Abstract
Peptidases play vital roles in physiology through the biosynthesis, degradation, and regulation of peptides. Prolyl endopeptidase-like (PREPL) is a newly described member of the prolyl peptidase family, with significant homology to mammalian prolyl endopeptidase (PEP) and the bacterial peptidase oligopeptidase B (OPDB). The biochemistry and biology of PREPL is of fundamental interest due to this enzyme’s homology to the biomedically important prolyl peptidases and its localization in the central nervous system (CNS). Furthermore, genetic studies of patients suffering from hypotonia-cystinuria syndrome (HCS) have revealed a deletion of a portion of the genome that includes the PREPL gene. HCS symptoms thought to be caused by lack of PREPL include neuromuscular and mild cognitive deficits. A number of complementary approaches, ranging from biochemistry to genetics, will be required to understand the biochemical, cellular, physiological, and pathological mechanisms regulated by PREPL. We are particularly interested in investigating physiological substrates and pathways controlled by PREPL. Here, we use a fluorescence polarization activity-based protein profiling (fluopol-ABPP) assay to discover selective small-molecule inhibitors of PREPL. Fluopol-ABPP is a substrate-free approach that is ideally suited for studying serine hydrolases for which no substrates are known, such as PREPL. After screening over 300,000 compounds using fluopol-ABPP, we employed a number of secondary assays to confirm assay hits and characterize a group of 3-oxo-1-phenyl-2,3,5,6,7,8-hexahydroisoquinoline-4-carbonitrile and 1-alkyl-3-oxo-3,5,6,7-tetrahydro-2H-cyclopenta[c]pyridine-4-carbonitrile PREPL inhibitors that are able to block PREPL activity in cells. Moreover, when administered to mice, 1-isobutyl-3-oxo-3,5,6,7-tetrahydro-2H-cyclopenta[c]pyridine-4-carbonitrile distributes to the brain, indicating that it crosses the blood-brain barrier, and may be useful for in vivo studies. The application of fluopol-ABPP has led to the first reported PREPL inhibitors, and these inhibitors will be of great value in studying the biochemistry of PREPL, and in eventually understanding the link between PREPL and HCS.
Keywords: Prolyl peptidases, activity-based proteomics, fluopol, high-throughput screening, chemical inhibitors, Prolyl endopeptidase-like
Introduction
The prolyl peptidases are a family of biomedically relevant enzymes (Figure 1A) that cleave peptides on the C-terminal side of proline residues.1–3 These enzymes participate in a variety of biological processes ranging from peptide biosynthesis, catabolism, and bioactive peptide regulation. For example, prolyl endopeptidase (PEP) regulates the production of the anti-fibrotic peptide Ac-SDKP.4–6 The prolyl peptidases have also been targeted in drug development. As an example, dipeptidyl peptidase 4 (DPP4) inhibitors form a mechanistically-distinct class of anti-diabetic drugs.2,7–9 These inhibitors slow down the DPP4-mediated inactivation of the insulinotropic peptide glucagon-like peptide 1 (GLP-1)10–12, resulting in raised insulin levels and improved physiological glucose tolerance.2,3,13 These findings have fueled the interest in the biochemistry and physiological functions of the prolyl peptidases.
Figure 1.
PREPL, a mammalian disease-associated peptidase. A) Dendrogram of Mus musculus PREPL, PEP, DPP4, FAP, DPP7, DPP8, DPP9, prolyl carboxypeptidase (PRCP), and aminopeptide hydrolase (APEH). B) Genetic studies of patients with hypotonia-cystinuria syndrome (HCS) revealed a deletion in the genome that includes PREPL. PREPL loss is thought to lead to hypotonia, but the substrates and biochemical pathways regulated by PREPL remain enigmatic.
The most recently discovered member of this family is prolyl endopeptidase-like (PREPL).14–19 Deletions in PREPL and the neighboring gene SLC3A1 have been identified in patients suffering from hypotonia-cystinuria syndrome (HCS).14,17,19 Since prior work has linked SLC3A1 deletion to cystinuria20,21, the data suggest that the loss of PREPL is associated with the low muscle tone (hypotonia) observed in these patients (Figure 1B).17 PREPL is primarily found within the nervous system22, specifically neurons23, which together with the hypotonia phenotype suggests that this peptidase might be involved in neuromuscular function. To date, however, no substrate for PREPL has been identified,16,18 and PREPL has not been shown to cleave any PEP substrates. Our current lack of knowledge about the substrates and pathways regulated by PREPL prevents any insight into the mechanistic connection between PREPL and HCS, despite the strong genetic association.
With no specific PREPL inhibitors and no PREPL knockout mice available, we decided to screen for small-molecule PREPL inhibitors, which would provide a valuable tool for investigating the catalytic functions of this enzyme. The first step in discovering a small-molecule inhibitor for an enzyme is the development of a high quality assay of enzyme activity24. This can be particularly challenging for targets like PREPL that do not have any known substrates. As a member of the serine hydrolase superfamily, however, PREPL has a catalytic serine nucleophile that can be labeled in an activity-dependent manner by fluorophosphonate activity-based probes.16,25 Fortunately, a platform has recently been introduced for high-throughput screening where compounds are assayed for their ability to block the increase in polarization signal observed upon fluorescent activity-based probe labeling of enzymes. This platform, referred to as fluorescence polarization activity-based protein profiling (fluopol-ABPP),26,27 has already been used to identify novel inhibitors for several enzymes from multiple mechanistic classes, including RBBP926, PME-127, GSTO126 and PAD428. Here, we use fluopol-ABPP to discover selective PREPL inhibitors.
Experimental Section
Materials
Fluorophosphonate-rhodamine (FP-Rh)29 and FP-polyethyleneglycol-rhodamine (FP-PEG-Rh)30 were synthesized following previously described protocols. Polyclonal antibodies were generated by Open Biosystems in rabbits using a peptide epitope (EELGLDSTDAFEALKKYLKF) derived from murine PREPL.
Cloning, Expression and Purification of PREPL
The Mus musculus Prepl (mPrepl) gene was PCR amplified from an Open Biosystems clone containing the full-length open reading frame (pCMV_mPrepl, Open Biosystems clone ID: 3585402) using forward primer AAA AGG ATC CCA TGG ATG CAT TTG AAA AAG TGA G and reverse primer AAA AGG TAC CTC AGA ACT TTA GGT ATT TCT TCA GC. The resulting insert was then ligated into the pTrcHisB expression vector (Invitrogen) using the BamHI and KpnI restriction sites. The resulting vector, pTrcHisB_mPrepl, was amplified in Top10 cells, purified, and sequenced to confirm the correct coding sequence.
