Abstract
The influenza A M2 protein forms a proton channel for virus infection and also mediates virus assembly and budding. The minimum protein length that encodes both functions contains the transmembrane (TM) domain (roughly residues 22 to 46) for the amantadine-sensitive proton-channel activity and an amphipathic cytoplasmic helix (roughly residues 45 to 62) for curvature induction and virus budding. However, structural studies involving the TM domain with or without the amphipathic helix differed on the drug-binding site. Here we use solid-state NMR spectroscopy to determine the amantadine binding site in the cytoplasmic-helix-containing M2(21–61). 13C-2H distance measurements of 13C-labeled protein and 2H-labeled amantadine showed that in DMPC bilayers, the first equivalent of drug bound S31 inside the M2(21–61) pore, similar to the behavior of M2TM in DMPC bilayers. The non-specific surface site of D44 observed in M2TM is disfavored in the longer peptide. Thus, the pharmacologically relevant drug-binding site in the fully functional M2(21–61) is S31 in the TM pore. Interestingly, when M2(21–61) was reconstituted into a virus-mimetic membrane containing 30% cholesterol, no chemical shift perturbation was observed for pore-lining residues, while M2TM in the same membrane exhibited drug-induced chemical shift changes. Reduction of the cholesterol level and the use of unsaturated phospholipids shifted the conformational equilibrium of M2TM fully to the bound state, but did not rescue drug binding to M2(21–61). These results suggest that the amphipathic helix, together with cholesterol, modulates the ability of the TM helices to bind amantadine. Thus, the M2 protein interacts with the lipid membrane and small-molecule inhibitors in a complex fashion, and a careful examination of the environmental dependence of the protein conformation is required to fully understand the structure-function relation of this protein.
Introduction
The M2 protein of the influenza A virus is important for the virus lifecycle. The well-studied proton channel activity of M2 manifests itself prominently in the early stage of infection, when virus endocytosis into the acidic host cell endosome opens the tetrameric proton channel and acidifies the virus interior, causing the release of the ribonucleoprotein into the host cell.1,2 Binding of the antiviral drugs amantadine (Amt) and rimantadine (Rmt) inhibits the proton channel activity. The transmembrane (TM) domain of the protein, roughly spanning residues 22 to 46, is the core of the proton channel function.3 It contains the drug-binding residue S31,4,5 the pH-sensing and proton-selective residue H37,6,7 and the channel-gating residue W41.8 In the second function of M2, the cytoplasmic tail C-terminal to the TM domain binds the matrix protein M1 and cholesterol, and mediates virus assembly and budding.9,10 Electron microscopy and mutagenesis data showed that an amphipathic helix in the cytoplasmic tail, roughly spanning residues 45 to 62, is both sufficient and necessary for membrane scission of the newly assembled virus from the host cell.11,12 Simultaneous mutation of five hydrophobic residues in this amphipathic helix to Ala inhibited filamentous virion formation, virus budding and membrane scission.11,12
While the functional role of M2 in virus assembly and budding is now clear, questions linger about the role of the amphipathic helix in M2’s proton channel function. While a construct containing both the TM and amphipathic helices showed the same single-channel conductivity as the full-length protein, the TM peptide had about half the activity.3 However, the M2 transmembrane peptide (M2TM) was poorly expressed in oocytes, giving high uncertainty to the conductivity result. Indeed, the penta-Ala mutant of the full-length protein exhibited the same proton conductivity as the wild-type protein, suggesting that the amphipathic helix was not required for the proton channel activity.3 Debate about the role of the amphipathic cytoplasmic helix in the proton channel function was also fueled by two opposing high-resolution structures of drug-complexed M2. A crystal structure of Amt-bound M2TM(22–46) found Amt electron densities in the N-terminal pore, suggesting a physical occlusion mechanism for inhibition.5 In contrast, a solution NMR structure of Rmt-bound M2(18–60) in DHPC micelles found drug-protein nuclear Overhauser (NOE) cross peaks for residues on the C-terminal lipid-facing surface of the TM helical bundle,13 suggesting an allosteric inhibition mechanism. A subsequent solid-state NMR study of M2TM in DMPC bilayers partly resolved this discrepancy, showing that the surface binding site was populated only by excess drugs from the membrane side, whereas the first equivalent of drug bound the N-terminal pore of the channel with much higher affinity,4 consistent with functional data.14,15 Nevertheless, it remains unclear why no pore-bound drug was detected in the solution NMR structure of M2(18–60), and the longer protein length used in that study was often cited to justify the relevance of the surface binding site.16,17 Recently, the orientation of M2(22–62) in lipid bilayers was determined by solid-state NMR 18 and EPR constraints:19 the amphipathic helix was found to lie parallel to the membrane and pack closely against the TM helix, suggesting interactions between the two domains.
