Abstract
Fanconi D2 (FANCD2) is monoubiquitinated on K561 (FANCD2-Ub) in response to DNA double-strand breaks (DSBs) to stimulate repair of these potentially lethal DNA lesions. FANCD2-Ub was upregulated in CD34+ chronic myeloid leukemia (CML) cells and in BCR-ABL1 kinase –positive cell lines in response to elevated levels of reactive oxygen species (ROS) and DNA cross-linking agent mitomycin C. Downregulation of FANCD2 and inhibition of FANCD2-Ub reduced the clonogenic potential of CD34+ CML cells and delayed BCR-ABL1 leukemogenesis in mice. Retarded proliferation of BCR-ABL1 -positive FANCD2−/− leukemia cells could be rescued by FANCD2 expression. BCR-ABL1 –positive FANCD2−/− cells accumulated more ROS-induced DSBs in comparison to BCR-ABL1 –positive FANCD2+/+ cells. Antioxidants diminished the number of DSBs and enhanced proliferation of BCR-ABL1 –positive FANCD2−/− cells. Expression of wild-type FANCD2 and FANCD2(S222A) phosphorylation-defective mutant (deficient in stimulation of intra-S phase checkpoint but proficient in DSB repair), but not FANCD2(K561R) monoubiquitination-defective mutant (proficient in stimulation of intra-S phase checkpoint but deficient in DSB repair) reduced the number of DSBs and facilitated proliferation of BCR-ABL1 –positive FANCD2−/− cells. We hypothesize that FANCD2-Ub plays an important role in BCR-ABL1 leukemogenesis due to its ability to facilitate the repair of numerous ROS-induced DSBs.
Keywords: BCR-ABL1, FANCD2-Ub, CML, transformation, DNA damage
Introduction
Chronic myeloid leukemia in chronic phase (CML-CP) and a cohort of acute lymphoblastic leukemia (ALL) are characterized by the presence of the Philadelphia chromosome (Ph) that results from a (9;22)(q34;q11) reciprocal translocation that juxtaposes a portion of the c-abl oncogene 1 (ABL1) gene on chromosome 9 with a fragment of the breakpoint cluster region (BCR) gene on chromosome 22, generating the BCR-ABL1 fusion oncogene 1. BCR-ABL1 kinase stimulates oxidative DNA damage such as 8-oxoguanine (8-oxoG) and increases the number of DNA double-strand breaks (DSBs) in CML-CP cells 2–4. In addition, BCR-ABL1-mediated stimulation of activation-induced cytidine deaminase (AID) is responsible for elevation of DNA damage in Ph-positive ALL and in CML lymphoblastic phase 5, 6. These potentially lethal DNA lesions may present a significant obstacle for leukemia cell survival. Thus, repair of the elevated levels of DNA lesions may play an essential role in survival of CML cells. In support of this hypothesis, it has been reported that cells undergo DNA damage-dependent apoptotic crisis during v-ABL1-mediated transformation 7. Similar mechanisms may operate in BCR-ABL1-induced leukemias allowing only survival of leukemia cells which can repair numerous DNA lesions 8.
BCR-ABL1 stimulates three major mechanisms to repair numerous DNA double-strand breaks (DSBs) in leukemia cells: homologous recombination repair (HRR), non-homologous end-joining (NHEJ), and single-strand annealing (SSA) 2, 4, 9–11. Among the various DNA repair proteins, Fanconi anemia D2 (FANCD2) has the unique ability to affect all of these DSB repair pathways 12–14. Accordingly, FANCD2 may play an important role in BCR-ABL1 –positive leukemias.
Inactivating mutations in Fanconi anemia (FA) genes cause congenital disorders characterized by chromosomal instability, hypersensitivity to DNA cross-linking agents, and increased predisposition to cancer 15. Thirteen FA genes have been identified and cloned (A, B, C, D1[BRCA2], D2, E, F, G, I, J, L, M and N); these gene products cooperate in a common cellular pathway to protect the genome from DNA damage 16. Following DNA damage, key events in the FA pathway are phosphorylation and monoubiquitination of FANCI and FANCD2, which form a heterodimer co-localizing in nuclear foci considered to be DNA repair structures 17–20.
FANCD2 is monoubiquitinated on K561 (FANCD2-Ub) after DNA damage induced by cross-linking agents, ionizing radiation and endogenous stresses such as reactive oxygen species (ROS) 16, 21, 22. FANCD2-Ub depends on a nuclear complex consisting of FANCA, B, C, E, F, G, L, and M, which harbors E3 ubiquitin ligase activity. FANCD2-Ub is then targeted to DSBs to play an essential role in their repair via HRR and SSA 12. Although FANCD2 appeared to inhibit the third major DSB repair pathway – DNA protein kinase (DNA-PK) -mediated NHEJ (D-NHEJ) – it may stimulate the PARP-1 -dependent back-up pathway (B-NHEJ) 13, 14. In addition, FANCD2 is phosphorylated on S222 (FANCD2-pS222) by ATM kinase 18. FANCD2-Ub and FANCD2-pS222 are independent post-translational modifications of the FANCD2 protein regulating discrete cellular signaling pathways. Monoubiquitination on K561 is essential for targeting FANCD2 to DSB repair sites, whereas phosphorylation on S222 is important for activation of the S phase checkpoint.
Due to its role in response to DNA damage, the FA pathway plays a pivotal role in protecting genome stability and cancer prevention. The cumulative incidence of tumors in persons with FA mutations was 39%–76% by the age of 40–48 years, 10%–37% of which were hematologic malignancies 23–25. Acute myeloid leukemia (AML), myelodysplastic syndrome (MDS) and ALL were most common in FA patients; conversely CML and Ph–positive ALL cases have not been detected.
Here we show that, in contrary to the common dogma that loss of FA pathway may contribute to carcinogenesis, FANCD2-Ub plays an important role in BCR-ABL1-mediated leukemogenesis by protecting leukemia cells from the lethal effect of excessive oxidative DNA damage.
Materials and methods
Cells
The SV40–transformed lymphoblast cell line lacking FANCD2 protein expression PD20(FA-D2) and PD20F, PD20F(S222A), PD20F(K561R) cells with restored FANCD2 wild-type (wt), S222A mutant or K561R mutant protein expression, respectively, were kindly provided by Dr Alan D. D’Andrea (Dana-Farber Cancer Institute, Boston, MA, USA) and cultured in RPMI-1640 medium supplemented with 10% fetal bovine serum (FBS) 26. Human hematopoietic cell lines Mo7e and UT7, and their p210BCR-ABL1-positive counterparts were described before and cultured in Iscove’s Modified Dulbecco medium (IMDM) (StemCell Technologies, Inc., Vancouver, Canada) supplemented with 10% FBS and 5ng/ml recombinant human granulocyte-macrophage colony-stimulating factor (GM-CSF) 27. CML cells were obtained from bone marrow after receiving informed consent according to institutional guidelines from the Stem Cell and Leukemia Core Facility of the University of Pennsylvania (Philadelphia, PA, USA) and the Institute of Hematology and Blood Transfusions in Warsaw (Warsaw, Poland). CD34+ cells were selected through the use of human CD34+ selection cocktail (StemCell Technologies). CD34+ normal bone marrow cells were commercially available (Cambrex Bio-Science Walkersville, MD, USA). CD34+ cells were maintained in IMDM supplemented with 10% FBS with the addition of growth factors: 10pg/ml GM-CSF, 10pg/ml G-CSF, 10ng/ml SCF, 10ng/ml LIF, 10ng/ml MIP-1α, 0.2 ng/ml IL-6 (PeproTech Inc., Rocky Hill, NJ, USA, and StemCell Technologies). Murine bone marrow cells from FANCD2 knockout (FANCD2−/−) or wild type (+/+) mice on a mixed 129S4 and C57BL/6J background (kindly provided by Dr Marcus Grompe, Oregon Health & Science University, Portland, OR, USA 28) were maintained in IMDM supplemented with 10% FBS in the presence of recombinant interleukin-3 (IL-3) and 10ng/ml SCF.