Expression was carried out in Rosetta 2(DE3)pLysS competent cells (EMD Biosciences), by growing a starter culture overnight, diluting 1:100 into fresh media the next morning, and inducing this culture with 1 mM IPTG at OD 0.5. After 12–15 hours at 37°C, cells were harvested and frozen. The pellets were suspended in 20 mM Na2HPO4, 0.75 M NaCl, pH 7.4 (lysis buffer) with 1% Triton X-100 and lysed by sonication at 4°C. The lysate was centrifuged at 5,000 x g for 10 minutes, whereupon the supernatant was applied to a Ni2+-charged IMAC Sepharose 6 Fast Flow resin (GE Healthcare). The resin was then washed with 20 mM Na2HPO4, 0.75 M NaCl, 30 mM imidazole, pH 7.4 (wash buffer) containing 1% Triton X-100. This was followed by a wash with wash buffer to which no Triton X-100 has been added. After these wash steps, PREPL was eluted from the solid support with 20 mM Na2HPO4, 0.75 M NaCl, 200 mM imidazole, pH 7.4 (elution buffer). The protein was concentrated and exchanged into PBS using Amicon Ultra-15 10 kDa molecular weight cutoff filters (Millipore). The PREPL concentration was determined by Bradford Assay and brought to 2 mg/ml in PBS and 10% glycerol. The protein was then frozen at −80°C. The activity of each enzyme batch was assessed through a labeling reaction with FP-Rh activity-based probe (ABP). The labeled enzyme was run out on an SDS-PAGE gel and analyzed using a Typhoon flatbed scanner (GE Healthcare Life Sciences). The presence of a fluorescent band at approximately 75 kDa, which corresponds to the molecular weight of PREPL, demonstrated that the enzyme batch was active. The gels were then Coomassie stained to determine the overall purity of the sample (see Supporting Information Figure S1 for representative examples of ABPP and Coomassie gels).
To obtain a negative control for the screen, catalytically inactive PREPL(S470A) was prepared by site-directed mutagenesis from the pTrcHisB_mPrepl vector using a QuickChange® site-directed mutagenesis kit (Stratagene). The following primers were used to introduce the mutation: CGC TGA GCG CTT TCG CTG CTG GAG GTG TGC TCG and CGA GCA CAC CTC CAG CAG CGA AAG CGC TCA GCG. The vector was sequenced to confirm the mutation. This construct, pTrcHisB_mPrepl(S470A), was used to express the inactive enzyme. PREPL(S470A) was purified in the same way as the wild-type (WT) enzyme and showed no activity by ABPP.
PREPL fluopol-ABPP Assay and Screening
Using the fluopol-ABPP assay26, an initial screen was carried out against 18,000 compounds from the Maybridge Hitfinder™ Collection and a validation fraction of National Institutes of Health (NIH) Molecular Libraries Small Molecule Repository. Prior to the start of the assay, 10 μL of assay buffer [0.01% Pluronic acid (Invitrogen), 50 mM Tris- HCl pH 8.0, 150 mM NaCl, 1 mM DTT (Invitrogen)] containing PREPL (33 nM, 30 nM final concentration) was dispensed into 384-well microtiter plates. Test compound (50 nL) in DMSO or DMSO alone (0.59% final concentration DMSO; 10 μM final compound concentration for the Maybridge library and 5 μM compound concentration for the NIH validation set) was added to the appropriate wells and incubated for 30 minutes at 25 °C. The assay was initiated by dispensing 1.0 μL FP-PEG-Rh probe (750 nM, 75 nM final concentration) in assay buffer to all wells. The reactions were incubated for 15 minutes at room temperature and read on the Perkin Elmer Envision with the Optimized Bodipy TMR FP Dual Emission Label 2100–8070 consisting of the following filters and Mirror Modules: Bodipy TMR FP Dial Mirror Module (2100–4080), Bodipy TMR FP Excitation Filter (2100–5050), Bodipy TMR FP Emission Filter S-pol (2100–5160) and Bodipy TMR FP Emission Filter P-pol (2100–5170). The percent inhibition for each compound was calculated as follows: Percent inhibition = (Test Compound mP - median NegativeControl mP)/(median PositiveControl mP – median NegativeControl mP) x 100. Test compound was defined as wells containing PREPL in the presence of test compound, negative controls were defined as wells containing PREPL and DMSO, and positive controls were defined as wells containing no PREPL protein. As an initial filter to remove non-selective serine hydrolase inhibitors, we selected compounds that inhibited PREPL >50% relative to control reactions and simultaneously showed <20% inhibition in previous fluopol-ABPP screens against serine hydrolases PME-1 and Cgi67.
MLPCN screen
This initial screen was followed up by a screening the 324,751-compound Molecular Libraries Probe Productions Centers Network (MLPCN) library in collaboration with the MLPCN at the Scripps Research Institute. Prior to the start of the assay, 4.0 μL of Assay Buffer [0.01% Pluronic acid (Invitrogen), 50 mM Tris- HCl pH 8.0, 150 mM NaCl, 1 mM DTT (Invitrogen)] containing 37.5 nM of PREPL was dispensed into 1536-well microtiter plates. Next, 30 nL of test compound in DMSO or DMSO alone (0.59% final concentration DMSO; 5.9 μM compound) was added to the appropriate wells and incubated for 30 minutes at 25 °C. The assay was started by dispensing 1.0 μL of 375 nM FP-PEG-Rh probe in assay buffer to all wells. Plates were centrifuged and after 15 minutes of incubation at 25 °C, fluorescence polarization was read on a Viewlux microplate reader (PerkinElmer) using a BODIPY TMR FP filter set and a BODIPY dichroic mirror (excitation = 525 nm, emission = 598 nm). Fluorescence polarization was read for 15 seconds for each polarization plane (parallel and perpendicular). The well fluorescence polarization value (mP) was obtained via the PerkinElmer Viewlux software. The percent inhibition for each compound was calculated as described above. A mathematical algorithm was used to determine nominally inhibiting compounds in the primary screen. Two values were calculated: (i) the average percent inhibition of all compounds tested, and (ii) three times their standard deviation. The sum of these two values was used as a cutoff parameter (i.e. any compound that exhibited greater inhibition than the cutoff parameter was declared active). The reported PubChem Activity Score (http://pubchem.ncbi.nlm.nih.gov/) has been normalized to 100% observed primary inhibition. Negative percent inhibition values are reported as activity score zero. The activity score range for active compounds is 100–11, for inactive 11–0.
Active compounds in the primary screen (AID 2751, PubChem’s BioAssay identifier) were followed up in a confirmation screen performed in triplicate (AID 2803). Active compounds from this confirmation screen and from the initial 18,000-compound screen were carried forward through a series of secondary assays to establish their ability to inhibit PREPL and their feasibility for use in cells and tissues.
Single concentration ABPP assays as a secondary screen to confirm PREPL inhibition
PREPL (17 μl, 117 nM, final concentration 100 nM) in assay buffer (PBS with 0.012% Pluronic (Invitrogen)) was preincubated with inhibitor (1 μl, 400 μM, final concentration 20 μM) for 15 minutes at room temperature. FP-Rh (2 μl, 1 μM, final concentration 100 nM) was then added and allowed to react for 7 minutes, whereupon the reaction was quenched by the addition of 10 μl SDS-PAGE loading buffer. The samples were run on SDS-PAGE, and visualized by in-gel fluorescence scanning. The percentage activity remaining was determined by measuring the integrated optical intensity of the bands using ImageQuant software. A reduction of the band intensity by more than 20% was taken to indicate that the compound was capable of inhibiting PREPL, confirming the result from the high-throughput screen.