In this work, we use solid-state NMR (SSNMR) spectroscopy to answer two questions. First, does Amt bind to the pore of the fully functional cytoplasmic-helix-containing M2 construct in DMPC bilayers? Second, does the lipid bilayer composition influence the conformational equilibrium of the long peptide with respect to drug binding? We directly measured protein-drug contacts in DMPC-reconstituted M2(21–61) to test the existence of the high-affinity binding site in the pore. We also investigated M2 conformation and drug binding in two virus-mimetic membranes containing varying amounts of cholesterol, sphingomyelin (SPM) and glycerophospholipids.20 Our results show that the virus-mimetic membranes shift the conformation equilibrium of the longer M2 construct to the unbound state while retaining the bound conformation of the shorter M2TM. Thus, the amphipathic helix, through a complex interplay with cholesterol, modulates the TM helix conformation to facilitate or weaken drug binding to the channel pore.
Materials and Methods
Membrane samples for SSNMR experiments
Two M2 constructs, M2TM (residues 22–46) and M2(21–61) were synthesized using Fmoc solid-phase chemistry (PrimmBiotech, Cambridge, MA) and purified to >95% purity. Uniformly 13C, 15N-labeled amino acids (Sigma-Aldrich and Cambridge Isotope Laboratories) were incorporated at residues V27, S31, G34 and D44. The first three labeled residues test the pore-binding site, whereas the labeled D44 tests the presence of the surface binding site. Most other residues implicated in surface binding by the solution NMR study13 showed longer distances to Rmt than D44, and thus were not labeled. Unlabeled peptides were used for static 2H quadrupolar echo experiments to determine the number of drugs bound to the channel and the effect of membrane composition on drug binding.
The M2 peptides were reconstituted into lipid membranes by detergent dialysis. For the 13C, 15N-labeled peptides, the peptide : lipid molar ratios were 1:8 for M2TM and 1:15 for M2(21–61), which corresponded to similar mass ratios of ~ 1 : 2. Three lipid membranes were used in this study: 1,2-dimyristoyl-sn-glycero-3-phosphocholine (DMPC) bilayer and two mixed membranes mimicking the virus envelope lipid composition to different extents. The virus-mimetic (VM) membrane is composed of 1,2-dipalmitoyl-sn-glycero-3-phosphocholine (DPPC), 1,2-dipalmitoyl-sn-glycero-3-phosphoethanolamine (DPPE), egg SPM, which contains predominantly saturated palmitoyl chains, and cholesterol at a molar ratio of 21% : 21% : 28% : 30%. The modified virus-mimetic (VM+) membrane contains 1-palmitoyl-2-oleoyl-sn-glycero-3-phosphocholine (POPC), 1-palmitoyl-2-oleoyl-sn-glycero-3-1-palmitoyl-2-oleoyl-sn-glycero-3-phosphoethanolamine (POPE), SPM and cholesterol at a molar ratio of 25.6% : 25.6% : 25.6% : 23%. Thus, the cholesterol mole fraction is moderately reduced in the VM+ membrane.
For the mixed membrane samples, the lipids were codissolved in chloroform and methanol and then dried under a stream of nitrogen gas to remove the bulk of the organic solvents. The film was redissolved in cyclohexane, frozen and lyophilized to obtain a completely dry homogeneous powder. This lipid powder was suspended in a pH 7.5 phosphate buffer (10 mM Na2HPO4/NaH2PO4, 1 mM EDTA, 0.1 mM NaN3) and freeze-thawed 6 times to produce a uniform vesicle suspension. The peptides were reconstituted into the lipid vesicles by dialysis using octylglucoside.21 The proteoliposome mixtures were centrifuged at 150,000 g to obtain ~40% hydrated membrane pellets, which were packed in 4 mm rotors for solid-state NMR experiments. Perdeuterated amantadine (d15-Amt) was directly titrated into the membrane pellet. After pellet formation and drug addition, samples for static 2H NMR experiments were lyophilized and rehydrated to ~40% with 2H-depleted water to ensure that d15-Amt was the only source of the 2H NMR signal. For 13C-2H REDOR experiments, d15-Amt was added at a ratio of 1 or 5 drugs per tetramer, corresponding to drug/lipid molar ratios of 1 : 60 or 1 : 12, respectively.
Solid-state NMR experiments
Solid-state NMR spectra were acquired on two spectrometers (Bruker Biospin) operating at field strengths of 14.1 Tesla and 9.4 Tesla. All static 2H experiments were conducted on the 14.1 Tesla spectrometer at a 2H Larmor frequency of 92.12 MHz. The quadrupolar-echo experiment used a pre-echo delay of 40–50 μs, an 8 μs shorter post-echo delay, and a 2H 90° pulse of 3.8 μs. The 2H time signal was left-shifted appropriately to capture the echo maximum before Fourier transformation to give spectra with a flat baseline. The spectra were measured at 303 K with 30,000 – 210,000 scans.