Chemicals
BCR-ABL1 tyrosine kinase inhibitor imatinib (Novartis Pharma AG, Basel, Switzerland), vitamin E (VE), N-acetylcysteine (NAC), mitomycin C (MMC) (all from Sigma, St. Louis, MO, USA), and 2,3-dichloro-5,8-dihydroxy-1,4-naphtoquinone (DDN) (Calbiochem, San Diego, CA, USA) were used.
Western analysis
Cells were lysed in 1×SDS sample buffer by boiling for 3 minutes, followed by sonication for 10 to 15 seconds. Cell lysates were examined by Western blotting using antibodies recognizing the following proteins: FANCD2 (Santa Cruz), FANCL, USP1, RAD51 (Upstate), tubulin, ABL1 (Calbiochem, San Diego, CA, USA) and Ku80 (AbD Serotec, Raleigh, NC, USA).
Retroviral infections
Infections were performed as previously described with modifications 29. In brief, helper-free retroviruses pMIG-IRES-GFP, pMIG-p210BCR-ABL1-IRES-GFP, and pMIG-p210BCR-ABL1(K1172)-IRES-GFP (kindly provided by Dr W. Pear, University of Pennsylvania, Philadelphia, PA, USA 30), and also pMMP-puro, pMMP-FANCD2(wt)-puro, pMMP-FANCD2(K561R)-puro and pMMP-FANCD2(S222A)-puro (kindly provided by Dr Alan D. D’Andrea 31) were generated in the Phoenix amphotropic packaging cell line. Supernatants containing viral particles were centrifuged for 3 hours at 20,000rpm and used to infect mononuclear bone marrow cells (BMCs) from 5-fluorouracil pre-treated (i.p. 150 mg/kg) FANCD2−/− or +/+ mice. In addition, these retroviral supernatants were used to infect PD20(FA-D2), PD20F, PD20F-S222A and PD20F-K561R lymphoblast cell lines. GFP+ cells were sorted using MoFlo High-speed Sorter (Dako Cytomation, CA, USA).
RNA interference
To downregulate FANCD2, siGENOME SMART pool M-016376-02-5 siRNA (accession number NM_001018115) was used (Thermo Fisher Scientific Lafayette, CO, USA); scrambled RNA was used as control. Transfection of siRNA was carried out using Amaxa nucleoporation system and Amaxa Human CD34+ Nucleofector Kit (Lonza, Cologne, Germany) according to manufacturer’s protocol. Downregulation of FANCD2 protein expression by siRNA was confirmed 48h later by Western blot.
Immunofluorescence
Nuclear localization of γ-H2AX, FANCD2 and RAD51 was detected by immunofluorescence, as previously described 32. Briefly, cytospins were fixed in PBS with 0.06% Triton X-100 and 4% formaldehyde, washed in PBS and 0.06% Triton X-100, and blocked in washing buffer supplemented with 1% bovine serum albumin (BSA). Cells were stained with first antibodies against FANCD2 (Santa Cruz Biotechnology Inc, Santa Cruz, CA, USA), γ-H2AX (Upstate Biotechnology, Lake Placid, NY, USA) or RAD51 (Oncogene Research Products, Cambridge, MA, USA). Secondary antibodies conjugated with Alexa Fluor 488 or Alexa Fluor 568 were applied (Molecular Probes, Eugene, OR, USA). Negative controls were performed without primary antibodies. DNA was counterstained with DAPI (4’,6’diamedino-2-phenylindole). Specific staining was visualized using an inverted Olympus IX70 fluorescence microscope equipped with 100 × UPlan Apo lens (numeric aperture 1.35), and a Cooke Sensicam QE camera (The Cooke Company, Auburn Hills, MI, USA). At least 25 individual cells were analyzed per experimental group. Images were acquired with Slidebook 3.0 (Intelligent Imaging Innovations, Denver, CO, USA). All graphic adjustments were performed using Adobe Photoshop (Adobe Systems, San Jose, CA, USA).
Leukemogenesis in SCID mice
Outbred SCID mice (Taconic Farms, Germantown, NY, USA) were injected intravenously with 5×104 PD20(FA-D2)-BCR-ABL1 or PD20F-BCR-ABL1 cells. Leukemia development was confirmed at necropsy. The experiments conform to the regulatory standards and were approved by the Institutional Animal Care and Use Committee at Temple University (Philadelphia, PA, USA).
Proliferation assays
GFP+ BCR-ABL1-positive FANCD2+/+, FANCD2−/− and FANCD2−/− (+) (FANCD2−/− cells transfected with pMMP-FANCD2(wt)-puro) BMCs, and Mo7e, Mo7e-BCR-ABL1, and CML CD34+ cells transfected with FANCD2-specific siRNA and scrambled RNA were plated in methylcellulose and colonies were counted as described before 29. MMC, NAC and VE were added when indicated. Proliferation of the parental and BCR-ABL1-positive PD20(FA-D2), PD20F, PD20F(S222A) and PD20F(K561R) was determined in trypan blue exclusion test. For the competition assay, 2×105 GFP+ PD20(FA-D2)-BCR-ABL1 or PD20F-BCR-ABL1 cells were mixed with equal number of their GFP- PD20(FA-D2) or PD20F counterparts. Total number of cells and number of GFP+ cells were counted using Olympus Ix70 fluorescence microscope and the ratio of GFP positive per total number of cells was calculated.
Comet assay
Comet assay was performed as described with modifications33, 34. Cells were drained in agarose and covered with enzyme buffer (control) or with the enzyme (1 µg/mL endonuclease III [EndoIII] or formamidopyrimidine-DNA glycosylase [(Fpg]) in buffer, and incubated for 30 minutes at 37°C. Comet tail moment was analyzed in 100 randomly selected images. Because our measurement system was not calibrated, tail moment was presented in arbitrary units. Data were analyzed using the Statistica (StatSoft, Tulsa, OK) statistical package. Results obtained with buffer only were subtracted from these obtained with enzyme.