Competitive ABPP Assays in Complex Proteomes
For in vitro experiments, brain proteomes were diluted to 1 mg/mL in PBS (pH 7.4) and spiked with 40 nM final concentration of recombinant PREPL. Pluronic was added to a final concentration of 0.01%. These proteomes were incubated with DMSO or 50 μM inhibitor (final concentration) for 30 minutes at room temperature. FP-Rh was then added at a final concentration of 0.5 μM. After 15 minutes, the reactions were quenched with SDS-PAGE loading buffer, separated by SDS-PAGE (10% acrylamide), and visualized in-gel with a Typhoon flatbed fluorescence scanner (GE Healthcare Life Sciences). Bands were quantified to assess the extent of labeling and determine whether or not a compound was able to inhibit PREPL in a complex proteome.
Determination of IC50 Values
For determination of in vitro IC50 values, compounds were preincubated with PREPL (100 nM final concentration in PBS, pH 7.4, with 0.01% final concentration of Pluronic) at the indicated concentrations for 15 minutes at room temperature. The reactions were performed in triplicate. The samples were then labeled with FP-Rh (100 nM final concentration) for 7 minutes, quenched, separated by SDS-PAGE, and visualized by in-gel fluorescence scanning. The percentage activity remaining was determined by measuring the integrated optical intensity of the bands using ImageQuant software. IC50 values were determined from a dose-response curve generated with Prism (GraphPad Software) using the following equation: Y = Bottom + (Top - Bottom)/(1+10^(X-LogIC50)).
PEP selectivity assay
PEP (0.70 μg/ml) was preincubated with 20 μM inhibitor for 15 minutes in PEP assay buffer (25 mM Na2HPO4, 0.5 mM EDTA, 2 mM DTT). Then, Z-Gly-Pro-AMC (Bachem) in PEP assay buffer was added to a final concentration of 133 μM. The cleavage of this fluorogenic substrate was monitored on a Spectramax Gemini XS Fluorescence plate reader (Molecular Devices), with excitation at 360 nm and emission at 460 nm.
In situ ABPP assays to determine cellular activity of PREPL inhibitors
Neuro2A cells were plated at 1.5 × 106 cells per well in 6-well plates. The next day, cells were transfected with pCMV_mPrepL (Open Biosystems Clone ID: 3585402) using Lipofectamine 2000 transfection reagent (Invitrogen). Two days post-transfection, inhibition and labeling were carried out in the intact cells before harvesting. First, the media covering the cells was removed and replaced with media containing 50 μM inhibitors. Cells were incubated in this media for 1.5 hours. A cell-permeable FP-alkyne probe was then added to the media to a final concentration of 10 μM and let incubate for 1 hour. Cells were washed 3 times with PBS and harvested with a cell scraper. The cell pellet was washed 3 times (by suspending in 1 mL PBS, centrifuging at 1400 x g for 3 minutes and then aspirating off PBS). After the final wash, the cell pellet was suspended in 80 μl PBS. Cells were lysed by 7 freeze-thaw cycles and centrifuged at 4 °C for 30 minutes (20,000 x g). The supernatant was transferred to a new tube and the protein concentration obtained by Bradford Assay. The samples were diluted to 4 mg/ml with PBS. To 43 μl of 4 mg/ml protein sample, 2 μl of 1.25 mM rhodamine-azide, TCEP (1 μl, 50 mM), ligand (3 μl, 1.67 mM) and CuSO4 (1 μl, 50 mM) were added.31,32 This reaction was allowed to proceed for 1 hour at room temperature, then quenched with SDS-PAGE loading buffer. Samples were separated by SDS-PAGE and visualized by in-gel fluorescence scanning. The percentage activity remaining was determined by measuring the integrated optical intensity of the PREPL bands using ImageQuant software.
Animal Studies
The C57BL/6J mice used in this study were either purchased (Jackson Labs, Bar Harbor, ME) or taken from a breeding colony. Animals were kept on a 12-hour light, 12-hour dark schedule and fed ad libitum. For brain collection, animals were first euthanized with CO2, followed by rapid isolation of the brain. The tissue was flash frozen in liquid N2 and stored at −80 °C. All animal care and use procedures were in strict accordance with the standing committee on the use of animals in research and teaching at Harvard University and the National Institute of Health guidelines for the humane treatment of laboratory animals.
In vivo distribution of PREPL inhibitors by LC-MS/MS
1-Isobutyl-3-oxo-2,3,5,6,7,8-hexahydroisoquinoline-4-carbonitrile (8) (20 μg) was delivered to a mouse (20 grams) via intraperitoneal injection (200 μL of 95/5 PBS/DMSO) for a final dose of 1 mg/kg. After 30 minutes, the mouse was sacrificed and the brain was rapidly isolated and flash frozen in liquid nitrogen. The sample was subsequently Dounce homogenized in chloroform: methanol: water (2: 1: 1) and this homogenate was centrifuged to separate organic and aqueous layers.33 The organic layer was isolated and concentrated under a stream of nitrogen. The residue was then dissolved in chloroform for liquid chromatography-tandem mass spectrometry (LC-MS/MS) analysis. Detection of inhibitor 8 was performed using an Agilent 6410 Triple Quad instrument with an electrospray ionization source operating in positive ionization mode. MS analysis was performed in multiple reaction monitoring (MRM) mode, which could specifically detect inhibitor 8 in biological matrices. The capillary voltage was set to 4 kV and the fragmentor voltage was 161 V. A drying gas temperature of 350 °C was used, with a flow rate of 8 L/min and a nebulizer pressure of 35 psi. Data were collected for two MRM transitions from the m/z 217.1 precursor ion: m/z 173.1 and m/z 155.1 with collision energies set to 36 V and 45 V, respectively. Liquid chromatography was performed using a Gemini C18 5 micron 110Å 50 × 4.60 mm column (Phenomenex). Mobile phases A and B were composed of GC-grade water and methanol (Burdick & Jackson), respectively, and both phases contained 0.1% formic acid and 5 mM ammonium formate. The mobile phase composition was held at 5% B with a flow rate of 0.1 mL/min for 5 min, was increased linearly from 5% to 100% B over 40 minutes during which the flow rate was 0.4 mL/min, was held at 100% B for 8 min at 0.5 mL/min, then held at 5% B for 7 min with a flow rate of 0.5 mL/min.
Results and Discussion
Expression and purification of PREPL for high-throughput screening
Murine Prepl (amino acids 1–638) was cloned into the pTrcHisB expression vector and expressed in Rosetta 2(DE3) pLysS competent cells. We found that Rosetta cells34, which are codon optimized for mammalian protein expression, were required for adequate protein expression. Recombinant PREPL was purified using standard immobilized metal affinity chromatography methods and analyzed by SDS-PAGE, followed by Coomassie staining, to assess the purity of the protein, which was typically >90% (Supporting Information Figure S1). FP-Rh labeling of recombinant PREPL was used to test the activity of the enzyme (Supporting Information Figure S1). Together, these data enabled us to conclude that the PREPL was folded and active, permitting the optimization of the fluopol-ABPP assay necessary for screening.