2D 13C-13C and 15N-13C correlation experiments were carried out using a 4 mm 1H/13C/15N magic-angle-spinning (MAS) probe. The temperatures were 243 K for DMPC samples and 273 K for virus-mimetic membrane samples. Typical MAS frequencies were 7 kHz. The 2D 13C-13C experiments used the DARR pulse sequence 22 while the 2D 15N-13C experiments used rotational-echo-double-resonance (REDOR) for polarization transfer between 15N and 13C.23
13C-detected 2H-dephased REDOR experiments on DMPC-bound M2 peptides 24 were carried out using a 4 mm 1H/13C/2H MAS probe on the 9.4 Tesla NMR spectrometer operating at 13C and 2H Larmor frequencies of 100.71 MHz and 61.48 MHz, respectively. The samples were spun at 4250 Hz at 243 K. At this temperature the protein was immobile but the drug remained dynamic.4 Most REDOR experiments involved a single selective 13C 180° pulse in the center of the mixing period and multiple 2H 180° pulses (12.4 μs long) spaced at half a rotor period apart. This experiment removes 13C-13C scalar coupling and gives long 13C T2 relaxation times to allow dipolar dephasing to be measured at long mixing times.25 An alternative REDOR version containing a single 2H composite 90°90°90° pulse and multiple 13C hard 180° pulses 4 was conducted for the spectrum in Fig. 3b. The mixing times and number of scans used for the REDOR experiments are listed in Table S1. The error bar ε for the REDOR dephasing S/S0 was propagated from the signal-to-noise ratios (sino) of the control (S0) and dephased (S) spectra using the equation .
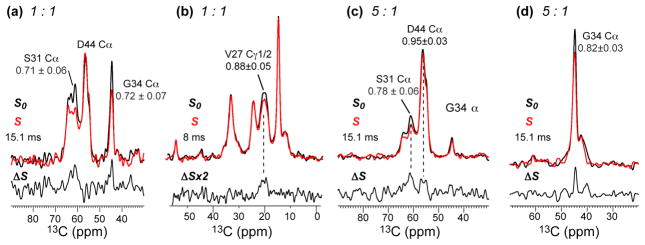
Fig. 3.
13C-2H REDOR spectra detecting the Amt binding site in DMPC-reconstituted M2(21–61). (a, b) 1 Amt per tetramer. (a) Cα region showing S31 dephasing and no D44 dephasing. The spectrum was measured using the single-13C-pulse REDOR. (b) Sidechain region showing V27 Cγ1 dephasing, measured using the single 2H-pulse REDOR. (c, d) 5 Amt per tetramer. (c) Cα region showing S31 dephasing and minimal D44 dephasing. (d) G34 Cα region showing significant dephasing. (c, d) were measured using the single 13C-pulse REDOR. S0 and S denote control and dephased spectra measured without and with the 2H pulses, while ΔS indicates the difference spectrum.
Results
Drug binding to M2(21–61) in DMPC bilayers
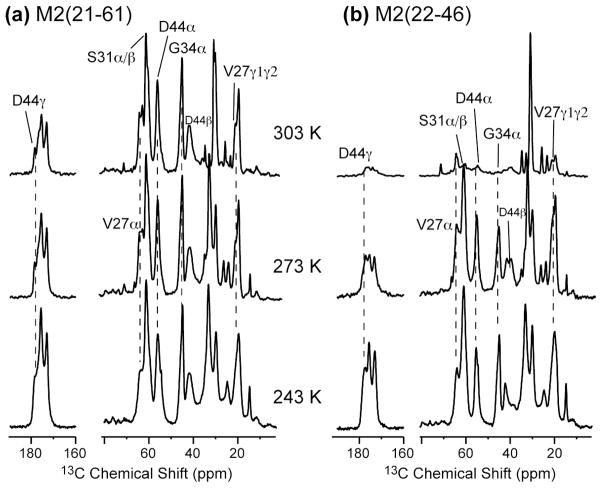
In low-melting one-component phosphocholine membranes such as DLPC and POPC bilayers, M2TM tetramers undergo uniaxial diffusion on an intermediate timescale at ambient temperature,26–29 severely broadening the NMR spectra. To determine if the longer M2(21–61) retains this fast rotational diffusion, we measured its 13C spectra as a function of temperature in the DMPC bilayer, whose main phase transition occurs at 296 K. Fig. 1 compares the 13C CP-MAS spectra of the short and long M2 constructs from 303 K to 243 K. At 303 K, M2TM only exhibited 6–10% of the 243 K intensities (Fig. 1b) due to uniaxial diffusion of the helical bundle, while M2(21–61) showed 60–70% of the intensities (Fig. 1a). The latter is only modestly lower than the Boltzmann factor of 80% expected between 303 K and 243 K, indicating that at 303 K the M2(21–61) backbone is immobilized in DMPC bilayers and only local small-amplitude motions remain. The backbone immobilization is expected because the membrane-parallel amphipathic helix significantly increases the radius of the tetrameric assembly and should slow down rotational diffusion.30 Potential interactions of the amphipathic helix with phospholipid headgroups may further restrict the tetramer mobility.
Fig. 1.
Variable temperature 13C MAS spectra of (a) M2(21–61) and (b) M2TM(22–46) in DMPC bilayers. M2TM intensities are significantly suppressed at 303 K due to intermediate-timescale rotational diffusion of the tetramers, while M2(21–61) intensities are largely retained at 303 K, indicating immobilization of the tetramers.
