Abstract
Introduction
Obstructive sleep apnea (OSA) is associated with increased risk for metabolic syndrome in both adults and children. In adults with OSA, serum levels of fatty acid binding protein 4 (FABP4) are elevated and associated with the degree of metabolic insulin resistance, independent of obesity. Therefore, we assessed plasma FABP4 levels and FABP4 allelic variants in obese and non-obese children with and without OSA.
Methods
A total of 309 consecutive children ages 5-8 years were recruited. Children were divided into those with OSA and without OSA (NOSA) based on the apnea-hypopnea index (AHI). Subjects were also subdivided into obese (OB) and non-obese (NOB) based on BMI z score). Morning fasting plasma FABP4 levels were assayed using ELISA, and 11 single-nucleotide polymorphisms (SNPs) within the FABP4 region were genotyped.
Results
Morning plasma FABP4 levels were increased in all children with OSA, even in NOB children. However, plasma FABP4 levels were strongly associated with BMI z score. Of the 11 SNPs tested, the frequency of rs1054135 (A/G) minor allele (A) was significantly increased in OSA. This SNP was also associated with increased plasma FABP4 levels in both OSA and obese subjects. The minor allele frequency of all other SNPs was similar in OSA and NOSA groups.
Conclusions
Childhood obesity and OSA are associated with higher plasma FABP4 levels and thus promote cardiometabolic risk. The presence of selective SNP (e.g., rs1054135) in the FABP4 gene may account for increased plasma FABP4 levels in the context of obesity and OSA in children.
Introduction
Obstructive sleep apnea (OSA), a serious health problem associated with obesity in children and adults, has reached epidemic proportions around the globe (1-3). Similar to obesity, the metabolic and cardiovascular complications of OSA in childhood include hyperlipidemia, metabolic syndrome and systemic low grade inflammation (4, 5), indicating that the clustering of OSA with cardiometabolic risk factors may involve shared molecular pathways. Furthermore, while the presence of OSA leads to increased levels of inflammatory mediators that promote development of endothelial and metabolic dysfunction in children (4-16), there is substantial variability of metabolic dysfunction or increased inflammatory markers among children with OSA, suggesting that individual genomic variance may account for such discrepancies.
Fatty acid binding protein 4 (FABP4), also known as aP2 or Adipocyte-FABP (A-FABP), is a cytosolic protein abundantly expressed in adipocytes and macrophages (17). A growing body of evidence from both animal and clinical studies suggests that FABP4 plays an important role in the interaction between lipid metabolism and inflammation, and may be involved in the development of the metabolic syndrome. Indeed, plasma FABP4 levels appear to correlate with the degree of metabolic dysfunction and obesity (17-19). Similar to adults, children who are obese are more likely to display elevations in plasma FABP4 levels, which will be reduced by interventions leading to weight loss (20-21). In a recent study, we showed that FABP4 genetic polymorphisms accounted for a substantial proportion of the variance in metabolic dysfunction in a community based pediatric cohort (22).
Based on aforementioned considerations, we hypothesized that the presence of OSA in children may be associated with increased plasma FABP4 levels and that the variance of such association may be explained by individual discrepancies in the presence of specific single nucleotide polymorphisms (SNPs) in the FABP4 gene.
Methods
Subjects
Consecutive children who were referred for evaluation of suspected sleep disordered breathing (SDB) to the University of Louisville Pediatric Sleep Medicine Center were recruited. The study was approved by the University of Louisville Human Research Committee, and informed consent was obtained from the legal caregiver of each participant. Clinical evaluation was performed in all these subjects and children with known diabetes, hypertension or another chronic disease condition associated with OSA and obesity were excluded from the study. As controls, we also invited children from the community who had no history of any chronic or acute disorder and who did not snore which was confirmed by physical examination. Of note, all children were otherwise healthy, and were representative of the demographic characteristics of the general population of the city of Louisville (http://ksdc.louisville.edu/sdc/census2000/cityprofiles/LouisvilleDP.pdf). Children were excluded if they were younger than 5 years or older than 8 years of age, had known diabetes or pre-diabetes (http://www.diabetes.org/pre-diabetes/pre-diabetes-symptoms.jsp), any defined genetic abnormality or underlying systemic disease including hypertension, or if they were within a month from any acute infectious process.
Overnight Polysomnography (NPSG)
All subjects underwent overnight polysomnography using standard techniques (23). Children were studied for up to 12 hours in a quiet, darkened room maintained at an ambient temperature of 24°C. No medication was used to induce sleep. The following parameters were measured: chest and abdominal wall movement by inductance plethysmography, heart rate by electrocardiography, air flow was triply monitored with a side stream end-tidal capnograph that also provides breath-by-breath assessment of end-tidal carbon dioxide levels (BCI SC-300, Menomonee Falls, WI), a nasal pressure cannula, and an oronasal thermistor. Arterial pulse oxygen saturation (SpO2) was assessed by pulse oximetry [Nellcor N 100 (Nellcor Inc, Hayward, CA)], with simultaneous recording of the pulse waveform. The bilateral electro-oculogram, 8 channels of electroencephalogram (2 frontal, 2 occipital, 2 temporal, and 2 central leads), chin and anterior tibial electromyograms, and analog output from a body position sensor were also monitored. All measures were digitized using a commercially available system (Sandman, Nellcor Puritan Bennett, Kanata, ON, Canada, or Stellate Instruments, Montreal, QC, Canada). Tracheal sound was monitored with a microphone sensor, and a digital time-synchronized video recording was performed. The sleep technician followed patient behavior and confirmed sleep position by the infrared camera inside the room.
