ST. Structure of PlcR: insights into virulence regulation and evolution of quorum sensing in Gram-positive bacteria. Proc Natl Acad Sci U S A. 2007;104:18490–18495. doi: 10.1073/pnas.0704501104. The first report of the existence of a conserved superfamily of peptide-responsive transcription factors structurally related to PrgX in gram positive bacteria. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Mashburn-Warren L, Morrison DA, Federle MJ. A novel double-tryptophan peptide pheromone controls competence in Streptococcus spp. via an Rgg regulator. Mol Microbiol. 2010;78:589–606. doi: 10.1111/j.1365-2958.2010.07361.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Comenge Y, Quintiliani R, Jr, Li L, Dubost L, Brouard JP, Hugonnet JE, Arthur M. The CroRS two-component regulatory system is required for intrinsic beta-lactam resistance in Enterococcus faecalis. J Bacteriol. 2003;185:7184–7192. doi: 10.1128/JB.185.24.7184-7192.2003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Le Breton Y, Muller C, Auffray Y, Rince A. New insights into the Enterococcus faecalis CroRS two-component system obtained using a differential-display random arbitrarily primed PCR approach. Appl Environ Microbiol. 2007;73:3738–3741. doi: 10.1128/AEM.00390-07. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Muller C, Le Breton Y, Morin T, Benachour A, Auffray Y, Rince A. The response regulator CroR modulates expression of the secreted stress-induced SalB protein in Enterococcus faecalis. J Bacteriol. 2006;188:2636–2645. doi: 10.1128/JB.188.7.2636-2645.2006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Giard JC, Riboulet E, Verneuil N, Sanguinetti M, Auffray Y, Hartke A. Characterization of Ers, a PrfA-like regulator of Enterococcus faecalis. FEMS Immunol Med Microbiol. 2006;46:410–418. doi: 10.1111/j.1574-695X.2005.00049.x. [DOI] [PubMed] [Google Scholar]
- 33.Riboulet-Bisson E, Sanguinetti M, Budin-Verneuil A, Auffray Y, Hartke A, Giard JC. Characterization of the ers regulon of Enterococcus faecalis. Infect Immun. 2008;76:3064–3074. doi: 10.1128/IAI.00161-08. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Riboulet-Bisson E, Le Jeune A, Benachour A, Auffray Y, Hartke A, Giard JC. Ers a Crp/Fnr-like transcriptional regulator of Enterococcus faecalis. Int J Food Microbiol. 2009;131:71–74. doi: 10.1016/j.ijfoodmicro.2008.06.022. [DOI] [PubMed] [Google Scholar]
- 35.Croucher NJ, Thomson NR. Studying bacterial transcriptomes using RNA-seq. Curr Opin Microbiol. 2010;13:619–624. doi: 10.1016/j.mib.2010.09.009. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Bensing BA, Meyer BJ, Dunny GM. Sensitive detection of bacterial transcription initiation sites and differentiation from RNA processing sites in the pheromone-induced plasmid transfer system of Enterococcus faecalis. Proc Natl Acad Sci U S A. 1996;93:7794–7799. doi: 10.1073/pnas.93.15.7794. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Greenfield TJ, Franch T, Gerdes K, Weaver KE. Antisense RNA regulation of the par post-segregational killing system: structural analysis and mechanism of binding of the antisense RNA, RNAII and its target, RNAI. Mol Microbiol. 2001;42:527–537. doi: 10.1046/j.1365-2958.2001.02663.x. [DOI] [PubMed] [Google Scholar]
- 38.Grundy FJ, Henkin TM. Regulation of gene expression by effectors that bind to RNA. Curr Opin Microbiol. 2004;7:126–131. doi: 10.1016/j.mib.2004.02.013. [DOI] [PubMed] [Google Scholar]
- 39.Fuchs RT, Grundy FJ, Henkin TM. The S(MK) box is a new SAM-binding RNA for translational regulation of SAM synthetase. Nat Struct Mol Biol. 2006;13:226–233. doi: 10.1038/nsmb1059. [DOI] [PubMed] [Google Scholar]
- 40.Grundy FJ, Henkin TM. The T box and S box transcription termination control systems. Front Biosci. 2003;8:d20–d31. doi: 10.2741/908. [DOI] [PubMed] [Google Scholar]
- 41••.Fox KA, Ramesh A, Stearns JE, Bourgogne A, Reyes-Jara A, Winkler WC, Garsin DA. Multiple posttranscriptional regulatory mechanisms partner to control ethanolamine utilization in Enterococcus faecalis. Proc Natl Acad Sci U S A. 2009;106:4435–4440. doi: 10.1073/pnas.0812194106. This papers presents compelling evidence for multiple novel forms of post-transcriptional control of the eut locus in E. faecalis. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Del Papa MF, Perego M. Ethanolamine activates a sensor histidine kinase regulating its utilization in Enterococcus faecalis. J Bacteriol. 2008;190:7147–7156. doi: 10.1128/JB.00952-08. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43••.Gao P, Pinkston KL, Nallapareddy SR, van Hoof A, Murray BE, Harvey BR. Enterococcus faecalis rnjB is required for pilin gene expression and biofilm formation. J Bacteriol. 2010;192:5489–5498. doi: 10.1128/JB.00725-10. The first report suggesting an important regulatory function for an rnj family RNAse in enterococci. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Ballering KS, Kristich CJ, Grindle SM, Oromendia A, Beattie DT, Dunny GM. Functional genomics of Enterococcus faecalis: multiple novel genetic determinants for biofilm formation in the core genome. J Bacteriol. 2009;191:2806–2814. doi: 10.1128/JB.01688-08. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Camejo A, Buchrieser C, Couve E, Carvalho F, Reis O, Ferreira P, Sousa S, Cossart P, Cabanes D. In vivo transcriptional profiling of Listeria monocytogenes and mutagenesis identify new virulence factors involved in infection. PLoS Pathog. 2009;5:e1000449. doi: 10.1371/journal.ppat.1000449. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Dunny GM. The peptide pheromone-inducible conjugation system of Enterococcus faecalis plasmid pCF10: cell–cell signalling, gene transfer, complexity and evolution. Philos Trans R Soc Lond B Biol Sci. 2007;362:1185–1193. doi: 10.1098/rstb.2007.2043. [DOI] [PMC free article] [PubMed] [Google Scholar]