To probe how Amt affects the conformation of M2(21–61) in DMPC bilayers, we measured 2D 13C-13C and 15N-13C correlation spectra in the absence and presence of the drug. Amt binding to M2TM has been shown to perturb the chemical shifts of pore-lining residues, especially V27, S31, G34, and H37.20,28,31 Fig. 2(a–c) shows that M2(21–61) exhibits similar chemical shift perturbations as M2(22–46): for example, the V27 Cα peak moved from 64.1 ppm to 61.5 ppm, the S31 15N signal shifted from 114.3 ppm to 120.3 ppm, and the G34 15N peak moved from 107.6 ppm to 110.0 ppm. The similarity of the chemical shift changes suggests that Amt binds at the same location in M2(21–61) as in M2TM.4,21 On the other hand, the conformational equilibrium of M2(21–61) is shifted more towards the drug-free state compared to M2TM. At 5 drugs per tetramer, about 18% of the V27 intensity and 11% of the S31 intensity remained at the unbound positions (Fig. 2c) while only ~5% of the intensities remained at the unbound positions for M2TM (Fig. 2d). This difference suggests that the amphipathic helix may partly interfere with the conformational changes of the TM helices that are necessary for drug binding. Fig. 2d shows that the shorter peptide in the absence of drug already exhibits a mixed S31/V27 peak (with an 15N chemical shift of 118 ppm) close to the bound position (120 ppm for 15N), suggesting that the structure distribution of drug-free M2TM already contains the conformation that resembles the drug-bound state. This observation is consistent with previous studies of M2TM backbone torsion angles 21,31 and MD simulations of the conformational heterogeneity of M2TM in lipid bilayers 32.
Fig. 2.
2D correlation spectra of M2(21–61) and M2TM(22–46) in DMPC bilayers without and with drug. (a–c) 15N-13C (top row) and 13C-13C (bottom row) correlation spectra of M2(21–61) at 243 K. (a) No Amt. (b) With 1 Amt per tetramer. (c) With 5 Amt per tetramer. Note the V27 and S31 chemical shift changes and the lack of D44 chemical shift change upon drug binding. (d) 2D 15N-13C correlation spectra of M2TM(22–46) without (top) and with Rmt (bottom). S31 exhibits large chemical shift changes.
Chemical shift perturbation is only an indirect indicator of the drug-binding site. To obtain definitive evidence for the drug location, we measured distance-dependent dipolar couplings between 13C-labeled M2(21–61) and perdeuterated Amt. These 13C-2H REDOR experiments were carried out under similar conditions to those of M2TM.4 Fig. 3 shows the 13C-2H REDOR spectra of M2(21–61) with 1 or 5 drugs per tetramer. For the 1:1 sample, we observed substantial dephasing for the S31 Cα/Cβ peak, with a normalized intensity ratio S/S0 of 0.71±0.06 at 15 ms (Fig. 3a). G34 Cα and V27 Cγ1 also showed significant dephasing (Fig. 3a, b), indicating that these residues constitute the boundaries of the pore binding site. No dephasing was observed for D44 at this drug concentration, indicating that the first equivalent of drug is not within atomic contact with the protein surface. These results are almost identical to those of M2TM,4 indicating that the amphipathic helix does not affect the drug binding behavior of M2(21–61) in DMPC bilayers.
When the drug concentration increased to 5 per tetramer, the pore-lining residues still showed REDOR dephasing (Fig. 3c, d), thus the pore binding site remains under excess drug.4 However, in contrast to M2TM, D44 Cα in M2(21–61) exhibited little dephasing: the S/S0 value was 0.95±0.03 at 15 ms, which was insignificant. In M2TM, D44 showed a much lower S/S0 value of 0.86±0.02 already at a shorter mixing time of 10.1 ms.4 Thus, Amt binds minimally, if at all, to D44 in the presence of the amphipathic helix. G34 Cα also retains its dephasing, but the S/S0 value is slightly higher in the 5:1 sample than the 1:1 sample (Fig. 3d), suggesting that the height of the drug inside the pore may differ slightly between the two samples due to modulation of the TM helix packing by excess drug 29. We did not attempt to quantify Amt distances to the individual residues because a significant fraction of the drug is outside the pore even at the 1:1 ratio, as shown by 2H spectra below (Fig. 4).
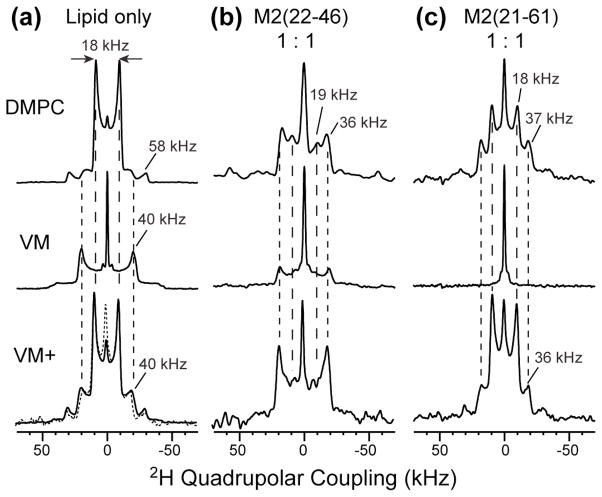
Fig. 4.