To optimize the fluopol-ABPP assay properly, it was necessary to generate a catalytically inactive mutant of PREPL. To accomplish this, we replaced the PREPL active site serine at position 47014,16 with an alanine by site-directed mutagenesis. PREPL(S470A) was expressed and purified in the same manner as the wild-type (WT) protein. This protein did not, however, react with FP-Rh, indicating that it was no longer catalytically active. Together, PREPL and PREPL(S470A) provided us with the necessary reagents for assay development and screening.
Optimization of the fluopol-ABPP assay
The fluopol-ABPP assay26,27 takes advantage of the fluorophosphonate labeling of serine hydrolases, in this case a candidate serine peptidase, to enable the substrate-free identification of enzyme inhibitors. In this assay, a purified enzyme and a compound are dispensed into the wells of a 384- or 1536-well microtiter plate. The assay is then initiated by the introduction of a fluorescent ABP, typically FP-Rh29, into each well. In the presence of inactive compounds, the enzyme is labeled with FP-Rh, resulting in a large increase in the fluorescence polarization signal, since the tumbling rate of a protein-bound fluorescent ABP is lower than that of an unbound ABP. An inhibitor, however, will block the interaction between the ABP and the enzyme, leading to a lower fluorescence polarization signal and the identification of a “hit” compound (Figure 2A). In this way, the fluopol-ABPP assay can be used to screen for inhibitors of probe-sensitive targets in the absence of a known natural substrate, which was essential in the case of PREPL.
Figure 2.
Fluopol-ABPP assay for the discovery of selective PREPL inhibitors. A) Recombinantly expressed and purified PREPL is added, along with a single compound, to individual wells in a microtiter plate. The FP-PEG-Rh ABP, which covalently labels serine hydrolases like PREPL, is then added to each sample. The presence of an inhibitor reduces PREPL labeling by FP-PEG-Rh, leading to a weaker fluorescence polarization signal (top). Inactive compounds will not reduce enzyme labeling with FP-PEG-Rh, which results in a much stronger fluorescence polarization signal. The assay can be used to identify active compounds during a high-throughput screen. B) The fluorescence polarization signal (mP) of PREPL(WT), PREPL(S470A), and no enzyme control with respect to time. C) Calculation of the Z-factor by comparing PREPL(WT) and PREPL(S470A) fluorescence polarization signals at different timepoints.
In order to detect reversible as well as irreversible inhibitors of PREPL in the fluopol-ABPP screen, it was necessary to identify a kinetically relevant timepoint for this particular enzyme. In a competitive labeling reaction, an irreversible inhibitor such as FP-Rh will always outcompete a reversible inhibitor given enough time. Therefore, it is crucial to pick conditions (timepoint, enzyme concentration, labeling reagent) where the enzyme is incompletely labeled and the effect of a reversible inhibitor can be detected.26,35 In addition, the Z-factor at the selected timepoint should be suitable for high-throughput screening (typically > 0.5)36.
The conditions for the PREPL fluopol-ABPP assay were established by testing various parameters. For example, several different concentrations of enzyme (5 μM, 1 μM, 0.5 μM) were incubated with 75 nM FP-Rh and labeling assessed at different timepoints. In all cases, however, complete labeling was observed already by 5 minutes (Supporting Information Figure S2). To slow the reaction down, we decided to use a different probe, FP-PEG-Rh, where a PEG linker connects the fluorophosphonate reactive group with the rhodamine dye (Supporting Information Figure S3). This probe modification often leads to slower reactions with serine hydrolases.30 With the FP-PEG-Rh probe, a time-dependent increase in fluorescence polarization signal was observed as the probe reacted with PREPL. Importantly, wells containing catalytically inactive PREPL(S470A) mutant, where the active site serine had been replaced with an alanine, or wells without enzyme showed no increase in fluorescence polarization upon addition of the FP-PEG-Rh probe (Figure 2B). For HTS analysis, we selected the kinetically relevant timepoint at 15 minutes (Figure 2B) where PREPL generated a high Z-factor (0.83) relative to control reactions (Figure 2C).
Screening for inhibitors of PREPL
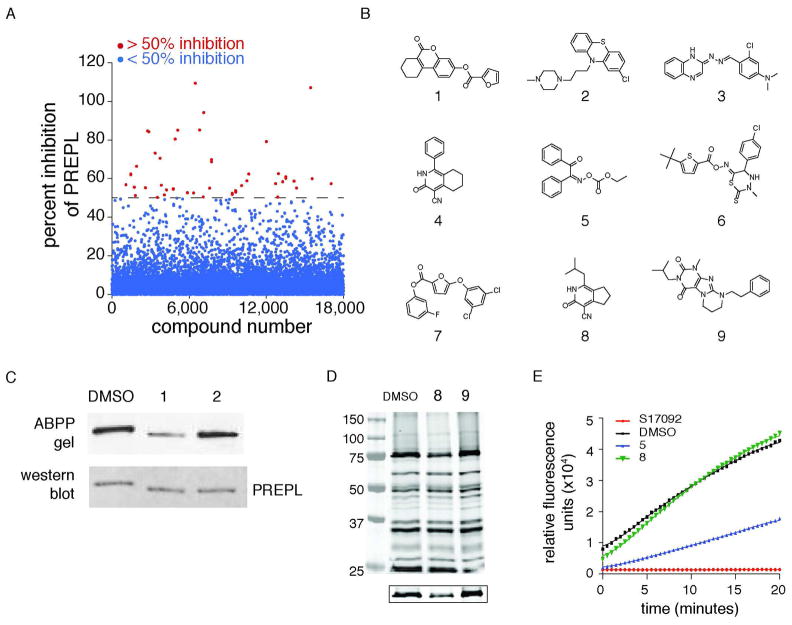
Prior to embarking on an HTS screen of a large (>100,000) compound library, we first wanted to demonstrate that our screening strategy could discover bona fide PREPL inhibitors. Therefore, we performed an initial screen against a pilot library containing 18,000 compounds. A total of 39 compounds were found that inhibited fluorescence polarization in the PREPL screen by at least 50% and simultaneously did not inhibit fluorescence polarization by more than 20% in two previously performed screens against the Maybridge Library, FAM108B (CGI67) and protein phosphatase methylesterase 1 (PME-1)27 (Figure 3A and Supporting Information Table S1, http://pubchem.ncbi.nlm.nih.gov/). Comparison of candidate PREPL inhibitors to hits in these other screens is intended to filter out promiscuous serine hydrolase inhibitors that are active against several enzymes.
Figure 3.