All of the studies were initially scored by a certified technician and were then reviewed by a physician who was experienced in pediatric PSG and underwent training in an accredited fellowship program using defined criteria (23-25). Blood was drawn the morning after the child completed the polysomnographic evaluation and after an overnight fast.
Demographics and definitions
All parents completed a detailed intake clinical questionnaire which included age, gender, ethnicity, and history of medication or of any other chronic disease. Height, weight and vital signs were recorded for each child, and body mass index (BMI) Z score was calculated using CDC 2000 growth standards (www.cdc.gov/growthcharts) and online software (www.cdc.gov/epiinfo). A BMI z-score>1.65 was considered as fulfilling obesity criteria. The diagnosis of OSA was defined by the presence of an obstructive apnea index ≥ 1 / hour of total sleep time and an obstructive apnea-hypopnea index (AHI) ≥ 5/hour of total sleep time and a nadir oxyhemoglobin saturation <92% (23). Control children had AHI < 1/hour of total sleep time and no oxygen desaturation events during sleep.
Plasma Assays
Plasma FABP4 levels were measured using commercial Enzyme Linked Immunosorbant Assay (ELISA) kits (ALPCO Diagnostics, Salem, NH) following the manufacturer's instructions. All assays were performed in duplicate and a calibration curve was included in each assay. This method has a detection level of 0.1ng/mL and exhibits linear behavior up to 250 ng/mL. The intra-assay and inter-assay coefficients of variability of plasma FABP4 were 5.3% and 6.8%, respectively.
Plasma insulin levels were measured using a commercially available radioimmunoassay kit (Coat-A-Count Insulin; Diagnostic Products Inc). This method has a detection level of 1.2 μIU/mL and exhibits linear behavior up to 350 μIU/mL, with intra-assay and interassay coefficients of variability of 3.1% and 4.9%, respectively. Plasma glucose level was measured using a commercial kit based on the hexokinase-glucose-6-phosphate dehydrogenase method (Flex Reagent Cartridges; Dade Behring, Newark, DE).
Serum lipids including total cholesterol, high-density lipoprotein (HDL-C) cholesterol, calculated low-density lipoprotein cholesterol (LDL-C), and triglycerides (TG) were also assessed using Flex Reagent Cartridges (Dade Behring).
DNA Extraction
Peripheral blood samples were collected in vacutainer tubes containing EDTA (Becton Dickinson, Franklin Lakes, NJ, USA). All DNA samples were extracted using QIAmp DNA blood kit (Qiagen, Valencia, CA) according to the manufacturer's protocol. The concentration and quality of the DNA were determined using a ND-1000 Spectrophotometer (Nanodrop Technologies, Wilmington, DE, USA). The purity of the DNA was determined by calculating the ratio of absorbance at 260/280 nm, and all DNA samples had a ratio of 1.8-1.9. The precise length of genomic DNA was determined by gel electrophoresis using 1% agarose gel. All the purified samples were stored at -80°C until further analyses.
Genotyping using Real-Time PCR
Genotyping was performed using the ABI PRISM 7500 Sequence Detection System for allelic discrimination following the manufacturer's instructions (Applied Biosystems). Eleven FABP4 single nucleotide polymorphisms (SNPs) were examined in this study. Polymorphisms were genotyped using TaqMan technology (Applied Biosystems, Inc.). Two fluorogenic minor groove binder probes were used for each locus using the dyes 6-carboxyfluorescein (FAM; excitation, 494 nm) and VIC (excitation, 538 nm) which are easily differentiated in RT-PCR system. Realtime PCR reaction was performed using 12.5 μl of TaqMan 2× universal master mix (Applied Biosystems, CA), 1.25 μl of each primer, 10.25 μl of RNase and DNase-free water (Ambion, Austin, TX), and 1 μl of sample DNA, in a total volume of 25 μl per single well reaction. Two wells of a 96 well-plate (Applied Biosystems, CA) were used for each sample for each of the 11 SNPs as well as a control, which did not include any DNA template, in each assay. Assay conditions were 2 min at 50°C, 10 min at 95°C, and 40 cycles of 95°C for 15 s and 60°C for 1 min. Initially, the SNP assay was set up using SDS, version 2.1, software (Applied Biosystems, CA) as an absolute quantification assay, but after assay completion the plate was read using the allelic discrimination settings. Post-assay analysis was performed using the SDS software. Hardy-Weinberg equilibrium (HWE) for each SNP in this population was verified using the equation p2+q2+2pq=1 where p and q represents the wild type and variant allele of a gene.
Statistical Analysis
Data were expressed as mean±SD. Significant differences within groups were analyzed using ANOVA for continuous variables and chi-square tests for categorical variables. Spearman's correlation analyses were conducted to examine potential associations between BMI and plasma concentrations of the various inflammatory mediators. We employed a general linear model to check standardized beta coefficient and to determine independent predictor for plasma FABP4 levels in these subjects. This model allowed us to test for the effects of categorical predictor variables as well as for continuous predictor variable. Statistical analyses were performed using SPSS software and Statistica (version 17.0; SPPS Inc., Chicago, IL., Version 9 StatSoft). All p-values reported are 2-tailed with statistical significance set at <0.05.
Results
A total of 309 children were recruited and subdivided into 4 sub-groups based on the presence or absence of obesity (OB and NOB) and OSA (OSA and NOSA), respectively. The subjects' demographic characteristics and lipid profiles are shown in Table 1. The cohort included 59.2% white non-hispanic, 27.2% African American, and 13.6% other ethnicities. The distribution of gender among groups was not statistically different (males: NOB-NOSA: 58.9%, OB-NOSA: 45.5%, NON-OSA: 56.5%, and OB-OSA: 62.4%, Chi2(3)=2.96, p=0.40).