Static 2H spectra of d15-Amt at 303 K. The spectra were measured on DMPC bilayer samples (top row), the 30%-cholesterol VM membrane samples (middle row), and the 23%-cholesterol VM+ membrane samples (bottom row). (a) Protein-free lipid bilayers. (b) Membranes containing M2TM at a ratio of 1 drug per tetramer. (c) Membranes containing M2(21–61) at a ratio of 1 drug per tetramer.
Amantadine orientation and dynamics in DMPC-reconstituted M2(21–61)
To further compare the behaviors of drug binding to M2TM versus M2(21–61), we investigated Amt orientation and dynamics using 2H NMR. The 2H spectra were measured at 303 K, near physiological temperature. As shown before, in protein-free DMPC bilayers, the Amt 2H spectrum contains an 18 kHz and 58 kHz splitting at an intensity ratio of 4:1 (Fig. 4a),4 indicating fast uniaxial diffusion or greater than 3-site jumps of the adamantane cage around its 3-fold molecular axis and around the bilayer normal. The molecular axis is tilted from the bilayer normal by either 37° or 80° based on the 2H order parameter. When a stoichiometric number of M2TM tetramers was present, a 36-kHz splitting accounted for ~76% of the total intensities (Fig. 4b), indicating that most drugs adopt a nearly upright orientation (~13° tilt angle) due to their confinement in the channel pore.4 The remaining spectral intensities are located at an 18-kHz splitting (14%) and an isotropic peak (10%) 4. When a stoichiometric number of M2(21–61) tetramers were present, the 2H spectrum became a 47% : 45% : 7% superposition of a 37-kHz splitting, the 18-kHz splitting, and an isotropic peak (Fig. 4c) (simulations not shown). Therefore, only about half of all drug molecules bind to the TM pore of M2(21–61), which is a lower fraction than to M2TM. The recent orientational structure of M2(22–62) showed that the TM helix in the long peptide has a similar tilt angle (32°) and packing as the TM helix in M2TM,18 indicating that the pore geometry is similar between the two constructs. Thus the 18 kHz splitting in the 2H spectrum of the M2(21–61) sample is unlikely to be due to very tilted drug molecules inside the pore, but most likely results from drugs in the lipid phase, similar to the case of the protein-free sample. Since about half of all drugs are located outside the pore and in the bilayer at this 1:1 ratio, the lack of D44 dephasing in the REDOR spectra (Fig. 3a) further supports the loss of the surface binding site in M2(21–61).
Effects of membrane composition on drug binding to M2TM and M2(21–61)
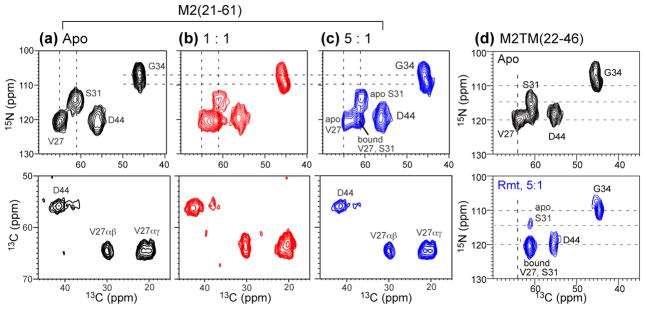
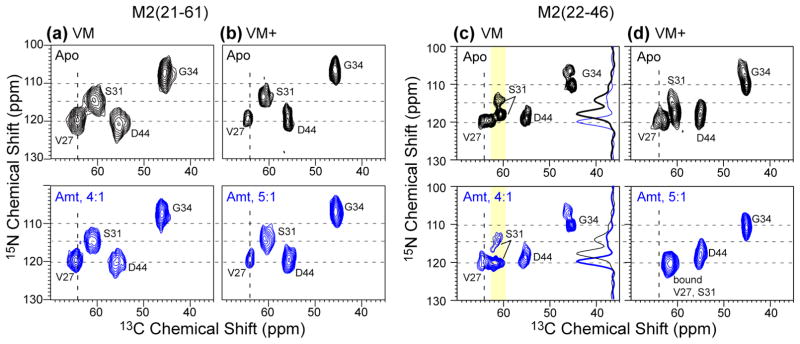
The above results were obtained in DMPC bilayers, whose viscosity near the physiological temperature is intermediate between that of cholesterol-rich membranes and low-melting phosphocholine bilayers. In a virus-mimetic (VM) lipid mixture containing 30% cholesterol, 28% SPM, and 42% saturated DPPC and DPPE lipids,20 we have reported before that M2TM exhibits similar average conformation as in phosphocholine bilayers but different dynamics and conformational equilibrium.31 Since the amphipathic helix is now known to interact with cholesterol to mediate filamentous virion formation11 and virus budding,12 we investigated whether M2(21–61) interacted with Amt differently between the DMPC and VM membranes. The conformation of M2(21–61) in the VM membrane is probed through 13C and 15N chemical shifts and their changes upon drug binding. Fig. 5a shows the 2D 15N-13C correlation spectra of VM-bound M2(21–61) without and with Amt. Strikingly, the chemical shifts were completely unaffected by the drug, in contrast to the shift changes of M2(21–61) in DMPC bilayers (Fig. 2a, c) and the shift changes of M2TM in the VM membrane (Fig. 5c). For example, in the VM membrane, drug-free M2TM showed two S31 15N peaks at 114.5 ppm and 118 ppm with an intensity ratio of ~1:2. Amt binding shifted most of the S31 intensities to 120 ppm, similar to the situation of M2TM in DMPC bilayers.