Identification of selective PREPL inhibitors. A) Screening 18,000 compounds using the fluopol-ABPP assay identified 39 candidate inhibitors with greater than 50% PREPL inhibition and less than 20% inhibition in two other serine hydrolase fluopol-ABPP screens (Cgi67 and PME-1). In the MLPCN screen, 725 such inhibitors were identified (this number was narrowed down to 556 by removing ester-containing compounds). B) Some representative hits from the fluopol-ABPP screen. C) Gel-based ABPP assays were then used to confirm PREPL inhibition by some of these compounds. D) A gel-based competitive ABPP with PREPL-spiked mouse brain lysates, inhibitors are assessed for their ability to selectively inhibit PREPL in a complex proteome. The PREPL band is located at 75kDa and shown in more detail below the main gel. E) Candidate inhibitors were also screened for crossreactivity against the highly related peptidase PEP. Examples of crossreactive (5) and non-crossreactive (8) compounds are shown. The control for 100% PEP inhibition, S17092, is a specific PEP inhibitor.
In order to identify additional scaffolds that could serve as selective PREPL inhibitors, we proceeded to screen PREPL against the MLPCN library, which contained 324,751 compounds at the time of the screen. The screen was performed essentially as described for the initial 18,000-compound library screen, with some minor modifications (see Experimental). This fluopol-ABPP screen initially identified 2221 (AID 2751) compounds that were active against PREPL. Retesting these compounds in triplicate against PREPL (AID 2803) removed a number of false positives and resulted in a total of 1333 hits.
To filter out compounds that were clearly not selective, the list was then narrowed down to compounds which inhibited PREPL by more than 50% and simultaneously did not show more than 20% inhibition in previous fluopol-ABPP screens against three other serine hydrolases (RBBP9, AID 1515; PME-1, AID 2130; LYPLA2, AID 2177) and a thiol-reactive glutathione S-transferase (GSTO1, AID 1974). This step removed an additional 608 molecules to afford 725 compounds. Finally, the list was narrowed down to 556 inhibitors by removing compounds containing activated esters, which we suspected would be unstable in cells or tissues (Supporting Information Table S2).
Secondary assays to validate the substrate confirmation workflow
Of the 595 compounds identified in the 18,000- and 324,751-compound screens, 590 were commercially available and purchased. These hits were subsequently confirmed using a competitive gel-based ABPP approach against purified, recombinant PREPL (Figure 3C).35 In this assay, recombinant PREPL was incubated with inhibitors at 20 μM or vehicle for 15 minutes, then treated with FP-Rh for 7 minutes prior to quenching with SDS-PAGE loading buffer. The samples were analyzed by SDS-PAGE and the activity of each compound determined based on the intensity of the PREPL band. This assay confirmed a number of compounds from both screens (Figure 3B) as efficient inhibitors of PREPL. We also made sure that these inhibitors were general inhibitors for the enzyme by testing the poorer PREPL substrate 4-Methylumbelliferyl-p-guanidinobenzoate (MUGB). Compound 8 also inhibited breakdown of MUGB by PREPL to indicate that these inhibitors are binding to and specifically inhibiting PREPL (Supporting Information Figure S4).
The identification of selective PREPL inhibitors
These candidate inhibitors were then assayed for ability to inhibit PREPL in a complex proteome using gel-based ABPP with FP-Rh (Figure 3D). By visualizing the labeling of all brain serine hydrolases simultaneously, these assays also enabled us to assess the selectivity of candidate inhibitors for PREPL over many mechanistically-related enzymes. Selective inhibitors should specifically reduce the intensity of the PREPL band with no effect on the signal intensity of any of the other labeling events in the proteome. There were no promiscuous inhibitors identified in this assay, however, several of the candidate inhibitors did not inhibit PREPL in the whole proteome and were excluded from future studies.
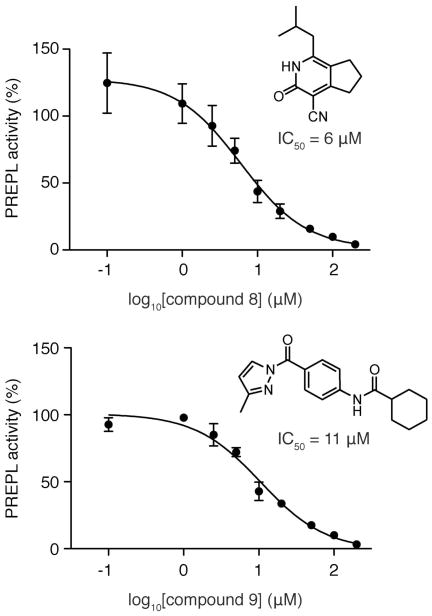
As further confirmation of selectivity, candidate inhibitors were also directly tested against the PREPL homolog PEP (Figure 1A) using a well-established fluorogenic PEP substrate assay with Z-Gly-Pro-AMC (Figure 3E).37 We considered inhibitors exhibiting >20% reduction in PEP activity as cross-reactive and discarded these compounds from future studies. The most promising inhibitors after these selectivity assays were then tested at different concentrations against FP-Rh to determine the IC50 values, which were typically in the low μM range (Figure 4). Together, these assays identified twelve selective PREPL inhibitors (compounds 4, 6–8, 10–18) (Figure 3B and Supporting Information Table S3).
Figure 4.
IC50 data for two of the representative inhibitors identified from the MLPCN screen that were selective for PREPL (over PEP), and were able to inhibit PREPL in a complex proteome (brain). The IC50 values of these compounds were determined using a gel-based ABPP approach and calculated using the Prism software (95% confidence interval for compound 8 is 3–12 μM and for compound 9 is 7–16 μM).
Cell-based ABPP assays
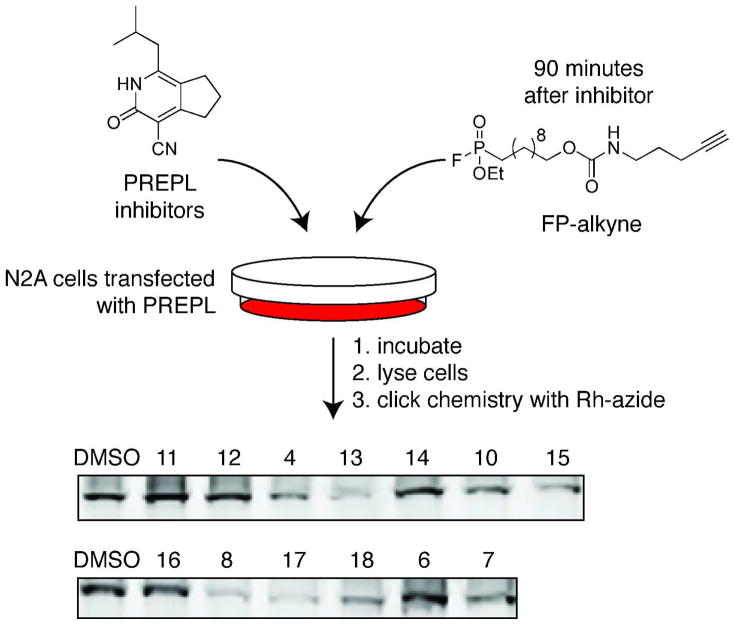
Having found inhibitors selective for PREPL in vitro, we wanted to explore the activity of these inhibitors in cells. To be able to detect reversible as well as irreversible inhibitors in this assay, we used a click chemistry-based approach where PREPL-transfected Neuro2A cells were first incubated with inhibitor, followed by addition of a cell-permeable FP-alkyne to the media.31,32,38 After washing and harvesting the cells, the labeled proteome was reacted with rhodamine-azide (Rh-N3) and separated by SDS-PAGE gel. The extent of inhibition was determined from the reduction in fluorescence of the PREPL band in samples containing inhibitor relative to vehicle-treated samples (Figure 5, Supporting Information Figure S5). All of the carbonitrile compounds (4, 8, 13, 17, 18) were able to cross the cell membrane and inhibit PREPL intracellularly, while the other inhibitors do not display significant inhibition of PREPL in situ. Compounds that were inactive in this assay were either unable to enter cells or may have been unstable (i.e. prone to hydrolysis) in intact cells (Supporting Information Figure S5). Together with the prior selectivity assays, these experiments revealed that inhibitors 4, 8, 13, 17, and 18 are all excellent tools for studying PREPL activity in cells.