Results
Enhanced expression of FANCD2-Ub in BCR-ABL1 leukemia cells responding to ROS and MMC
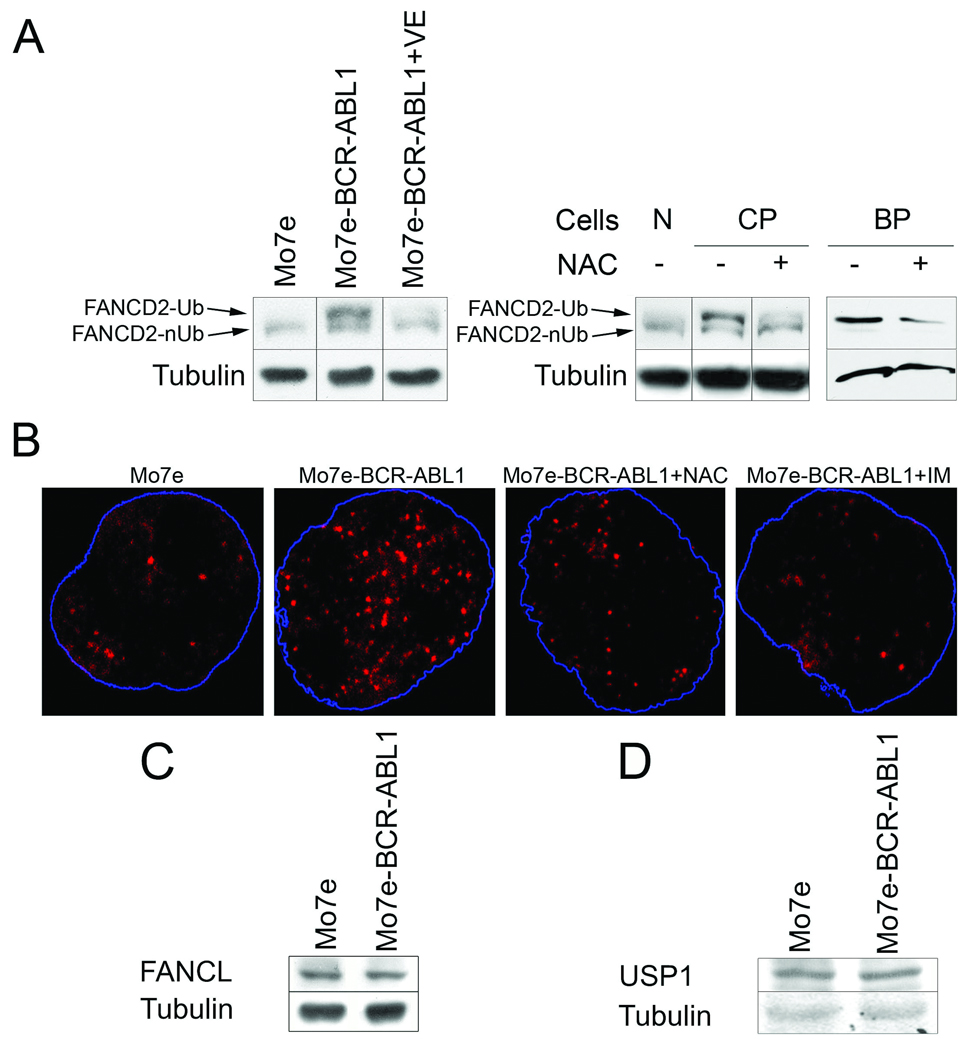
Western analysis indicated that Mo7e-BCR-ABL1 leukemia cells and CD34+ cells from CML-CP and CML-BP patient cells contained more FANCD2-Ub protein than normal counterparts (Fig. 1A), which was associated with enhanced nuclear foci formation by FANCD2-Ub (Fig. 1B) (nuclear foci are generated exclusively by FANCD2-Ub 35). These effects in Mo7e-BCR-ABL1 leukemia cells were not associated with their enhanced proliferation (data not shown) nor caused by deregulated expression of FANCD2 ubiquitinase FANCL or down-regulation of FANCD2 deubiquitinase USP1 (Fig. 1C and D, respectively), but were reversed after inhibition of BCR-ABL1 kinase with imatinib and reduction of ROS with antioxidants VE and NAC (Fig. 1A and B, and Suppl. Fig. 1).
Figure 1.
BCR-ABL1 kinase stimulated the expression of FANCD2-Ub in a kinase– and ROS– dependent manner. (A) Expression of FANCD2 monoubiquitinated on K561 (FANCD2-Ub) and non-ubiquitinated FANCD2 (FANCD2-nUb) was examined by Western analysis in Mo7e parental cells and BCR-ABL1 –positive counterparts and in CD34+ cells from healthy donors (N), CML-CP (CP) and CML-BP (BP) patients; tubulin served as loading control. When indicated, cells were treated for 24h with VE (200µM), NAC (50µM), and IM (1µM) in the presence of growth factors. (B) Nuclear localization of FANCD2 was determined by immunofluorescence in Mo7e and Mo7e-BCR-ABL1 cells and in cells treated for 24h with 50µM NAC and 1µM IM. Nuclei borders are marked in blue. Western analysis of FANCL (C) and USP1 (D); tubulin served as loading control. Panels of Western analyses presented here are from the same membranes.
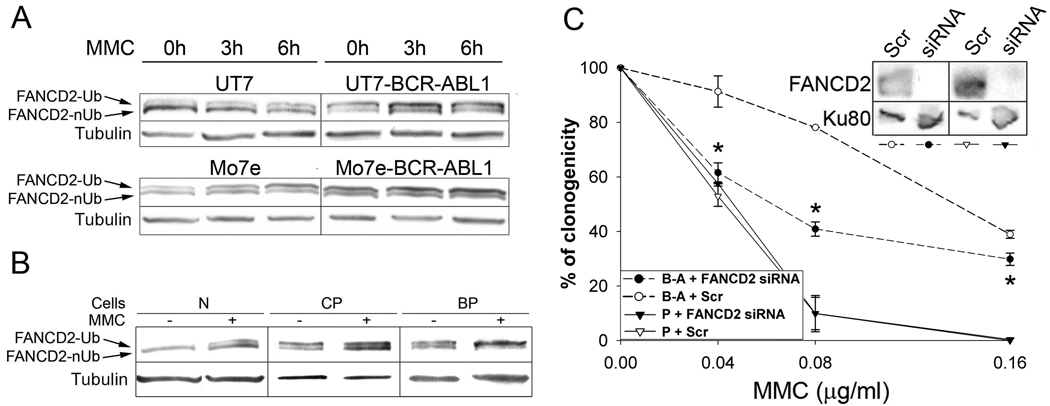
MMC is being routinely used to diagnose FA (FA patient cells are over-sensitive to cross-linking agents such as MMC) and to test the function of Fanconi anemia protein network resulting in FANCD2-Ub 22. MMC-treated UT7-BCR-ABL1 and Mo7e-BCR-ABL1 cells displayed bigger increase of FANCD2-Ub in comparison to parental UT7 and Mo7e cells (Fig. 2A). MMC treatment enhanced also the expression of FANCD2-Ub in CD34+ CML-CP and CML-BP cells (Fig. 2B). Downregulation of FANCD2 by siRNA increased the sensitivity Mo7e-BCR-ABL1 cells to MMC (Fig. 2C) suggesting that FANCD2 plays an essential role in response to DNA damage in CML.
Figure 2.
BCR-ABL1 leukemia cells displayed an enhancement of MMC-induced FANCD2-Ub: role in cell survival. (A) FANCD2-Ub expression was examined by Western analysis in total cell lysates obtained at the indicated times after 0.5µg/ml MMC treatment; tubulin served as loading control. (B) CD34+ bone marrow cells obtained from healthy volunteers (N) or from CML-CP (CP) and CML-BP (BP) patients were treated for 6h with 0.5µg/ml MMC and the presence of FANCD2-Ub was examined by Western analysis; tubulin served as loading control. (C) FANCD2 was downregulated by specific siRNA in Mo7e (P) and Mo7e-BCR-ABL1 (B-A) cells; Scr served as a negative control (inset). Clonogenic activity was examined in the presence of the indicated concentrations of MMC; *p<0.05 in comparison to B-A+Scr using Student t test.