Fig. 5.
2D 15N-13C correlation spectra of M2(21–61) and M2TM(22–46) in cholesterol-containing membranes without (top row, black) and with drug (bottom row, blue). (a) M2(21–61) in VM membranes. (b) M2(21–61) in VM+ membranes. (c) M2TM in VM membranes. (d) M2TM in the modified VM+ membrane. The cytoplasmic-helix-containing M2(21–61) does not show chemical shift changes by Amt in either membrane, while M2TM shows chemical shift changes in both membranes, and is more significantly perturbed in the VM+ membrane than in the VM membrane.
To test whether the lack of chemical shift changes of VM-bound M2(21–61) indeed results from the lack of drug binding, we measured the 2H spectra of d15-Amt in the VM membrane at 303 K. In the protein-free membrane, the spectrum is dominated (~82%) by a 40 kHz splitting (Fig. 4a), indicating that most drugs are upright while undergoing uniaxial rotation around the bilayer normal. This upright orientation contrasts with the 37° or 80° orientation of the drug in DMPC bilayers, and can be attributed to the high viscosity of the VM membrane. The 40-kHz splitting appears to be distinct from the ~36 kHz splitting of the pore-bound drug, which corresponds to a ~13° tilt angle.4 The protein-free VM membrane also shows a sharp isotropic peak (18% of the total intensity) with a linewidth of < 0.8 kHz, suggesting that a small fraction of Amt partitions into either the inter-lamellar aqueous solution or lipid phases with isotropic symmetry, such as micelles, small vesicles, or the cubic phase.
When a stoichiometric number of M2TM tetramers was present, the 2H spectrum was very similar to the protein-free spectrum, except that the 40-kHz splitting accounted for a smaller fraction (~75%) of the total intensity (Fig. 4b). In dramatic contrast, when a stoichiometric number of M2(21–61) tetramers was present, the 40-kHz splitting disappeared altogether, leaving only the isotropic peak in the spectrum (Fig. 4c), indicating that the drug no longer lies within the anisotropic environment of the lamellar bilayer. Thus, the lack of chemical shift perturbation of VM-bound M2(21–61) is correlated with the exclusion of the drug from the lamellar bilayer (Table S2). Since the same protein : lipid molar ratio (1:15) was used for the VM and DMPC samples, the absence of chemical shift changes of the VM-bound M2(21–61) cannot be attributed to the lack of protein-free membrane surface. Indeed, when the protein concentration was reduced by a factor of two (P/L = 1:30), we observed the same isotropic 2H spectrum (Fig. S1), confirming that drug exclusion from the lamellar bilayer occurs even when there is sufficient free membrane surface.
To determine whether the loss of drug binding to M2(21–61) is due to the high viscosity of the VM membrane, we changed the VM membrane composition by replacing DPPC and DPPE with unsaturated POPC and POPE, and reducing the cholesterol concentration from 30% to 23%. In the protein-free VM+ membrane, the 2H spectra of the drug resumed the 18-kHz splitting seen in DMPC bilayers (Fig. 4a), indicating that the drug is able to adopt the same tilted orientation in the VM+ membrane as in the fluid DMPC bilayers. When M2TM tetramers were present at the 1:1 ratio, the 2H spectrum was a 82% : 11% : 7% superposition of a 37 kHz, 14 kHz splitting and an isotropic peak (Fig. 4b). Thus, the drug is predominantly upright, indicating its binding to the pore of the VM+ bound M2TM channels. This conclusion is further supported by the 2D correlation spectra, which showed clear drug-induced chemical shift changes, with almost no unbound signals of S31, V27 and G34 left (Fig. 5d, S2b). In contrast, when a stoichiometric number of M2(21–61) tetramers were present in the VM+ membrane, the 2H spectrum showed only a low intensity (16% of the whole spectrum) of the 36-kHz splitting, and 2D correlation spectra exhibited very little chemical shift changes (Fig. 5b, S2a). Therefore, while the fluid VM+ membrane facilitated drug binding to M2TM, it did not rescue drug binding to M2(21–61) (Table S4), suggesting that cholesterol may have site-specific interactions with the amphipathic helix to affect the drug binding equilibrium to the TM pore.