Table 1. Demographic characteristics and fasting morning plasma concentrations of lipids, glucose, insulin, and FABP4 in subgroups of obese and OSA subjects.
| Variables | NOB-NOSA (n=90) | NOB-OSA (n=92) | p value | OB-NOSA (n=33) | OB-OSA (n=94) | p value | Overall p value OB vs NOB |
|---|---|---|---|---|---|---|---|
| Age (yrs) | 7.0 ± 0.68 | 7.1 ± 0.93 | 0.85 | 7.2 ± 1.2 | 7.4 ± 1.2 | 0.54 | 0.002 |
| BMI z Score | 0.24 ± 0.86 | 0.07 ± 1.05 | 0.26 | 2.3 ±0.5 | 2.4 ± 0.4 | 0.20 | <0.001 |
| AHI (/h TST) | 0.35 ± 0.23 | 4.2 ± 6.2 | <0.001 | 0.34 ± 0.2 | 5.6 ± 6.7 | <0.001 | 0.005 |
| FABP4 level (ng/mL) | 7.4 ± 1.67 | 15.3 ± 1.61 | <0.001 | 13.2 ± 2.9 | 23.4 ± 4.7 | <0.001 | <0.001 |
| TC (mg/dL) | 155.3 ± 31.9 | 165.1 ± 24.7 | <0.04 | 173.7 ± 25.4 | 186.8 ± 26.5 | <0.05 | 0.56 |
| Triglycerides (mg/dL) | 83.2 ± 36.0 | 77.2 ± 41.6 | 0.47 | 110.8 ± 36.1 | 118.5 ± 46.9 | 0.55 | 0.17 |
| HDL-C (mg/dL) | 59.1 ± 12.4 | 52.1 ± 9.3 | <0.04 | 48.5 ± 7.5 | 40.6 ± 10.7 | <0.01 | 0.06 |
| VLDL-C (mg/dL) | 16.6 ± 12.6 | 19.6 ± 8.4 | <0.01 | 22.4 ± 21.3 | 27.6 ± 9.4 | <0.01 | 0.16 |
| LDL-C (mg/dL) | 85.6 ± 27.3 | 94.8 ± 22.6 | <0.01 | 102.9 ± 16.8 | 112.4 ± 21.2 | <0.01 | 0.36 |
| Glucose (mg/dL) | 76.8 ± 16.1 | 77.1 ± 15.1 | 0.95 | 83.7 ± 9.8 | 89.8 ± 11.6 | <0.01 | <0.04 |
| Insulin (μIU/mL) | 6.5 ± 12.4 | 7.6 ± 13.2 | 0.44 | 11.4 ± 13.6 | 16.2 ± 9.4 | <0.01 | <0.02 |
| HOMA-IR | 1.23 ± 0.49 | 1.45 ± 0.49 | 0.003 | 2.36 ± 0.33 | 3.6 ± 0.27 | <0.001 | <0.001 |
BMI, Body mass index; AHI, Obstructive apnea hypopnea index; FABP, Fatty acid binding protein; TC, total cholesterol; HDL-C, High density lipoprotein Cholesterol; VLDL-C, Very low density lipoprotein- Cholesterol; LDL-C, Low density lipoprotein-Cholesterol; P-value was calculated using one way ANOVA.
Children with OSA had significantly higher increased variant allele frequency of rs1054135 than NOSA. The variant allele frequency of all other studied SNPs was not statistically different in OSA and NOSA groups (Table 2). The frequency of heterozygous genotype was higher in NOB-OSA when compared with NOB-NOSA (p=0.003) but was not statistically different in OB-OSA and OB-NOSA (p=0.47). Of note, the variant allele frequency of rs1054135 was higher in OB as compared to NOB through increased “G” [standardized β coefficient (SE); -0.51(0.05)] allele, in contrast to having the heterozygous or “A” allele [standardized β coefficient (SE); 0.31(0.05); p<0.001; Variance 51.4%]. The variant allele frequencies of all other SNPs were similar between OB and NOB groups (Table 3). However, the variant allele frequency of rs1054135 was higher in OB-NOSA than in NOB-NOSA (Table 4). The distribution of rs1054135 showed that NOB-NOSA group had increased frequency of the “GG” genotype with decreased “AA” and heterozygous genotype frequency. Similarly, distribution of rs2303519 showed that heterozygous genotype frequency was decreased in NOB-NOSA and increased in OB-NOSA. The variant allele frequency of all other genotypes was not statistically different in subgroups of OSA and obesity (Table 4). Plasma FABP4 levels were significantly higher in OSA than in NOSA, and OB children had significantly higher plasma FABP4 levels than NOB subjects (Figure 1). Furthermore, BMI z score and OSA severity are predictive of the FABP4 level (0.48*BMI z-score + 0.27*AHI= FABP 4 level (R2=0.33, p<0.0001). Subjects with the variant allele of rs1054135 had significantly higher plasma FABP4 levels (mean±SD; 19.7±6.7 ng/mL) compared to subjects without the variant allele (11.9±4.7 ng/mL, p<0.001 after adjusting for potential confounders including age, gender, ethnicity, AHI, and BMI z score). Among the other SNPs, there was no statistically significant difference between plasma FABP levels.
Table 2.