FANCD2 supports the growth of BCR-ABL1 leukemia cells
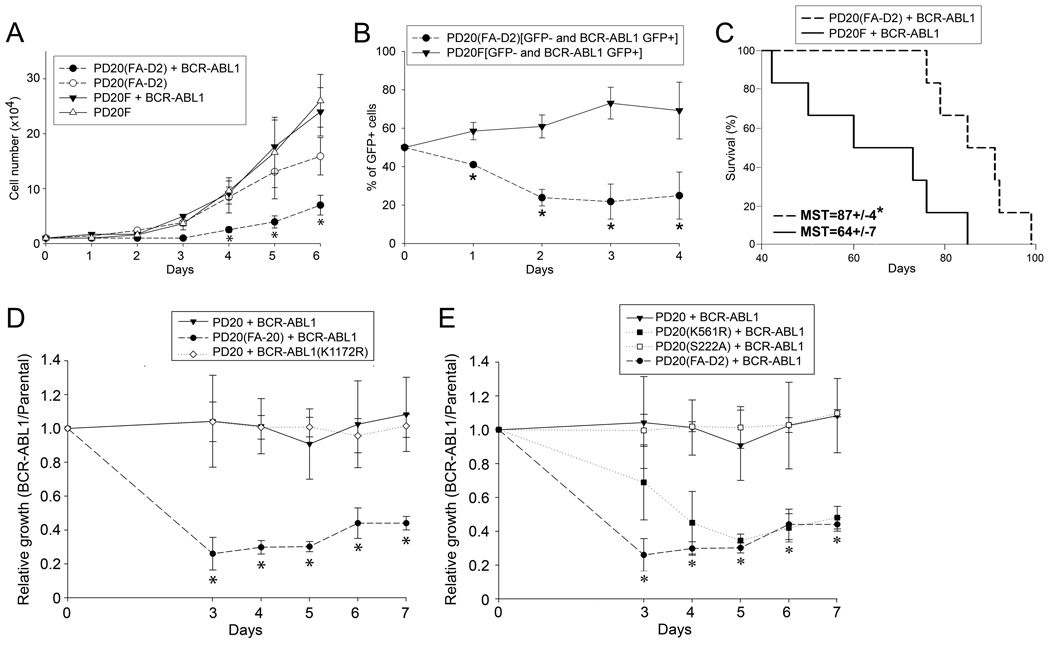
To directly test the role of FANCD2 in BCR-ABL1 –induced leukemogenesis the SV40–transformed lymphoblast cell line lacking FANCD2 protein expression PD20(FA-D2) and PD20F cells with restored expression of FANCD2 wild-type (wt) protein were transfected with p210BCR-ABL1 (Suppl. Fig. 2). BCR-ABL1 kinase –positive PD20(FA-D2) cells displayed reduced proliferation rate in comparison to PD20(FA-D2) parental cells and BCR-ABL1 kinase –positive PD20F cells (Fig. 3A); this effect was associated with an increase of the percentage of sub-G1 apoptotic cells in PD20(FA-D2)+BCR-ABL1 cells in comparison to PD20+BCR-ABL1 cells (10.7±6.1 and 1.7±0.9, respectively, p=0.027).
Figure 3.
FANCD2-Ub is required for the proliferation of BCR-ABL1 –positive cells. (A) Proliferation of parental PD20(FA-D2) and PD20F lymphoblast cell lines and their BCR-ABL1 –positive counterparts was assessed by trypan blue exclusion test. (B) 1 : 1 mixed populations of GFP+ BCR-ABL1-positive PD20(FA-D2) : GFP- of PD20(FA-D2) cells and GFP+ BCR-ABL1-positive PD20F : GFP- of PD20F cells were prepared. The percentage of GFP+ cells (BCR-ABL1-positive) in each population was counted during in vitro culture using a fluorescent microscope. (C) Survival of SCID mice injected with 105 BCR-ABL1-positive PD20(FA-D2) cells or BCR-ABL1-positive PD20F cells. Leukemia development was confirmed at necropsy. (D) PD20(FA-D2) and PD20F cells were infected with retroviral constructs containing p210BCR-ABL1-IRES-GFP, p210BCR-ABL1(K1172R)-IRES-GFP or IRES-GFP (control). Proliferation of GFP+ cells was assessed in trypan blue. Relative growth represents a fold of change of GFP+ BCR-ABL1 –positive cell number in comparison to the number of GFP+ BCR-ABL1 -negative counterparts (control). (E) PD20(FA-D2), PD20F, PD20F(S222A), and PD20F(K561R) cells were infected with retroviral constructs containing p210BCR-ABL1-IRES-GFP or IRES-GFP (control). Relative growth represents a fold of change of GFP+ BCR-ABL1 –positive cell number in comparison to the number of GFP+ BCR-ABL1 -negative counterparts. Results in A, B, D and E represent mean ± SD, *p<0.05 using Student t test; results in C show MST = median survival time; *p=0.012 using Kaplan-Meier LogRank test.
Next, equal numbers of GFP- PD20(FA-D2) or PD20F cells and GFP+ BCR-ABL1 –transformed counterparts were mixed together and cultured. BCR-ABL1 kinase-positive FANCD2-negative cells displayed a clear growth disadvantage in comparison to their BCR-ABL1 kinase –negative counterparts as indicated by decreasing percentage of GFP+ cells (Fig. 3B). At the same time BCR-ABL1 kinase did not exert an anti-proliferative effect in PD20F cells expressing FANCD2. The in vitro proliferative defect of BCR-ABL1 kinase-positive FANCD2-negative lymphoblast cells was accompanied by delayed leukemogenesis in SCID mice (Fig. 3C). BCR-ABL1 –mediated growth inhibition in the absence of FANCD2 was depended upon BCR-ABL1 kinase activity because BCR-ABL1(K1172R) kinase-dead mutant did not exert this effect (Fig. 3D).
Next we examined which function of FANCD2, K561 ubiquitination-dependent DSB repair and/or S222 phosphorylation-induced S phase checkpoint is required to support proliferation of BCR-ABL1 –positive cells. Expression of FANCD2 wild-type (proficient in DNA repair and S phase checkpoint) and FANCD2 phosphorylation-less S222A mutant (proficient in DNA repair, but deficient in S phase checkpoint) rescued the growth of BCR-ABL1 –positive FANCD2-deficient cells (Fig. 3E). In contrast, expression of FANCD2 monoubiquitination-deficient K561R mutant (defective in DSB repair, but proficient in S phase checkpoint) did not restore the growth of BCR-ABL1 –positive FANCD2-deficient cells. Altogether, these results strongly suggest that FANCD2-Ub plays a significant role in the growth of BCR-ABL1 –positive leukemia cells.
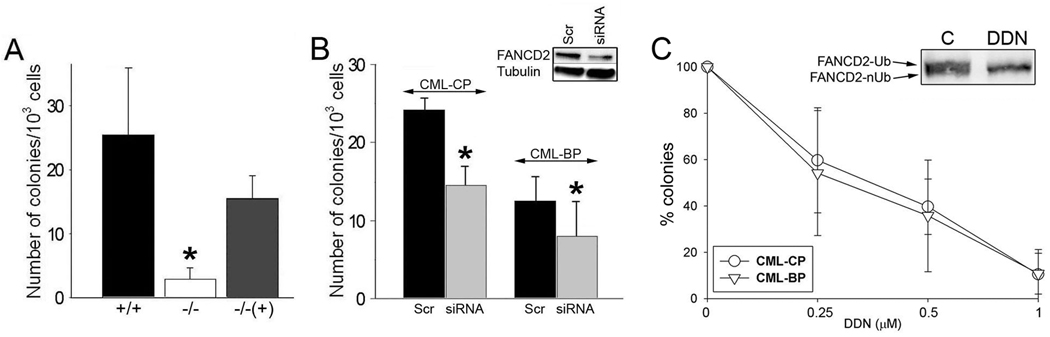
In support for this hypothesis the clonogenic potential of BCR-ABL1-positive FANCD2−/− bone marrow cells was reduced by approximately 10-fold in comparison to BCR-ABL1-positive FANCD2+/+ counterparts (Fig. 4A). The growth defect was dependent on BCR-ABL1; non-transformed −/− and +/+ bone marrow cells displayed similar clonogenic activities stimulated by SCF + GM-CSF (data not shown). Restoration of FANCD2 expression “rescued” the impaired clonogenic activity of BCR-ABL1-positive FANCD2−/− cells. The presence or absence of FANCD2 did not affect the population of BCR-ABL1 –positive stem cell-enriched Lin-Kit+Sca1+ and early progenitor-enriched Lin-Kit+Sca1− populations in clonogenic cells (Suppl. Fig. 3).
Figure 4.