Discussion
The above SSNMR data not only resolve the lingering issue about how Amt interacts with the influenza M2 protein but also reveal complex interactions among the membrane composition, drug, and protein length. Direct 13C-2H REDOR distance data of drug-complexed M2(21–61) in DMPC bilayers show unambiguously that Amt binds S31 inside the TM pore, similar to M2TM in the same membrane. The presence of this amphipathic helix shifted the drug-binding equilibrium more towards the unbound state compared to M2TM: 2H spectra of the drug indicate that about 47% of the first equivalent of drug bound M2(21–61) while 76% of the drug bound M2TM (Fig. 4b, c). The different binding equilibria are consistent with the higher remaining intensities of the unbound peaks in the spectra of the longer peptide (Fig. 2c). The reduced affinity of the drug for the M2(21–61) pore compared to M2TM can be attributed to the coverage of the membrane surface by the amphipathic helix. The recent orientational study of M2(22–62) showed that the four cytoplasmic helices together cover an area of ~1000 Å2.18 Given the phosphocholine headgroup area of ~65 Å2, the P : L molar ratio of 1:15 used for our samples, and the two bilayer surfaces, about 50% of each bilayer surface is covered by the cytoplasmic helices. Thus, drug access to the N-terminal pore should be more restrictive for M2(21–61) than for M2TM(22–46). Despite this reduced access, the 13C-2H REDOR data indicate that Amt is still able to partition into the N-terminal pore, and is within atomic contacts (< ~6 Å) with S31, V27 and G34, while the lipid-associated drug no longer has significant contact with D44 (Fig. 3c).
A recent solid-state NMR study of M2(18–60) in POPC bilayers also showed two sets of peaks that correspond to the unbound and bound conformations, whose relative intensities depended on the Rmt concentration: with 1 Rmt per tetramer, only ~25% of the intensities lie in the bound state.16 In comparison, the equilibrium of Amt-complexed M2(21–61) in DMPC bilayers is shifted much more towards the bound state: at the 1:1 drug : tetramer ratio, 50–70% of the protein is in the bound state based on the S31 signals (Fig. 2b). In addition, D44 in M2(21–61) did not exhibit noticeable chemical shift perturbation by Amt (Fig. 2) but was perturbed by Rmt in the previous study. These differences cannot be attributed to chain length difference between DMPC and POPC bilayers 33, because previous studies of the effects of the membrane thickness on M2 conformational equilibria 20,31 indicate that the thicker POPC bilayer should favor drug binding than the thinner DMPC bilayer. Instead, the Amt-Rmt binding difference may reflect a lower affinity of Rmt to the pharmacologically relevant pore binding site but a higher affinity to the surface site. This proposal is supported by the 2H spectra of d15-Rmt versus d15-Amt bound to DMPC-reconstituted M2TM at the 1:1 ratio: ~27% of Rmt was found in the lipid-bound orientation 34 while only 14% of Amt partitioned into the lipid bilayer 4.
The REDOR spectra (Fig. 3) and the drug 2H spectra (Fig. 4) of the DMPC samples indicate that Amt has little affinity to the surface D44 site in M2(21–61), in contrast to M2TM. This observation is consistent with surface plasmon resonance data of full-length M2, which showed that the drug affinity for the surface site is ~400 fold weaker than for the channel pore.4,35 This ratio is much larger than the 40-fold affinity difference for M2TM estimated from the 2H NMR spectra. 4 As suggested by the orientation study of M2(22–62), 18 the lower drug affinity for D44 in the longer construct is most likely due to the position and orientation of the amphipathic helix. By packing closely to the TM helix, this amphipathic helix may block drug access to D44 at the end of the TM domain. Thus, solid-state NMR data and biochemical evidence both indicate that D44 is unrelated to drug inhibition in the functionally intact M2 protein.
Given the strong affinity of Amt to the channel pore of both short and long M2 constructs in DMPC bilayers, the loss of drug binding to M2(21–61) in the VM and VM+ membranes (Fig. 4, 5) is striking, and gives an example of the dramatic influence of the lipid environment on the functional structures of membrane proteins (Table S4). We attribute the absence of drug binding to a combination of the high viscosity of the membrane and specific interactions of cholesterol with the amphipathic helix. High viscosity of the VM membrane reduces protein conformational plasticity, as evidenced by the much larger order parameters of M2TM in VM than in phosphocholine membranes.20 The importance of conformational plasticity for M2 function and drug binding has been well documented 20,32,37–39 and is evidenced here by the fact that M2TM binds drug more fully in the fluid VM+ membrane than in the VM membrane (Fig. 4, 5).20,31,36–38 However, the VM+ membrane did not rescue drug binding to M2(21–61) (Fig. 5a, b): the scenario that the drug may bind the pore but not cause chemical shift changes is unlikely. Thus, conformational rigidity alone is insufficient for explaining the loss of drug binding to M2(21–61). Rather, specific interactions between cholesterol and the amphipathic helix appears to be present that perturb the TM helix packing and hence the drug binding equilibrium. The N-terminal half of the cytoplasmic helix contains a cholesterol recognition amino acid consensus (CRAC) motif 39, and M2 preparations from influenza virus-infected cells were found to contain 0.5–0.9 molecules of cholesterol per monomer. Functional studies in cholesterol-free E. coli and cholesterol-free liposomes showed that drug-sensitive proton channel activity does not require cholesterol 39. The current data suggest that not only is cholesterol not required for proton transport, but high cholesterol may interfere with the drug sensitivity of the channel activity. In liposomes composed of POPC, POPG and cholesterol at a 4:1:2 mole ratio, M2(18–60) exhibits Amt-sensitive proton fluxes.3 Thus, at sufficiently low cholesterol levels, the conformational equilibrium of the cytoplasmic-helix-containing M2 may change to allow Amt and Rmt binding to the pore. The ability of the cytoplasmic helix, through interactions with the membrane, to impact TM helix packing sheds light on the possible reason for the lack of pore-bound drug in the solution NMR structure of M2(18–60) in DHPC micelles.13 Although the detergent environment differs drastically from the virus-mimetic membranes, the cytoplasmic helix may be affected by DHPC micelles in such a way as to similarly alter the TM helix packing and abolish drug binding.