Distribution of allele and genotype frequencies of FABP4 SNPs in children with OSA and NOSA.
| SNPs | Genotype | NOSA (n=123) n (%) | OSA (n=186) n (%) | p value |
|---|---|---|---|---|
| rs1054135 | GG | 90 (73.2) | 88 (47.8) | <0.001 |
| AA | 10 (8.2) | 42 (22.8) | ||
| Both | 23 (18.7) | 54 (29.3) | ||
| rs2303519 | CC | 90 (73.2) | 126 (68.1) | 0.34 |
| Both | 33 (26.8) | 59 (31.9) | ||
| rs16909233 | CC | 96 (78.05) | 137 (73.7) | 0.38 |
| Both | 27 (21.95) | 49 (26.3) | ||
| rs7821186 | AA | 74 (64.3) | 103 (62.8) | 0.95 |
| GG | 12 (10.4) | 17 (10.4) | ||
| Both | 29 (25.2) | 44 (26.8) | ||
| rs16909187 | GG | 86 (74.8) | 109 (66.1) | 0.26 |
| AA | 3 (2.6) | 4 (2.4) | ||
| Both | 26 (22.6) | 52 (31.5) | ||
| rs16909192 | AA | 90 (78.3) | 117 (71.8) | 0.39 |
| CC | 2 (1.7) | 2 (1.2) | ||
| Both | 23 (20) | 44 (27) | ||
| rs10808846 | CC | 58 (50.9) | 66 (41) | 0.27 |
| AA | 15 (13.2) | 25 (15.5) | ||
| Both | 41 (36) | 70 (43.5) | ||
| rs7018409 | GG | 87 (75.6) | 111 (67.7) | 0.32 |
| AA | 3 (2.6) | 4 (2.4) | ||
| Both | 25 (21.7) | 49 (29.9) | ||
| rs2290201 | TT | 54 (46.9) | 64 (39.3) | 0.43 |
| CC | 21 (18.3) | 36 (22.1) | ||
| Both | 40 (34.8) | 63 (38.6) | ||
| rs6992708 | GG | 15 (13.0) | 24 (14.6) | 0.40 |
| AA | 57 (49.6) | 68 (41.5) | ||
| Both | 43 (37.4) | 72 (43.9) |
Table 3. Distributions of allele and genotype frequencies of FABP4 SNPs in children categorized as obese or non-obese.
| SNPs | Genotype | NOB (n=180) | OB (n=127) | Overall p value |
|---|---|---|---|---|
| rs1054135 | GG | 135 (75) | 43 (33.9) | <0.001 |
| AA | 19 (10.6) | 33 (25.9) | ||
| Both | 26 (14.4) | 51 (40.2) | ||
| rs2303519 | CC | 135 (74.6) | 81 (63.8) | 0.04 |
| Both | 46 (25.4) | 46 (36.2) | ||
| rs16909233 | GG | 143 (78.6) | 90 (70.9) | 0.12 |
| Both | 39 (21.4) | 37 (29.1) | ||
| rs7821186 | AA | 99 (60) | 78 (68.4) | 0.35 |
| GG | 19 (11.5) | 10 (8.8) | ||
| Both | 47 (28.5) | 26 (22.8) | ||
| rs16909187 | GG | 114 (69.1) | 81 (70.4) | 0.61 |
| AA | 3 (1.8) | 4 (3.5) | ||
| Both | 48 (29.1) | 30 (26.1) | ||
| rs16909192 | AA | 124 (75.2) | 83 (73.5) | 0.90 |
| CC | 2 (1.2) | 2 (1.8) | ||
| Both | 39 (23.6) | 28 (24.8) | ||
| rs10808846 | CC | 73 (45.1) | 51 (45.1) | 0.13 |
| AA | 29 (17.9) | 11 (9.7) | ||
| Both | 60 (37.0) | 51 (45.1) | ||
| rs7018409 | GG | 116 (70.3) | 82 (71.9) | 0.58 |
| AA | 3 (1.8) | 4 (3.5) | ||
| Both | 46 (27.9) | 28 (24.6) | ||
| rs2290201 | TT | 68 (41.5) | 50 (43.9) | 0.12 |
| CC | 40 (24.4) | 17 (14.9) | ||
| Both | 56 (34.1) | 47 (41.2) | ||
| rs6992708 | GG | 26 (15.8) | 13 (11.4) | 0.58 |
| AA | 73 (44.2) | 52 (45.6) | ||
| Both | 66 (40) | 49 (43) |
Table 4.
Distributions of allele and genotype frequencies of FABP4 SNPs as a function of the presence or absence of OSA and obesity.