Downregulation of FANCD2 inhibited BCR-ABL1 –mediated leukemogenesis in primary cells. (A) Bone marrow cells from FANCD2+/+ and −/− littermates were co-infected with retroviral constructs containing p210BCR/ABL-IRES-GFP and IRES-puro (+/+ and −/−, respectively) or FANCD2-IRES-puro (−/− (+)). GFP+ cells were plated in methylcellulose in the presence of puromycin, and colonies were counted after 7 days. *p<0.05 in comparison to +/+ and −/− (+) groups. (B) CD34+ cells from CML-CP and CML-BP patients were transfected with FANCD2-specific siRNA (siRNA) and scrambled (Scr), plated in methylcellulose, and colonies were counted after 10 days. Downregulation of FANCD2 by siRNA was confirmed by Western analysis (inset); *p<0.05 in comparison to corresponding Scr group. (C) CD34+ cells from CML-CP and CML-BP patients were plated in methylcellulose in the presence of with various concentrations of FANCD2 inhibitor DDN, and colonies were counted after 10 days. Results represent mean percentage of colonies ± SD in comparison to untreated control group. Donwregulation of monoubiquitinated (FANCD2-Ub), but not non-ubiquitinated (FANCD2-nUb) form of FANCD2 by DDN was detected (inset).
CML-CP and CML-BP patient cells were used to determine the role of FANCD2 in primary leukemia cells. Downregulation of FANCD2 by siRNA in CD34+ cells from CML-CP and CML-BP inhibited their clonogenic potential (Fig. 4B).
In addition, FANCD2-Ub inhibitor, DDN, reduced the clonogenic potential of CD34+ CML-CP and CML-BP cells (Fig. 4C). DDN inhibits accumulation of FANCD2-Ub and formation of FANCD2 nuclear foci without disruption of the upstream FA core complex 36. Although DDN may have other targets, these data implicate FANCD2-Ub in BCR-ABL1 –induced leukemias.
FANCD2-Ub protects BCR-ABL1 –positive cells from ROS-induced DSBs to support leukemogenesis
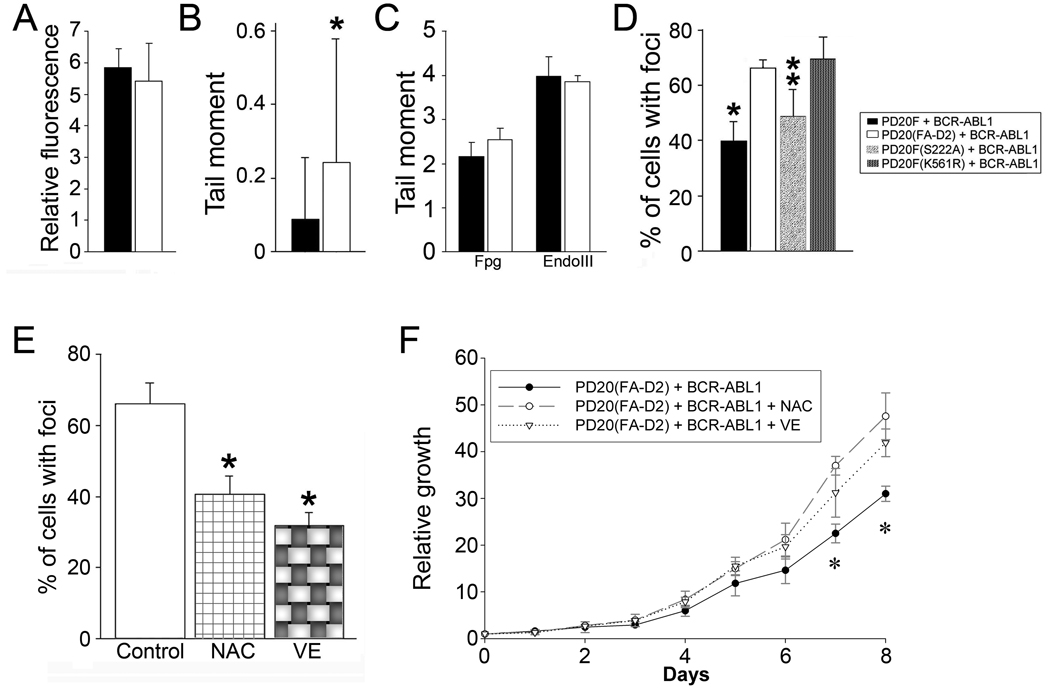
BCR-ABL1 kinase stimulates repair of numerous ROS-induced DSBs 2, 4, 37. Since FANCD2 plays a major role in DSB repair, it may be involved in facilitation of DSB repair to protect leukemia cells from the anti-proliferative and/or lethal effect of DSBs 16. To test this hypothesis we measured ROS and oxidative DNA damage in BCR-ABL1 –positive FANCD2-deficient cells (PD20(FA-D2)+BCR-ABL1) and in BCR-ABL1 –positive FANCD2-proficient cells (PD20F+BCR-ABL1). Although the presence or absence of FANCD2 did not have an impact on ROS (Fig. 5A), PD20(FA-D2)+BCR-ABL1 cells accumulated more DNA lesions than PD20F+BCR-ABL1 cells as detected by comet assay (Fig. 5B). More specifically, PD20(FA-D2)+BCR-ABL1 cells did not contain more oxidized bases (Fig. 5C), but they accumulated an excess of DSBs (Fig. 5D) in comparison to PD20F+BCR-ABL1 cells. In addition, the number of DSBs in PD20(FA-D2)+BCR-ABL1 cells was reduced after expression of FANCD2 wild-type and the cell-cycle regulation defective FANCD2(S222A) mutant, but not of the DSB repair-deficient FANCD2(K561R) mutant (Fig. 5D).
Figure 5.
FANCD2-Ub protected BCR-ABL1 –positive leukemia cells from the toxic effect of ROS-induced DSBs. (A–D) ROS and DNA damage were measured in BCR-ABL1 –positive PD20(FA-D2), PD20F, PD20F(S222A), and PD20F(K561R) cells. (A) ROS levels were measured by DCFDA. (B) Spontaneous DNA damage was assessed by the comet assay; *p<0.05 in comparison to PD20F + BCR-ABL1 cells. (C) Levels of oxidative damage were measured using endonucleases EndoIII and Fpg, which convert oxidative lesions into gaps detectable by comet assay (D) Spontaneous DSBs were detected by γ-H2AX immunostaining. Results represent % of cells containing at least 5 γ-H2AX nuclear foci. *p<0.05 in comparison to PD20(FA-D2) + BCR-ABL1 cells, and **p<0.05 in comparison to PD20(FA-D2) + BCR-ABL1 cells and PD20F(K561R) + BCR-ABL1 cells using Student t test. (E, F) PD20(FA-D2)+BCR/ABL cells were cultured without (Control) or with the addition of antioxidants, NAC (50µM) or VE (200µM). (E) Spontaneous DSBs were detected by γ-H2AX foci. Results are shown as percentage of cells containing at least five γ-H2AX foci per nucleus; *p<0.05 in comparison to Control using Student t test. (F) Relative growth, results represent an increase of cell number in comparison to day 0; *p<0.05 in comparison to the other two groups using Student t test.
These results clearly implicated the role of FANCD2-Ub in protection of BCR-ABL1 –positive leukemia cells from the potential lethal effect of an excess of ROS-induced DSBs. This speculation is further supported by the data showing that inhibition of ROS by VE and NAC (Suppl. Fig. 4) reduced accumulation of DSBs in PD20(FA-D2)+BCR-ABL1 cells (Fig. 5E), which was associated with their accelerated proliferation (Fig. 5F).
Discussion
Oncogene activation is often associated with changes in metabolic parameters including enhanced generation of ROS 38. ROS are well known for playing a dual role as both beneficial and deleterious species. The “two-faced” character of ROS is supported by numerous evidence that ROS can act as secondary messenger in intracellular signaling cascades and epigenetic modifications, which induce and maintain the oncogenic phenotype of cancer cells; however, ROS can also damage DNA, proteins, and lipids to induce cellular senescence and apoptosis 39. Thus, tumor cells need to protect themselves from an excess of ROS in order to survive. In concordance, hematopoietic cells may undergo p53-mediated apoptosis and/or p38 MAPK-dependent senescence shortly after expression of oncogenes such as BCR-ABL1, v-Abl, CBFB-MYH11, and RUNX1-ETO 7, 40. Presumably, only clones capable of dealing with the ”crisis” emerge as leukemic populations and become tolerant to higher levels of oxidative DNA lesions, for example 8-oxoG and DSBs 3, 4. Accordingly, genes regulating DNA repair mechanisms may be important for leukemogenesis induced by BCR-ABL1 and other oncogenic tyrosine kinases.