Figure 5.
Inhibition of PREPL in cells was determined with a competitive ABPP assay using the cell-permeable FP-alkyne ABP. Inhibitors were added to the media of PREPL-transfected Neuro2A cells at 50μM final concentration, followed by the addition of the FP-alkyne probe. Inhibitors that are active in situ will reduce labeling by the activity-based probe, resulting in a lighter band on the gel, as seen for compounds 8 and 17, for example.
CNS delivery of 1-isobutyl-3-oxo-3,5,6,7-tetrahydro-2H-cyclopenta[c]pyridine-4-carbonitrile (8)
We selected 1-isobutyl-3-oxo-3,5,6,7-tetrahydro-2H-cyclopenta[c]pyridine-4-carbonitrile (8), the most potent carbonitrile inhibitor in the brain proteome ABPP assay, and tested whether this compound could cross the blood-brain barrier in mice.39 Thirty minutes after an intraperitoneal injection of inhibitor 8, the mouse was sacrificed and the brain collected and frozen. The sample was subsequently extracted and analyzed to determine whether compound 8 was present in the brain. The analysis was performed using the highly selective multiple reaction monitoring (MRM) detection method on a triple quadrupole (QQQ) LC-MS system,40 which relied on characteristic transitions that occurred during the fragmentation of inhibitor 8 (Supporting Information Figure S6). We saw a strong signal for compound 8 in the inhibitor-treated mice, while there was no signal in the untreated control, as expected (Figure 6 and Supporting Information Figure S7). Since inhibitor 8 had already been shown to selectively inhibit PREPL in brain proteomes, this data suggests that we may be able to use this inhibitor to study PREPL in vivo. Physiological experiments will require the optimization of the dosing, but the discovery of a selective and bioavailable inhibitor is a promising first step.
Figure 6.
Compound 8 is able to cross the blood-brain barrier. Brain extracts from mice treated with 8 and untreated mice were analyzed on a triple quadrupole mass spectrometer in the multiple reaction monitoring (MRM) mode. Two characteristic transitions for compound 8 (m/z 217.1-to-173.1 and m/z 217.1-to-155.1) were monitored and chromatograms are shown for these transitions for mice treated with 8 and untreated mice.
Proposed mechanism of inhibition for identified PREPL inhibitors
The fluopol-ABPP screen and subsequent selectivity experiments resulted in a set of selective PREPL inhibitors (Supporting Information Tables S3 and S4). Mechanistically, some of these compounds may be inhibiting PREPL by reacting or binding to the nucleophilic serine (Ser470) in the PREPL active site (Figure 7). Carbonitriles have emerged as a potent chemotype in the design of DPP4 inhibitors.41–43 For example, saxagliptin (BMS-477118), a FDA-approved DPP4 inhibitor, contains a carbonitrile.41 Structural studies with carbonitrile-containing compounds demonstrate that the carbonitrile is electrophilic, and reacts with the serine nucleophile to form a covalent imidate adduct.43 We propose that the carbonitrile compounds identified in our screen as PREPL inhibitors likely operate through an analogous mechanism. In this model, Ser470 attacks the carbonitrile to form a covalent imidate adduct (Figure 7A). Importantly, crystallographic studies have shown that such covalent imidate adducts can be removed from the active site by soaking in a non-covalent binder, which demonstrates that these carbonitriles are covalent-reversible inhibitors.43
Figure 7.
Potential mechanisms of PREPL inhibition by the different inhibitor chemotypes. A) In analogy to known DPP4 inhibitors9,41, we suspect that the carbonitrile inhibitors are likely functioning through the reversible attack of Ser470 of PREPL to form an imidate adduct. B) Similarly, the carbonyl groups (ester and oxime esters) are electrophilic and can react with PREPL to form an acyl-enzyme intermediate, which are likely removed by hydrolysis of the acyl-enzyme intermediate to regenerate PREPL.
In the second group of inhibitors we identified, compounds possess electrophilic acyl groups that could react with Ser470 to form acyl-enzyme intermediates. Two of the compounds from the initial screen are activated esters, a 3-fluorophenol ester (7) and an oxime ester (6). The oxime ester functional group has recently been shown to be a covalent inhibitor of serine hydrolases, and the acyl-enzyme intermediate has been confirmed by mass spectrometry.44 In our case, compounds 6 and 7 likely interact with PREPL through the nucleophilic addition of Ser470 to the electrophilic carbonyl of these inhibitors to form an acyl-enzyme intermediate (Figure 7B). This group can then be removed through subsequent hydrolysis of the acyl-enzyme intermediate with water (Figure 7B).45 It is also likely that the acyl pyrazole group in compound 9 is electrophilic enough to react with an activated serine nucleophile. Studies with organic compounds have demonstrated that alcohols and amines are able to hydrolyze acyl pyrazoles46–48, but no studies with macromolecules, such as serine hydrolases, have to our knowledge been reported. While the available evidence supports the inhibition mechanisms proposed above, these will need to be verified through additional mechanistic and structural studies.
Comparison of inhibitors to identify structural elements necessary for PREPL inhibition
At first glance, the structures of the PREPL inhibitors seemed remarkably different (Figure 3B and Figure 8). To gain insight into features that may favor PREPL inhibition, we analyzed the relative activities of the carbonitrile compounds in the MLPCN screen. We compared the percent inhibition of carbonitriles that differed by a single R group in the 1 position (Figure 8A). The data for methyl (22%), ethyl (30%), propyl (55%), and isopropyl (74%) groups clearly showed a trend that PREPL inhibition correlated with increased hydrophobicity (and branching) at the 1 position. Based on this data, we propose a model for the key elements necessary for PREPL inhibition. In our model, the structural features shared by PREPL inhibitors include an electrophilic carbonyl or carbonitrile (red), an aromatic ring (green), and a bulky hydrophobic group (blue) (Figure 8B). These hypotheses will need to be confirmed by targeted modification of PREPL at the critical sites in future structure-activity relationship studies.