| Group 1 | Group 2 | 1 vs. 2 | Group 3 | Group 4 | 3 vs. 4 | 1 vs. 3 | 2 vs. 4 | ||
|---|---|---|---|---|---|---|---|---|---|
| SNPs | Genotype | NOB-NOSA (n=90; %) | NOB-OSA (n=92; %) | p value | OB-NOSA (n=33; %) | OB-OSA (n=94; %) | p value | p value | p value |
| rs105135 | GG | 77 (85.6) | 58 (64.4) | 0.003 | 13 (39.4) | 30 (31.9) | 0.47 | <0.001 | <0.001 |
| AA | 4 (4.4) | 15 (16.7) | 6 (18.2) | 27 (28.7) | |||||
| Both | 9 (10) | 17 (18.9) | 14 (42.4) | 37 (39.4) | |||||
| rs2303519 | CC | 73 (81.1) | 62 (68.1) | 0.05 | 17 (51.5) | 64 (68.1) | 0.89 | 0.001 | 0.99 |
| Both | 17 (18.9) | 29 (31.9) | 16 (48.5) | 30 (31.9) | |||||
| rs16909233 | GG | 76 (84.4) | 67 (72.8) | 0.06 | 20 (60.6) | 70 (74.5) | 0.13 | 0.005 | 0.79 |
| Both | 14 (15.6) | 25 (27.2) | 13 (39.4) | 24 (25.5) | |||||
| rs7821186 | AA | 53 (63.1) | 46 (56.8) | 0.62 | 21 (67.7) | 57 (68.7) | 0.59 | 0.64 | 0.22 |
| GG | 8 (9.5) | 11 (13.6) | 4 (12.9) | 6 (7.2) | |||||
| Both | 23 (27.4) | 24 (29.6) | 6 (19.4) | 20 (24.1) | |||||
| rs16909187 | GG | 63 (75) | 51 (62.9) | 0.03 | 23 (74.2) | 58 (69.1) | 0.46 | 0.52 | 0.06 |
| AA | 3 (3.6) | 0 (0) | 0 (0) | 4 (4.8) | |||||
| Both | 18 (21.4) | 30 (37.0) | 8 (25.8) | 22 (26.2) | |||||
| rs16909192 | AA | 67 (79.8) | 57 (70.4) | 0.09 | 23 (74.2) | 60 (73.2) | 0.68 | 0.46 | 0.29 |
| CC | 2 (2.4) | 0 (0) | 0 (0) | 2 (2.4) | |||||
| Both | 15 (17.9) | 24 (29.6) | 8 (25.8) | 20 (24.4) | |||||
| rs10808846 | CC | 43 (51.8) | 30 (37.9) | 0.14 | 15 (48.4) | 36 (43.9) | 0.63 | 0.93 | 0.44 |
| AA | 11 (13.3) | 18 (22.8) | 4 (12.9) | 7 (8.5) | |||||
| Both | 29 (34.9) | 31 (39.2) | 12 (38.7) | 39 (47.6) | |||||
| rs7018409 | GG | 64 (76.2) | 52 (64.2) | 0.03 | 23 (74.2) | 59 (71.1) | 0.46 | 0.48 | 0.05 |
| AA | 3 (3.6) | 0 (0) | 0 (0) | 4 (4.8) | |||||
| Both | 17 (20.2) | 29 (35.8) | 8 (25.8) | 20 (24.1) | |||||
| rs2290201 | TT | 40 (47.6) | 28 (35) | 0.22 | 14 (45.2) | 36 (43.4) | 0.93 | 0.52 | 0.13 |
| CC | 17 (20.2) | 23 (28.7) | 4 (12.9) | 13 (15.7) | |||||
| Both | 27 (32.1) | 29 (36.3) | 13 (41.9) | 34 (40.9) | |||||
| rs6992708 | GG | 11 (13.1) | 15 (18.5) | 0.18 | 4 (12.9) | 9 (10.8) | 0.95 | 0.82 | 0.29 |
| AA | 43 (51.2) | 30 (37.0) | 14 (45.2) | 38 (45.8) | |||||
| Both | 30 (35.7) | 36 (44.4) | 13 (41.9) | 36 (43.4) |
Figure 1.
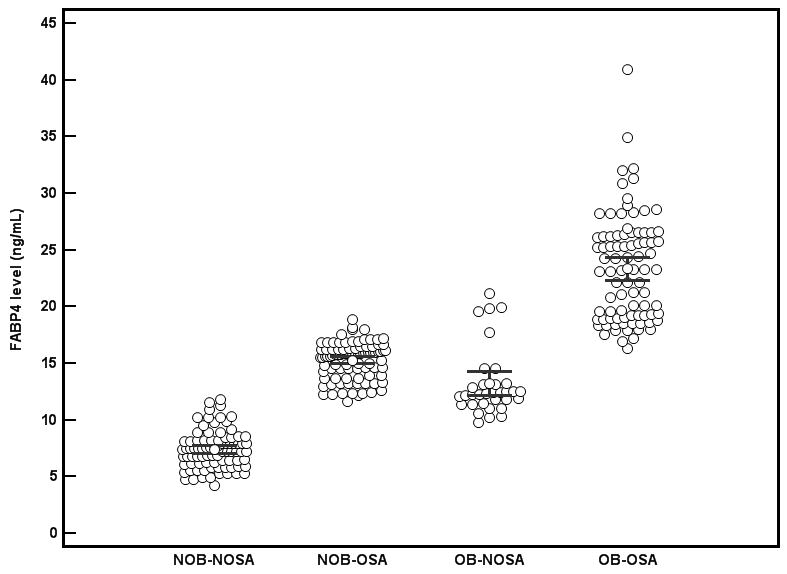
Fasting morning plasma concentrations of FABP4 in subgroups of obese and OSA subjects. Individual levels, means and 95% confidence interval are shown. (NOB-NOSA vs. NOB-OSA, OB-NOSA vs. NOB-NOSA, and OB-NOSA vs. OB-OSA were all significantly different, p<0.001).
Discussion
In this study we found that plasma FABP4 levels are significantly higher in subjects with OSA when compared with NOSA. After categorizing subjects based on BMI z score, we found that increased plasma FABP4 levels not only correlated with obesity but also that the effect of OSA persists even among obese subjects. In this study we included SNPs whose minor allele frequency was greater than 0.05 in the general population so that heterozygous or rare forms of an allele can be detected in studied samples. The 11 SNPs considered in this study were all found to have minor allele frequencies greater that 0.05. For example according to HapMap data, the minor allele frequency for rs1054135 is 0.133 for European, 0.68 for Asian and 0.31 for Sub-Sahara Africans. In addition, all the SNPs used in this study were in Hardy-Weinberg equilibrium. Furthermore, the frequency of the rs1054135, a SNP that locates to the 3′ untranslated region (3′UTR) in the FABP4 gene, was higher in OSA compared to NOSA, and seemingly contributed to higher plasma FABP4 levels, while the frequencies of all other SNPs tested for the FABP4 gene were not statistically different in OSA and NOSA.