FANCD2, a member of FA protein family, plays a central role in DSB repair, because it regulates HRR, SSA and possibly also B-NHEJ 12–14. These mechanisms may be seminal for protection of BCR-ABL1 leukemia cells from the potentially lethal effect of elevated levels of endogenous (ROS-dependent) and exogenous (genotoxic agent-induced) DSBs. Therefore, FANCD2 may protect cells from apoptosis induced by DNA damage and facilitate DSB repair in Ph-positive leukemias. This hypothesis is supported by data showing that downregulation of FANCD2 expression either by siRNA or genetic knockdown reduced the in vitro growth rate and in vivo leukemogenic potential of BCR-ABL1 kinase –positive cell lines, BCR-ABL1 –transformed FANCD2−/− murine bone marrow cells, and CML-CP and CML-BP CD34+ cells. We showed that reduced clonogenic potential of BCR-ABL1 –transformed FANCD2−/− cells in comparison to BCR-ABL1 –positive FANCD2+/+ and FANCD2−/− (+FANCD2) counterparts was not associated with any specific alterations in stem cell enriched Lin-Sca1+Kit+ and early progenitor enriched Lin-Sca1-Kit+ populations. However, Zhang et al. reported that non-transformed FANCD2−/− cells displayed a decline in the size of Lin-Sca1+Kit+ pool associated with proliferation disturbances 41, 42. This discrepancy could be explained by the impact of BCR-ABL1 kinase on Lin-Sca1+Kit+ cells and also by fact that bone marrow cells used in our studies were obtained from the mice pre-treated with 5-FU to enrich the population with stem cells whereas those used by Zhang and colleagues were harvested from untreated mice.
Although the growth defect in BCR-ABL1 –positive FANCD2-deficient cells was not associated with enhanced ROS and increased numbers of oxidized DNA bases, accumulation of excessive DSBs was readily detectable. This suggests that high number of DSBs in BCR-ABL1 –positive FANCD2-deficient cells was not dependent on enhanced generation of ROS-induced DSBs, but rather due to inefficient DSB repair. In concordance with this speculation, expression of FANCD2 wild-type and FANCD2(S222A) mutant (proficient in DNA repair, but deficient in S phase checkpoint), but not FANCD2(K561R) mutant (deficient in DNA repair, but proficient in S phase checkpoint 18) prevented accumulation of DSBs and restored proliferative potential of BCR-ABL1 –positive FANCD2−/− cells. These results strongly suggest that FANCD2-Ub is required to catalyze DSB repair and protect BCR-ABL1 –positive leukemia cells from the detrimental effect of excessive unrepaired DSBs.
FA cells exhibit hypersensitivity to oxidative stress, which indicates that FA proteins may play a major role in protection against oxidative damage 43. For example, FANCC and FANCG are found to associate and possibly modulate a variety of proteins such as cytochrome P450, glutathione S-transferase, Cu/Zn superoxide dismutase and peroxiredoxin-3 to diminish oxidative stress and number of DSBs 43, 44. ROS also causes multimerization and nuclear complex formation by the core FA proteins necessary for the appearance of FANCD2-Ub 45. In CML cells, which contain high numbers of ROS-induced DSBs, FANCD2-Ub may become an important player in the protection of leukemia cells from an excess of potentially lethal DNA lesions. In concordance, CML primary cells and BCR-ABL1 –transformed cell lines overexpressed FANCD2-Ub in response to endogenous oxidative stress and genotoxic treatment, and ROS scavengers, such as vitamin E and N-acetylcysteine not only reduced the accumulation of DSBs, but also enhanced the growth of BCR-ABL1 –positive FANCD2-deficient cells. Our data are seemingly in contrast to these recently reported by Valeri and colleagues suggesting that BCR-ABL1 disrupts FANCD2-Ub 46. However, most of their measurements were performed in BCR-ABL1 –positive CD34+ umbilical cord blood cells 16h after MMC treatment thus addressing late stages of DNA damage response; at that time most MMC-induced DSBs are already repaired in BCR-ABL1 –positive cells whereas parental cells usually display slower and sometimes inefficient repair rate eventually leading to apoptosis 47.
FANCD2-Ub is also required during DNA replication stress, for the stability of common fragile sites and to protect ultrafine DNA bridges linking sister chromatids 48, 49. However, none of these functions appear to play a critical role in Ph-positive leukemias because the growth of BCR-ABL1 –positive FANCD2−/− cells was restored, at least partially by reduction of the number of ROS-induced DSBs. Moreover, the role of FANCD2 in protection from telomere erosion is probably irrelevant here because BCR-ABL1 –positive leukemia cells tolerate short telomeres 50, 51. In addition, since BCR-ABL1 inhibits FOXO3a transcription factor, it is rather unlikely that the interaction between FANCD2-Ub and FOXO3a is relevant for Ph-positive leukemia cells 52, 53. In summary, we believe that FANCD2-Ub is required to facilitate the repair of numerous ROS-induced DSBs which, if unrepaired may impair the growth of BCR-ABL1 –positive leukemia cells. This hypothesis is supported by the observation that restoration of the reading frame of inactivated FANCD1/BRCA2 gene caused acceleration of AML 54.
Supplementary Material
Acknowledgements
We thank Dr. Alan D. D’Andrea (Dana-Farber Cancer Institute, Boston, MA, USA) for providing FANCD2 expression plasmids and FANCD2 cell lines, and Dr. Marcus Grompe (Oregon Health & Science University, Portland, OR, USA) for the knockout mice. We also thank Elisabeth Bolton for careful reading of the manuscript.
Financial support: This work was supported by NIH/NCI CA89052 and CA123014 (T. Skorski), by 1M19/NK1W/2009 and 1M19/NK1D/2009 from Medical University of Warsaw (T. Stoklosa and E. Glodkowska-Mrowka) and by N N401 039037 from Polish Ministry of Education and Science (G. Hoser). We thank E. Bolton for critical reading of the manuscript and K. Piwocka and M. Kucia-Kobialko for their help with nucleofection.
Footnotes
Supplementary Information accompanies the paper on the Leukemia website (http://www.nature.com.leu)
Conflict of interest
The Authors declare no conflict of interest.