Figure 8.
Analysis of the common structural features found among different inhibitor classes. A) Comparison of the percent inhibition of different carbonitrile derivatives in the original fluopol-ABPP screen demonstrates that increased hydrophobicity opposite from the carbonitrile group on the aromatic ring leads to improved inhibition. B) Comparison of the different inhibitor classes indicate that the three key elements in PREPL inhibitors identified in this study are an electrophilic carbonyl or carbonitrile (red), aromatic ring (green), and a hydrophobic group (blue).
Conclusions
This study identified the first set of PREPL inhibitors, and these compounds will find immediate use as chemical probes to investigate the biochemistry and biology of this enzyme. In addition, this study highlights the distinct advantage of the fluopol-ABPP methodology, which enabled the identification of chemical inhibitors of PREPL using a substrate-free format. The ability to screen for inhibitors without needing substrates is of paramount importance when studying novel and uncharacterized enzymes, such as in the case of the mammalian peptidase PREPL, which is the first reported use of the fluopol-ABPP assay to identify peptidase inhibitors. We anticipate that these compounds will be useful in structural studies of PREPL, by helping to define key elements of the PREPL active site. Furthermore, we specifically plan to use these inhibitors to study the biochemical function of PREPL by coupling inhibition of this enzyme to mass spectrometry approaches for substrate discovery that we have developed.6,33,49,50 In doing so, these inhibitors will prove to be useful chemical probes in further exploring the biochemistry of this uncharacterized peptidase, and might eventually help reveal the connection between PREPL and HCS.
Supplementary Material
Acknowledgments
We thank Pierre Baillargeon, Lina DeLuca (Lead Identification Division, Scripps Florida) for compound management. This work was supported by a Forris Jewitt Moore Fellowship sponsored by Amherst College (A.M.L.), an NIH training grant (GM007598) (A.M.L), Searle Scholar Award (A.S.), Burroughs Wellcome Fund Career Award in the Biomedical Sciences (A.S.), National Institutes of Health Grants 1DP2OD002374 (A.S.), CA132630 (B.F.C.), the National Science Foundation (predoctoral fellowships to D.A.B.), the California Breast Cancer Research Program (predoctoral fellowship to D.A.B.), and The Skaggs Institute for Chemical Biology. The efforts the National Institutes of Health Molecular Libraries Probe Production Center Network (MH084512) supported the research efforts of the above authors.
Footnotes
Supporting Information Available. Please refer to the supporting information for experimental results for PrepL expression and purification, comparison of labeling kinetics for FP-Rh and FP-PEG-Rh, structures of these probes, full gels for the in situ inhibition experiment, mass spectrometry data for characterization and quantitation of compound 8 inhibition with compound 8 using an alternative assay, full lists of hits from the two screens, further data on the activities of the carbonitrile compounds and a complete list of inhibitors described in this study. Complete references 7, 8, 41 and 43 can also be found in the supporting information. This information is available free of charge via the Internet at http://pubs.acs.org/.
References
- 1.Rosenblum JS, Kozarich JW. Curr Opin Chem Biol. 2003;7:496. doi: 10.1016/s1367-5931(03)00084-x. [DOI] [PubMed] [Google Scholar]
- 2.Thornberry NA, Weber AE. Curr Top Med Chem. 2007;7:557. doi: 10.2174/156802607780091028. [DOI] [PubMed] [Google Scholar]
- 3.Marguet D, Baggio L, Kobayashi T, Bernard AM, Pierres M, Nielsen PF, Ribel U, Watanabe T, Drucker DJ, Wagtmann N. Proc Natl Acad Sci U S A. 2000;97:6874. doi: 10.1073/pnas.120069197. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Cavasin MA, Liao TD, Yang XP, Yang JJ, Carretero OA. Hypertension. 2007;50:130. doi: 10.1161/HYPERTENSIONAHA.106.084103. [DOI] [PubMed] [Google Scholar]
- 5.Cavasin MA, Rhaleb NE, Yang XP, Carretero OA. Hypertension. 2004;43:1140. doi: 10.1161/01.HYP.0000126172.01673.84. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Nolte WM, Tagore DM, Lane WS, Saghatelian A. Biochemistry. 2009;48:11971. doi: 10.1021/bi901637c. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Edmondson SD, et al. J Med Chem. 2006;49:3614. doi: 10.1021/jm060015t. [DOI] [PubMed] [Google Scholar]
- 8.Lankas GR, et al. Diabetes. 2005;54:2988. doi: 10.2337/diabetes.54.10.2988. [DOI] [PubMed] [Google Scholar]
- 9.Villhauer EB, Brinkman JA, Naderi GB, Burkey BF, Dunning BE, Prasad K, Mangold BL, Russell ME, Hughes TE. J Med Chem. 2003;46:2774. doi: 10.1021/jm030091l. [DOI] [PubMed] [Google Scholar]
- 10.Kreymann B, Williams G, Ghatei MA, Bloom SR. Lancet. 1987;2:1300. doi: 10.1016/s0140-6736(87)91194-9. [DOI] [PubMed] [Google Scholar]
- 11.Turton MD, O’Shea D, Gunn I, Beak SA, Edwards CM, Meeran K, Choi SJ, Taylor GM, Heath MM, Lambert PD, Wilding JP, Smith DM, Ghatei MA, Herbert J, Bloom SR. Nature. 1996;379:69. doi: 10.1038/379069a0. [DOI] [PubMed] [Google Scholar]
- 12.Doyle ME, Egan JM. Recent progress in hormone research. 2001;56:377. doi: 10.1210/rp.56.1.377. [DOI] [PubMed] [Google Scholar]
- 13.Weber AE. Journal of medicinal chemistry. 2004;47:4135. doi: 10.1021/jm030628v. [DOI] [PubMed] [Google Scholar]
- 14.Jaeken J, Martens K, Francois I, Eyskens F, Lecointre C, Derua R, Meulemans S, Slootstra JW, Waelkens E, de Zegher F, Creemers JW, Matthijs G. Am J Hum Genet. 2006;78:38. doi: 10.1086/498852. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Martens K, Derua R, Meulemans S, Waelkens E, Jaeken J, Matthijs G, Creemers JW. Biol Chem. 2006;387:879. doi: 10.1515/BC.2006.111. [DOI] [PubMed] [Google Scholar]