Several roles in gene expression have been attributed to the UTRs, including mRNA stability, mRNA localization, and translational efficiency. The function of a UTR depends on its sequence and can differ between mRNAs. What changes does rs1054135 makes in UTR and how it affects the function of FABP4 gene needs further investigation.
Before discussing the potential significance of our findings some methodological issues deserve comment. We a priori excluded any child with known diabetes, hypertension or another chronic disease condition associated with OSA and obesity. This methodological approach may have artificially reduced the impact of such conditions on plasma FABP4 levels as well as the magnitude of the association of any given FABP4 allelic variant with the degree of metabolic dysfunction or with plasma FABP4 levels. Therefore, it will be important to explore such possibilities in a larger and non-restrictive clinical referral pediatric population being assessed for OSA. Second, we restricted the age range of our cohort to a relatively narrow age window, which corresponds to a period that coincides with significant changes in food consumption patterns (26). This decision aimed to reduce as much as possible the presence of confounding factors across a wide age spectrum and the potential confounder of adolescence. Finally, we identified and recruited closely matching control children to nullify as best as possible some of the known potential confounding factors that could be introduced in the process of subject selection.
FABP4 has been proposed as a bridge between inflammatory processes and other biological pathways related to the metabolic syndrome (27, 28). In this study, plasma FABP4 levels were increased in OSA when compared to NOSA, and such effect persisted when their BMI z score was incorporated into the analyses. However, we found no association between the severity of OSA and plasma FABP4 levels even though we confirmed here the previously reported correlation between BMI z score and FABP4 concentrations (22), a finding that has also been reported by other groups on pediatric cohorts (20, 21).
These findings differ from the only other published study in adults in which the presence of OSA was not only a risk factor for elevated plasma FABP4 levels but the latter also correlated with degree of severity in OSA (29, 30). Indeed, Lam et al (2009) reported that plasma FABP4 levels were significantly associated with sleep hypoxemia parameters, including duration of oxygen desaturation and minimal oxygen saturation, independent of age and obesity. Plasma FABP4 levels were also associated with increased insulin resistance, independent of age and adiposity (29), a finding that we have also recently corroborated in children (22).
In this study, we found that of the 11 FABP4 SNPs examined, which were selected to cover the whole genomic sequence of FABP4, only the rs1054135 polymorphism was significantly more prevalent among OSA, as well as among OB children. In a previous study assessing genomic variability in the FABP4 gene in a population, those individuals carrying the T-87C polymorphism, which reduced the transcriptional efficiency of the FABP4 gene among the 7,899 adult participants, had lower plasma triglyceride levels and significantly reduced risk for coronary heart disease and type 2 diabetes compared with subjects homozygous for the wild-type allele (31). Damcott et al also examined the presence of the SNP (-376) in FABP4 as well as a classic PPAR-gamma SNP in males, and found that FABP4 and PPARgamma appeared to interact to influence insulin sensitivity and body composition (32). We should emphasize, however, that FABP4 is also expressed in activated macrophages (33-34) and that high FABP4 concentrations are detected in human atherosclerotic lesions (35). Thus, allelic variants that reduce plasma FABP4 expression or activity should be metabolically protective, while those associated with higher plasma FABP4 may increase susceptibility. In this regard, the genetic variability at the FABP4, as evidenced by rs1054135, increased plasma FABP4 levels in both OSA and OB children, suggesting an interaction between OSA and FABP4 alleles to increase plasma FABP4 levels. But no significant difference was observed in fasting glucose, triglycerides and total cholesterol in subjects with rs1054135 variant allele when compared to those without rs1054135 variant allele. Furthermore, the allelic variants of all other SNPs were not differentially distributed in the several subgroups and did not appear to play any significant role in the variation of plasma FABP4 levels. Taken together, these initial findings in a pediatric subjects with OSA suggest that FABP4 rs1054135 genomic variance may be an important determinant for altered serum FABP4 levels in these subjects.
In summary, we have shown that young school-age children with OSA and/or obesity exhibit higher circulating levels of the pro-inflammatory FABP4, which in turn may contribute to the risk for systemic inflammation and metabolic dysfunction (36). Furthermore, selective variants in FABP4 gene appear to contribute to the proinflammatory or diabetogenic potential of OSA and obesity during childhood.
Future plans
The specific aim of this study was to check the frequency of various known genotypes in subjects with and without OSA. We further attempted to check if there is any correlation between any FABP4 gene polymorphism and plasma FABP4 levels. We did not emphasize any of the functional aspects of the FABP4 SNP in this study since this would be beyond the scope of the study. Future steps in this study will be to assess the distribution of rs1054135 in a much larger population with and without OSA. If we find similar results to those reported in this study it will then be mandatory to assess the functional aspects of this polymorphism in relation to transcriptional regulation of the gene.
Acknowledgments
Funding Sources: DG is supported by National Institutes of Health grants HL-065270 and HL-086662.
Footnotes
Authors' Contributions: BB performed genotype analyses and drafted the initial version of the manuscript; AK performed genomic analyses and contributed to manuscript revisions for intellectual content; KS conducted statistical analyses; LKG, OSC, RB and JK recruited subjects, processed samples and conducted ELISA assays; BK and HH contributed to genomic polymorphism analyses and critical review of the manuscript; DG initiated the conceptual framework of the project, supervised its performance, contributed to funding and data analyses, and was involved in all aspects of manuscript writing including its final version. All authors have reviewed and approved the final version of the manuscript.
Publisher's Disclaimer: This is a PDF file of an unedited manuscript that has been accepted for publication. As a service to our customers we are providing this early version of the manuscript. The manuscript will undergo copyediting, typesetting, and review of the resulting proof before it is published in its final citable form. Please note that during the production process errors may be discovered which could affect the content, and all legal disclaimers that apply to the journal pertain.