REFERENCES
- 1.Jones D, Luthra R, Cortes J, Thomas D, O'Brien S, Bueso-Ramos C, et al. BCR-ABL fusion transcript types and levels and their interaction with secondary genetic changes in determining the phenRotype of Philadelphia chromosome-positive leukemias. Blood. 2008;112:5190–5192. doi: 10.1182/blood-2008-04-148791. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Cramer K, Nieborowska-Skorska M, Koptyra M, Slupianek A, Penserga ET, Eaves CJ, et al. BCR/ABL and other kinases from chronic myeloproliferative disorders stimulate single-strand annealing, an unfaithful DNA double-strand break repair. Cancer Res. 2008;68:6884–6888. doi: 10.1158/0008-5472.CAN-08-1101. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Koptyra M, Falinski R, Nowicki MO, Stoklosa T, Majsterek I, Nieborowska-Skorska M, et al. BCR/ABL kinase induces self-mutagenesis via reactive oxygen species to encode imatinib resistance. Blood. 2006;108:319–327. doi: 10.1182/blood-2005-07-2815. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Nowicki MO, Falinski R, Koptyra M, Slupianek A, Stoklosa T, Gloc E, et al. BCR/ABL oncogenic kinase promotes unfaithful repair of the reactive oxygen species-dependent DNA double-strand breaks. Blood. 2004;104:3746–3753. doi: 10.1182/blood-2004-05-1941. [DOI] [PubMed] [Google Scholar]
- 5.Klemm L, Duy C, Iacobucci I, Kuchen S, von Levetzow G, Feldhahn N, et al. The B cell mutator AID promotes B lymphoid blast crisis and drug resistance in chronic myeloid leukemia. Cancer Cell. 2009;16:232–245. doi: 10.1016/j.ccr.2009.07.030. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Feldhahn N, Henke N, Melchior K, Duy C, Soh BN, Klein F, et al. Activation-induced cytidine deaminase acts as a mutator in BCR-ABL1-transformed acute lymphoblastic leukemia cells. J Exp Med. 2007;204:1157–1166. doi: 10.1084/jem.20062662. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Radfar A, Unnikrishnan I, Lee HW, DePinho RA, Rosenberg N. p19(Arf) induces p53-dependent apoptosis during abelson virus-mediated pre-B cell transformation. Proc Natl Acad Sci U S A. 1998;95:13194–13199. doi: 10.1073/pnas.95.22.13194. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Wang PY, Young F, Chen CY, Stevens BM, Neering SJ, Rossi RM, et al. The biologic properties of leukemias arising from BCR/ABL-mediated transformation vary as a function of developmental origin and activity of the p19ARF gene. Blood. 2008;112:4184–4192. doi: 10.1182/blood-2008-02-142190. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Slupianek A, Nowicki MO, Koptyra M, Skorski T. BCR/ABL modifies the kinetics and fidelity of DNA double-strand breaks repair in hematopoietic cells. DNA Repair (Amst) 2006;5:243–250. doi: 10.1016/j.dnarep.2005.10.005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Fernandes MS, Reddy MM, Gonneville JR, DeRoo SC, Podar K, Griffin JD, et al. BCR-ABL promotes the frequency of mutagenic single-strand annealing DNA repair. Blood. 2009;114:1813–1819. doi: 10.1182/blood-2008-07-172148. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Brady N, Gaymes TJ, Cheung M, Mufti GJ, Rassool FV. Increased error-prone NHEJ activity in myeloid leukemias is associated with DNA damage at sites that recruit key nonhomologous end-joining proteins. Cancer Res. 2003;63:1798–1805. [PubMed] [Google Scholar]
- 12.Nakanishi K, Yang YG, Pierce AJ, Taniguchi T, Digweed M, D'Andrea AD, et al. Human Fanconi anemia monoubiquitination pathway promotes homologous DNA repair. Proc Natl Acad Sci U S A. 2005;102:1110–1115. doi: 10.1073/pnas.0407796102. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Adamo A, Collis SJ, Adelman CA, Silva N, Horejsi Z, Ward JD, et al. Preventing nonhomologous end joining suppresses DNA repair defects of Fanconi anemia. Mol Cell. 2010;39:25–35. doi: 10.1016/j.molcel.2010.06.026. [DOI] [PubMed] [Google Scholar]
- 14.Lundberg R, Mavinakere M, Campbell C. Deficient DNA end joining activity in extracts from fanconi anemia fibroblasts. J Biol Chem. 2001;276:9543–9549. doi: 10.1074/jbc.M008634200. [DOI] [PubMed] [Google Scholar]
- 15.D'Andrea AD, Grompe M. The Fanconi anaemia/BRCA pathway. Nat Rev Cancer. 2003;3:23–34. doi: 10.1038/nrc970. [DOI] [PubMed] [Google Scholar]
- 16.Moldovan GL, D'Andrea AD. How the fanconi anemia pathway guards the genome. Annu Rev Genet. 2009;43:223–249. doi: 10.1146/annurev-genet-102108-134222. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Garcia-Higuera I, Taniguchi T, Ganesan S, Meyn MS, Timmers C, Hejna J, et al. Interaction of the Fanconi anemia proteins and BRCA1 in a common pathway. Mol Cell. 2001;7:249–262. doi: 10.1016/s1097-2765(01)00173-3. [DOI] [PubMed] [Google Scholar]
- 18.Taniguchi T, Garcia-Higuera I, Xu B, Andreassen PR, Gregory RC, Kim ST, et al. Convergence of the fanconi anemia and ataxia telangiectasia signaling pathways. Cell. 2002;109:459–472. doi: 10.1016/s0092-8674(02)00747-x. [DOI] [PubMed] [Google Scholar]
- 19.Smogorzewska A, Matsuoka S, Vinciguerra P, McDonald ER, 3rd, Hurov KE, Luo J, et al. Identification of the FANCI protein, a monoubiquitinated FANCD2 paralog required for DNA repair. Cell. 2007;129:289–301. doi: 10.1016/j.cell.2007.03.009. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Ishiai M, Kitao H, Smogorzewska A, Tomida J, Kinomura A, Uchida E, et al. FANCI phosphorylation functions as a molecular switch to turn on the Fanconi anemia pathway. Nat Struct Mol Biol. 2008;15:1138–1146. doi: 10.1038/nsmb.1504. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Rothfuss A, Grompe M. Repair kinetics of genomic interstrand DNA cross-links: evidence for DNA double-strand break-dependent activation of the Fanconi anemia/BRCA pathway. Mol Cell Biol. 2004;24:123–134. doi: 10.1128/MCB.24.1.123-134.2004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Alpi AF, Patel KJ. Monoubiquitylation in the Fanconi anemia DNA damage response pathway. DNA Repair (Amst) 2009;8:430–435. doi: 10.1016/j.dnarep.2009.01.019. [DOI] [PubMed] [Google Scholar]
- 23.Alter BP. Cancer in Fanconi anemia, 1927–2001. Cancer. 2003;97:425–440. doi: 10.1002/cncr.11046. [DOI] [PubMed] [Google Scholar]
- 24.Kutler DI, Singh B, Satagopan J, Batish SD, Berwick M, Giampietro PF, et al. A 20-year perspective on the International Fanconi Anemia Registry (IFAR) Blood. 2003;101:1249–1256. doi: 10.1182/blood-2002-07-2170. [DOI] [PubMed] [Google Scholar]
- 25.Rosenberg PS, Greene MH, Alter BP. Cancer incidence in persons with Fanconi anemia. Blood. 2003;101:822–826. doi: 10.1182/blood-2002-05-1498. [DOI] [PubMed] [Google Scholar]
- 26.Garcia-Higuera I, Taniguchi T, Ganesan S, Meyn MS, Timmers C, Hejna J, et al. Interaction of the Fanconi anemia proteins and BRCA1 in a common pathway. Mol Cell. 2001;7:249–262. doi: 10.1016/s1097-2765(01)00173-3. [DOI] [PubMed] [Google Scholar]