- 16.Szeltner Z, Alshafee I, Juhasz T, Parvari R, Polgar L. Cellular and molecular life sciences : CMLS. 2005;62:2376. doi: 10.1007/s00018-005-5262-5. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Chabrol B, Martens K, Meulemans S, Cano A, Jaeken J, Matthijs G, Creemers JW. Journal of medical genetics. 2008;45:314. doi: 10.1136/jmg.2007.055475. [DOI] [PubMed] [Google Scholar]
- 18.Boonen K, Regal L, Jaeken J, Creemers JW. CNS & neurological disorders drug targets. 2011 doi: 10.2174/187152711794653760. [DOI] [PubMed] [Google Scholar]
- 19.Martens K, Heulens I, Meulemans S, Zaffanello M, Tilstra D, Hes FJ, Rooman R, Francois I, de Zegher F, Jaeken J, Matthijs G, Creemers JW. European journal of human genetics : EJHG. 2007;15:1029. doi: 10.1038/sj.ejhg.5201881. [DOI] [PubMed] [Google Scholar]
- 20.Endsley JK, Phillips JA, 3rd, Hruska KA, Denneberg T, Carlson J, George AL., Jr Kidney international. 1997;51:1893. doi: 10.1038/ki.1997.258. [DOI] [PubMed] [Google Scholar]
- 21.Lee WS, Wells RG, Sabbag RV, Mohandas TK, Hediger MA. The Journal of clinical investigation. 1993;91:1959. doi: 10.1172/JCI116415. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Huang CC, Chang WS. BMC molecular biology. 2009;10:67. doi: 10.1186/1471-2199-10-67. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Oldham MC, Konopka G, Iwamoto K, Langfelder P, Kato T, Horvath S, Geschwind DH. Nature neuroscience. 2008;11:1271. doi: 10.1038/nn.2207. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Moss ML, Rasmussen FH. Analytical biochemistry. 2007;366:144. doi: 10.1016/j.ab.2007.04.043. [DOI] [PubMed] [Google Scholar]
- 25.Bachovchin DA, Ji T, Li W, Simon GM, Blankman JL, Adibekian A, Hoover H, Niessen S, Cravatt BF. Proceedings of the National Academy of Sciences of the United States of America. 2010;107:20941. doi: 10.1073/pnas.1011663107. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Bachovchin DA, Brown SJ, Rosen H, Cravatt BF. Nature biotechnology. 2009;27:387. doi: 10.1038/nbt.1531. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Bachovchin DA, Mohr JT, Speers AE, Wang C, Berlin JM, Spicer TP, Fernandez-Vega V, Chase P, Hodder PS, Schurer SC, Nomura DK, Rosen H, Fu GC, Cravatt BF. Proceedings of the National Academy of Sciences of the United States of America. 2011. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Knuckley B, Jones JE, Bachovchin DA, Slack J, Causey CP, Brown SJ, Rosen H, Cravatt BF, Thompson PR. Chem Commun (Camb) 2010;46:7175. doi: 10.1039/c0cc02634d. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Patricelli MP, Giang DK, Stamp LM, Burbaum JJ. Proteomics. 2001;1:1067. doi: 10.1002/1615-9861(200109)1:9<1067::AID-PROT1067>3.0.CO;2-4. [DOI] [PubMed] [Google Scholar]
- 30.Kidd D, Liu Y, Cravatt BF. Biochemistry. 2001;40:4005. doi: 10.1021/bi002579j. [DOI] [PubMed] [Google Scholar]
- 31.Speers AE, Adam GC, Cravatt BF. Journal of the American Chemical Society. 2003;125:4686. doi: 10.1021/ja034490h. [DOI] [PubMed] [Google Scholar]
- 32.Speers AE, Cravatt BF. Chemistry & biology. 2004;11:535. doi: 10.1016/j.chembiol.2004.03.012. [DOI] [PubMed] [Google Scholar]
- 33.Saghatelian A, Trauger SA, Want EJ, Hawkins EG, Siuzdak G, Cravatt BF. Biochemistry. 2004;43:14332. doi: 10.1021/bi0480335. [DOI] [PubMed] [Google Scholar]
- 34.Choi AH, Basu M, McNeal MM, Bean JA, Clements JD, Ward RL. Protein expression and purification. 2004;38:205. doi: 10.1016/j.pep.2004.08.009. [DOI] [PubMed] [Google Scholar]
- 35.Leung D, Hardouin C, Boger DL, Cravatt BF. Nature biotechnology. 2003;21:687. doi: 10.1038/nbt826. [DOI] [PubMed] [Google Scholar]
- 36.Zhang JH, Chung TD, Oldenburg KR. Journal of biomolecular screening : the official journal of the Society for Biomolecular Screening. 1999;4:67. doi: 10.1177/108705719900400206. [DOI] [PubMed] [Google Scholar]
- 37.Yoshimoto T, Ogita K, Walter R, Koida M, Tsuru D. Biochimica et biophysica acta. 1979;569:184. doi: 10.1016/0005-2744(79)90053-6. [DOI] [PubMed] [Google Scholar]
- 38.Tully SE, Cravatt BF. Journal of the American Chemical Society. 2010;132:3264. doi: 10.1021/ja1000505. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Schinkel AH, Smit JJ, van Tellingen O, Beijnen JH, Wagenaar E, van Deemter L, Mol CA, van der Valk MA, Robanus-Maandag EC, te Riele HP, et al. Cell. 1994;77:491. doi: 10.1016/0092-8674(94)90212-7. [DOI] [PubMed] [Google Scholar]
- 40.Kitteringham NR, Jenkins RE, Lane CS, Elliott VL, Park BK. Journal of chromatography B, Analytical technologies in the biomedical and life sciences. 2009;877:1229. doi: 10.1016/j.jchromb.2008.11.013. [DOI] [PubMed] [Google Scholar]
- 41.Augeri DJ, et al. Journal of medicinal chemistry. 2005;48:5025. doi: 10.1021/jm050261p. [DOI] [PubMed] [Google Scholar]
- 42.Villhauer EB, Brinkman JA, Naderi GB, Burkey BF, Dnning BE, Prasad K, Mangold BL, Russell ME, Hughes TE. Journal of medicinal chemistry. 2003;46:2774. doi: 10.1021/jm030091l. [DOI] [PubMed] [Google Scholar]
- 43.Longenecker KL, et al. Biochemistry. 2006;45:7474. doi: 10.1021/bi060184f. [DOI] [PubMed] [Google Scholar]
- 44.Bachovchin DA, Wolfe MR, Masuda K, Brown SJ, Spicer TP, Fernandez-Vega V, Chase P, Hodder PS, Rosen H, Cravatt BF. Bioorganic & medicinal chemistry letters. 2010;20:2254. doi: 10.1016/j.bmcl.2010.02.011. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Kraut J. Annual review of biochemistry. 1977;46:331. doi: 10.1146/annurev.bi.46.070177.001555. [DOI] [PubMed] [Google Scholar]
- 46.Bernatowicz MS, Wu Y, Matsueda GR. The Journal of Organic Chemistry. 1992;57:2497. [Google Scholar]
- 47.Johnson AL, Sweetser PB. The Journal of Organic Chemistry. 1976;41:110. [Google Scholar]
- 48.Jursic BS, Zdravkovski Z. Journal of Molecular Structure: THEOCHEM. 1994;303:177. [Google Scholar]
- 49.Tagore DM, Nolte WM, Neveu JM, Rangel R, Guzman-Rojas L, Pasqualini R, Arap W, Lane WS, Saghatelian A. Nat Chem Biol. 2009;5:23. doi: 10.1038/nchembio.126. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Tinoco AD, Tagore DM, Saghatelian A. J Am Chem Soc. 2010;132:3819. doi: 10.1021/ja909524e. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.