References
- 1.Gozal D. Obstructive sleep apnea in children: implications for the developing central nervous system. Semin Pediatr Neurol. 2008;15(2):100–6. doi: 10.1016/j.spen.2008.03.006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Capdevila OS, Dayyat E, Kheirandish-Gozal L, Gozal D. Prevalence of epileptiform activity in healthy children during sleep. Sleep Med. 2008 Mar;9(3):303–9. doi: 10.1016/j.sleep.2007.03.024. [DOI] [PubMed] [Google Scholar]
- 3.Dayyat E, Kheirandish-Gozal L, Gozal D. Childhood Obstructive Sleep Apnea: One or Two Distinct Disease Entities? Sleep Med Clin. 2007 Sep;2(3):433–44. doi: 10.1016/j.jsmc.2007.05.004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Gozal D, Capdevila OS, Kheirandish-Gozal L. Metabolic alterations and systemic inflammation in obstructive sleep apnea among nonobese and obese prepubertal children. Am J Respir Crit Care Med. 2008 May 15;177(10):1142–9. doi: 10.1164/rccm.200711-1670OC. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Kwok KL, Ng DK, Chan CH. Cardiovascular changes in children with snoring and obstructive sleep apnoea. Ann Acad Med Singapore. 2008 Aug;37(8):715–21. [PubMed] [Google Scholar]
- 6.Gozal D. Sleep, sleep disorders and inflammation in children. Sleep Med. 2009 Sep;10 1:S12–6. doi: 10.1016/j.sleep.2009.07.003. [DOI] [PubMed] [Google Scholar]
- 7.Suheyl Ezgu F, Hasanoglu A, Tumer L, Ozbay F, Aybay C, Gunduz M. Endothelial activation and inflammation in prepubertal obese Turkish children. Metabolism. 2005 Oct;54(10):1384–9. doi: 10.1016/j.metabol.2005.05.003. [DOI] [PubMed] [Google Scholar]
- 8.Tauman R, Ivanenko A, O'Brien LM, Gozal D. Plasma C-reactive protein levels among children with sleep-disordered breathing. Pediatrics. 2004 Jun;113(6):e564–9. doi: 10.1542/peds.113.6.e564. [DOI] [PubMed] [Google Scholar]
- 9.Tauman R, O'Brien LM, Gozal D. Hypoxemia and obesity modulate plasma C-reactive protein and interleukin-6 levels in sleep-disordered breathing. Sleep Breath. 2007 Jun;11(2):77–84. doi: 10.1007/s11325-006-0085-7. [DOI] [PubMed] [Google Scholar]
- 10.O'Brien LM, Serpero LD, Tauman R, Gozal D. Plasma adhesion molecules in children with sleep-disordered breathing. Chest. 2006 Apr;129(4):947–53. doi: 10.1378/chest.129.4.947. [DOI] [PubMed] [Google Scholar]
- 11.Kheirandish-Gozal L, Capdevila OS, Tauman R, Gozal D. Plasma C-reactive protein in nonobese children with obstructive sleep apnea before and after adenotonsillectomy. J Clin Sleep Med. 2006 Jul 15;2(3):301–4. [PMC free article] [PubMed] [Google Scholar]
- 12.Gozal D, Crabtree VM, Sans Capdevila O, Witcher LA, Kheirandish-Gozal L. C-reactive protein, obstructive sleep apnea, and cognitive dysfunction in school-aged children. Am J Respir Crit Care Med. 2007 Jul 15;176(2):188–93. doi: 10.1164/rccm.200610-1519OC. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Gozal D, Serpero LD, Sans Capdevila O, Kheirandish-Gozal L. Systemic inflammation in non-obese children with obstructive sleep apnea. Sleep Med. 2008 Mar;9(3):254–9. doi: 10.1016/j.sleep.2007.04.013. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Kim J, Bhattacharjee R, Snow AB, Capdevila OS, Kheirandish-Gozal L, Gozal D. Myeloid-related protein 8/14 levels in children with obstructive sleep apnoea. Eur Respir J. 2010 Apr;35(4):843–50. doi: 10.1183/09031936.00075409. [DOI] [PubMed] [Google Scholar]
- 15.Khalyfa A, Capdevila OS, Buazza MO, Serpero LD, Kheirandish-Gozal L, Gozal D. Genome-wide gene expression profiling in children with non-obese obstructive sleep apnea. Sleep Med. 2009 Jan;10(1):75–86. doi: 10.1016/j.sleep.2007.11.006. [DOI] [PubMed] [Google Scholar]
- 16.Kim J, Lee S, Bhattacharjee R, Khalyfa A, Kheirandish-Gozal L, Gozal D. Leukocyte Telomere Length and Plasma Catestatin and Myeloid Related Protein 8/14 Concentrations in Children with Obstructive Sleep Apnea. Chest. 2010 Mar 18; doi: 10.1378/chest.09-2832. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Zimmerman AW, Veerkamp JH. New insights into the structure and function of fatty acid-binding proteins. Cell Mol Life Sci. 2002 Jul;59(7):1096–116. doi: 10.1007/s00018-002-8490-y. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Furuhashi M, Hotamisligil GS. Fatty acid-binding proteins: role in metabolic diseases and potential as drug targets. Nat Rev Drug Discov. 2008 Jun;7(6):489–503. doi: 10.1038/nrd2589. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Furuhashi M, Fucho R, Gorgun CZ, Tuncman G, Cao H, Hotamisligil GS. Adipocyte/macrophage fatty acid-binding proteins contribute to metabolic deterioration through actions in both macrophages and adipocytes in mice. J Clin Invest. 2008 Jul;118(7):2640–50. doi: 10.1172/JCI34750. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Yun KE, Kim SM, Choi KM, Park HS. Association between adipocyte fatty acid-binding protein levels and childhood obesity in Korean children. Metabolism. 2009 Jun;58(6):798–802. doi: 10.1016/j.metabol.2009.01.017. [DOI] [PubMed] [Google Scholar]