- 27.Rink L, Slupianek A, Stoklosa T, Nieborowska-Skorska M, Urbanska K, Seferynska I, et al. Enhanced phosphorylation of Nbs1, a member of DNA repair/checkpoint complex Mre11-RAD50-Nbs1, can be targeted to increase the efficacy of imatinib mesylate against BCR/ABL-positive leukemia cells. Blood. 2007;110:651–660. doi: 10.1182/blood-2006-08-042630. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Houghtaling S, Timmers C, Noll M, Finegold MJ, Jones SN, Meyn MS, et al. Epithelial cancer in Fanconi anemia complementation group D2 (Fancd2) knockout mice. Genes Dev. 2003;17:2021–2035. doi: 10.1101/gad.1103403. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Skorski T, Bellacosa A, Nieborowska-Skorska M, Majewski M, Martinez R, Choi JK, et al. Transformation of hematopoietic cells by BCR/ABL requires activation of a PI-3k/Akt-dependent pathway. Embo J. 1997;16:6151–6161. doi: 10.1093/emboj/16.20.6151. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Pear WS, Miller JP, Xu L, Pui JC, Soffer B, Quackenbush RC, et al. Efficient and rapid induction of a chronic myelogenous leukemia-like myeloproliferative disease in mice receiving P210 bcr/abl-transduced bone marrow. Blood. 1998;92:3780–3792. [PubMed] [Google Scholar]
- 31.Taniguchi T, Garcia-Higuera I, Xu B, Andreassen PR, Gregory RC, Kim ST, et al. Convergence of the fanconi anemia and ataxia telangiectasia signaling pathways. Cell. 2002;109:459–472. doi: 10.1016/s0092-8674(02)00747-x. [DOI] [PubMed] [Google Scholar]
- 32.Canitrot Y, Falinski R, Louat T, Laurent G, Cazaux C, Hoffmann JS, et al. p210 BCR/ABL kinase regulates nucleotide excision repair (NER) and resistance to UV radiation. Blood. 2003;102:2632–2637. doi: 10.1182/blood-2002-10-3207. [DOI] [PubMed] [Google Scholar]
- 33.Singh NP, McCoy MT, Tice RR, Schneider EL. A simple technique for quantitation of low levels of DNA damage in individual cells. Exp Cell Res. 1988;175:184–191. doi: 10.1016/0014-4827(88)90265-0. [DOI] [PubMed] [Google Scholar]
- 34.Majsterek I, Blasiak J, Mlynarski W, Hoser G, Skorski T. Does the bcr/abl-mediated increase in the efficacy of DNA repair play a role in the drug resistance of cancer cells? Cell Biol Int. 2002;26:363–370. doi: 10.1006/cbir.2002.0865. [DOI] [PubMed] [Google Scholar]
- 35.Montes de Oca R, Andreassen PR, Margossian SP, Gregory RC, Taniguchi T, Wang X, et al. Regulated interaction of the Fanconi anemia protein, FANCD2, with chromatin. Blood. 2005;105:1003–1009. doi: 10.1182/blood-2003-11-3997. [DOI] [PubMed] [Google Scholar]
- 36.Landais I, Sobeck A, Stone S, LaChapelle A, Hoatlin ME. A novel cell-free screen identifies a potent inhibitor of the Fanconi anemia pathway. Int J Cancer. 2009;124:783–792. doi: 10.1002/ijc.24039. [DOI] [PubMed] [Google Scholar]
- 37.Slupianek A, Schmutte C, Tombline G, Nieborowska-Skorska M, Hoser G, Nowicki MO, et al. BCR/ABL regulates mammalian RecA homologs, resulting in drug resistance. Mol Cell. 2001;8:795–806. doi: 10.1016/s1097-2765(01)00357-4. [DOI] [PubMed] [Google Scholar]
- 38.Finkel T. Oxidant signals and oxidative stress. Curr Opin Cell Biol. 2003;15:247–254. doi: 10.1016/s0955-0674(03)00002-4. [DOI] [PubMed] [Google Scholar]
- 39.Martindale JL, Holbrook NJ. Cellular response to oxidative stress: signaling for suicide and survival. J Cell Physiol. 2002;192:1–15. doi: 10.1002/jcp.10119. [DOI] [PubMed] [Google Scholar]
- 40.Wajapeyee N, Wang SZ, Serra RW, Solomon PD, Nagarajan A, Zhu X, et al. Senescence induction in human fibroblasts and hematopoietic progenitors by leukemogenic fusion proteins. Blood. 2010;115:5057–5060. doi: 10.1182/blood-2009-09-245928. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Zhang QS, Marquez-Loza L, Eaton L, Duncan AW, Goldman DC, Anur P, et al. Fancd2−/− mice have hematopoietic defects that can be partially corrected by resveratrol. Blood. 2010;116:5140–5148. doi: 10.1182/blood-2010-04-278226. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Zhang QS, Marquez-Loza L, Eaton L, Duncan AW, Goldman DC, Anur P, et al. Fancd2−/− mice have hematopoietic defects that can be partially corrected by resveratrol. Blood. 2010;8 doi: 10.1182/blood-2010-04-278226. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Pang Q, Andreassen PR. Fanconi anemia proteins and endogenous stresses. Mutat Res. 2009;668:42–53. doi: 10.1016/j.mrfmmm.2009.03.013. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Futaki M, Igarashi T, Watanabe S, Kajigaya S, Tatsuguchi A, Wang J, et al. The FANCG Fanconi anemia protein interacts with CYP2E1: possible role in protection against oxidative DNA damage. Carcinogenesis. 2002;23:67–72. doi: 10.1093/carcin/23.1.67. [DOI] [PubMed] [Google Scholar]
- 45.Park SJ, Ciccone SL, Beck BD, Hwang B, Freie B, Clapp DW, et al. Oxidative stress/damage induces multimerization and interaction of Fanconi anemia proteins. J Biol Chem. 2004;279:30053–30059. doi: 10.1074/jbc.M403527200. [DOI] [PubMed] [Google Scholar]
- 46.Valeri A, Alonso-Ferrero ME, Rio P, Pujol MR, Casado JA, Perez L, et al. Bcr/Abl interferes with the Fanconi anemia/BRCA pathway: implications in the chromosomal instability of chronic myeloid leukemia cells. PLoS One. 2010;5:e15525. doi: 10.1371/journal.pone.0015525. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Nieborowska-Skorska M, Stoklosa T, Datta M, Czechowska A, Rink L, Slupianek A, et al. ATR-Chk1 axis protects BCR/ABL leukemia cells from the lethal effect of DNA double-strand breaks. Cell Cycle. 2006;5:994–1000. doi: 10.4161/cc.5.9.2722. [DOI] [PubMed] [Google Scholar]
- 48.Vinciguerra P, Godinho SA, Parmar K, Pellman D, D'Andrea AD. Cytokinesis failure occurs in Fanconi anemia pathway-deficient murine and human bone marrow hematopoietic cells. J Clin Invest. 2010;120:3834–3842. doi: 10.1172/JCI43391. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Howlett NG, Taniguchi T, Durkin SG, D'Andrea AD, Glover TW. The Fanconi anemia pathway is required for the DNA replication stress response and for the regulation of common fragile site stability. Hum Mol Genet. 2005;14:693–701. doi: 10.1093/hmg/ddi065. [DOI] [PubMed] [Google Scholar]
- 50.Keller G, Brassat U, Braig M, Heim D, Wege H, Brummendorf TH. Telomeres and telomerase in chronic myeloid leukaemia: impact for pathogenesis, disease progression and targeted therapy. Hematol Oncol. 2009;27:123–129. doi: 10.1002/hon.901. [DOI] [PubMed] [Google Scholar]
- 51.Callen E, Samper E, Ramirez MJ, Creus A, Marcos R, Ortega JJ, et al. Breaks at telomeres and TRF2-independent end fusions in Fanconi anemia. Hum Mol Genet. 2002;11:439–444. doi: 10.1093/hmg/11.4.439. [DOI] [PubMed] [Google Scholar]
- 52.Li J, Du W, Maynard S, Andreassen PR, Pang Q. Oxidative stress-specific interaction between FANCD2 and FOXO3a. Blood. 115:1545–1548. doi: 10.1182/blood-2009-07-234385. 1545. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Ghaffari S, Jagani Z, Kitidis C, Lodish HF, Khosravi-Far R. Cytokines and BCR-ABL mediate suppression of TRAIL-induced apoptosis through inhibition of forkhead FOXO3a transcription factor. Proc Natl Acad Sci U S A. 2003;100:6523–6528. doi: 10.1073/pnas.0731871100. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Ikeda H, Matsushita M, Waisfisz Q, Kinoshita A, Oostra AB, Nieuwint AW, et al. Genetic reversion in an acute myelogenous leukemia cell line from a Fanconi anemia patient with biallelic mutations in BRCA2. Cancer Res. 2003;63:2688–2694. [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.