- 21.Reinehr T, Stoffel-Wagner B, Roth CL. Retinol-binding protein 4 and its relation to insulin resistance in obese children before and after weight loss. J Clin Endocrinol Metab. 2008 Jun;93(6):2287–93. doi: 10.1210/jc.2007-2745. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Khalyfa A, Bhushan B, Hegazi M, Kim J, Kheirandish-Gozal L, Bhattacharjee R, et al. Fatty-acid binding protein 4 gene variants and childhood obesity: potential implications for insulin sensitivity and CRP levels. Lipids Health Dis. 2010;9:18. doi: 10.1186/1476-511X-9-18. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Montgomery-Downs HE, O'Brien LM, Gulliver TE, Gozal D. Polysomnographic characteristics in normal preschool and early school-aged children. Pediatrics. 2006 Mar;117(3):741–53. doi: 10.1542/peds.2005-1067. [DOI] [PubMed] [Google Scholar]
- 24.Standards and indications for cardiopulmonary sleep studies in children. American Thoracic Society. 1996 doi: 10.1164/ajrccm.153.2.8564147. Am J Respir Crit Care Med. [DOI] [PubMed] [Google Scholar]
- 25.Silber MH, Ancoli-Israel S, Bonnet MH, Chokroverty S, Grigg-Damberger MM, Hirshkowitz M, et al. The visual scoring of sleep in adults. J Clin Sleep Med. 2007 Mar 15;3(2):121–31. [PubMed] [Google Scholar]
- 26.Maeda K, Cao H, Kono K, Gorgun CZ, Furuhashi M, Uysal KT, et al. Adipocyte/macrophage fatty acid binding proteins control integrated metabolic responses in obesity and diabetes. Cell Metab. 2005 Feb;1(2):107–19. doi: 10.1016/j.cmet.2004.12.008. [DOI] [PubMed] [Google Scholar]
- 27.Cabre A, Lazaro I, Girona J, Manzanares JM, Marimon F, Plana N, et al. Fatty acid binding protein 4 is increased in metabolic syndrome and with thiazolidinedione treatment in diabetic patients. Atherosclerosis. 2007 Nov;195(1):e150–8. doi: 10.1016/j.atherosclerosis.2007.04.045. [DOI] [PubMed] [Google Scholar]
- 28.Karakas SE, Almario RU, Kim K. Serum fatty acid binding protein 4, free fatty acids, and metabolic risk markers. Metabolism. 2009 Jul;58(7):1002–7. doi: 10.1016/j.metabol.2009.02.024. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Lam DC, Xu A, Lam KS, Lam B, Lam JC, Lui MM, et al. Serum adipocyte-fatty acid binding protein level is elevated in severe OSA and correlates with insulin resistance. Eur Respir J. 2009 Feb;33(2):346–51. doi: 10.1183/09031936.50075408. [DOI] [PubMed] [Google Scholar]
- 30.Reinehr T, Stoffel-Wagner B, Roth CL. Adipocyte fatty acid-binding protein in obese children before and after weight loss. Metabolism. 2007 Dec;56(12):1735–41. doi: 10.1016/j.metabol.2007.07.019. [DOI] [PubMed] [Google Scholar]
- 31.Tuncman G, Erbay E, Hom X, De Vivo I, Campos H, Rimm EB, et al. A genetic variant at the fatty acid-binding protein aP2 locus reduces the risk for hypertriglyceridemia, type 2 diabetes, and cardiovascular disease. Proc Natl Acad Sci U S A. 2006 May 2;103(18):6970–5. doi: 10.1073/pnas.0602178103. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Damcott CM, Moffett SP, Feingold E, Barmada MM, Marshall JA, Hamman RF, et al. Genetic variation in fatty acid-binding protein-4 and peroxisome proliferator-activated receptor gamma interactively influence insulin sensitivity and body composition in males. Metabolism. 2004 Mar;53(3):303–9. doi: 10.1016/j.metabol.2003.10.010. [DOI] [PubMed] [Google Scholar]
- 33.Fu Y, Luo N, Lopes-Virella MF. Oxidized LDL induces the expression of ALBP/aP2 mRNA and protein in human THP-1 macrophages. J Lipid Res. 2000 Dec;41(12):2017–23. [PubMed] [Google Scholar]
- 34.Liu QY, Quinet E, Nambi P. Adipocyte fatty acid-binding protein (aP2), a newly identified LXR target gene, is induced by LXR agonists in human THP-1 cells. Mol Cell Biochem. 2007 Aug;302(1-2):203–13. doi: 10.1007/s11010-007-9442-5. [DOI] [PubMed] [Google Scholar]
- 35.Boord JB, Maeda K, Makowski L, Babaev VR, Fazio S, Linton MF, et al. Adipocyte fatty acid-binding protein, aP2, alters late atherosclerotic lesion formation in severe hypercholesterolemia. Arterioscler Thromb Vasc Biol. 2002 Oct 1;22(10):1686–91. doi: 10.1161/01.atv.0000033090.81345.e6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Visser M, Bouter LM, McQuillan GM, Wener MH, Harris TB. Low-grade systemic inflammation in overweight children. Pediatrics. 2001 Jan;107(1):E13. doi: 10.1542/peds.107.1.e13. [DOI] [PubMed] [Google Scholar]