The sensitive dependence of M2 conformation and drug interaction on the lipid composition raises the question of what is the best membrane to use that reproduces the full panel of M2’s functions. The current data suggest that reproducing the average lipid composition of the virus envelope 40,41 may not be sufficient for M2(21–61). The spatial distribution of lipids in the virus envelope is heterogeneous, and functional studies have shown that M2 is localized to the boundary of lipid rafts,39 which may have intermediate viscosities not unlike those of DMPC bilayers. Indeed, the results here indicate that DMPC bilayers are surprisingly apt in producing TM helix conformations that are competent for drug binding in both lengths of the M2 protein.3 Thus, our data validate the use of DMPC-based membranes in many solid-state NMR studies of M2TM and M2(21–61) so far.4,21,42 The fact that M2(21–61) does not bind Amt in the VM membrane and is unlikely to bind Amt in the VM+ membrane, indicates a complex interplay between cholesterol and the amphipathic helix, which affects the structure and assembly of the TM helices. Future work directly interrogating the lipid interactions of M2(21–61) and the detailed phase properties of cholesterol-containing membranes will be useful for elucidating the lipid-dependent conformational equilibria of the M2 protein.
Supplementary Material
Acknowledgments
This work is supported by NIH grant GM088204. The authors thank Dr. Jun Wang (UPenn) for providing d15-Amt.
Footnotes
Supporting Information Available
Tables of REDOR experimental conditions, estimates of the fraction of bound drug, 31P chemical shift principal values, and additional NMR spectra. This information is available free of charge via the Internet at http://pubs.acs.org/.
References
- 1.Cady SD, Luo WB, Hu FH, Hong M. Biochemistry. 2009;48:7356–7364. doi: 10.1021/bi9008837. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Pinto LH, Lamb RA. J Biol Chem. 2006;281:8997–9000. doi: 10.1074/jbc.R500020200. [DOI] [PubMed] [Google Scholar]
- 3.Ma C, Polishchuk AL, Ohigashi Y, Stouffer AL, Schoffin A, Magavern E, Jing X, Lear JD, Freire E, Lamb RA, DeGrado WF, Pinto LH. Proc Natl Acad Sci USA. 2009;106:12283–12288. doi: 10.1073/pnas.0905726106. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Cady SD, Schmidt-Rohr K, Wang J, Soto CS, DeGrado WF, Hong M. Nature. 2010;463:689–692. doi: 10.1038/nature08722. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Stouffer AL, Acharya R, Salom D, Levine AS, Di Costanzo L, Soto CS, Tereshko V, Nanda V, Stayrook S, DeGrado WF. Nature. 2008;451:596–599. doi: 10.1038/nature06528. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Wang C, Lamb RA, Pinto LH. Biophys J. 1995;69:1363–1371. doi: 10.1016/S0006-3495(95)80003-2. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Hu F, Luo W, Hong M. Science. 2010;330:505–509. doi: 10.1126/science.1191714. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Tang Y, Zaitseva F, Lamb RA, Pinto LH. J Biol Chem. 2002;277:39880–39886. doi: 10.1074/jbc.M206582200. [DOI] [PubMed] [Google Scholar]
- 9.McCown MF, Pekosz A. J Virol. 2006;80:8178–8189. doi: 10.1128/JVI.00627-06. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Zhang J, Leser GP, Pekosz A, Lamb RA. Virology. 2000;269:325–334. doi: 10.1006/viro.2000.0228. [DOI] [PubMed] [Google Scholar]
- 11.Rossman JS, Jing X, Leser GP, Balannik V, Pinto LH, Lamb RA. J Virol. 2010;84:5078–5088. doi: 10.1128/JVI.00119-10. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Rossman JS, Jing X, Leser GP, Lamb RA. Cell. 2010;142:902–913. doi: 10.1016/j.cell.2010.08.029. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Schnell JR, Chou JJ. Nature. 2008;451:591–595. doi: 10.1038/nature06531. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Jing X, Ma C, Ohigashi Y, Oliveira FA, Jardetzky TS, Pinto LH, Lamb RA. Proc Natl Acad Sci U S A. 2008;105:10967–10972. doi: 10.1073/pnas.0804958105. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Ohigashi Y, Ma C, Jing X, Balannick V, Pinto LH, Lamb RA. Proc Natl Acad Sci U S A. 2009;106:18775–18779. doi: 10.1073/pnas.0910584106. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Andreas LB, Eddy MT, Pielak RM, Chou JJ, Griffin RG. J Am Chem Soc. 2010;132:10958–10960. doi: 10.1021/ja101537p. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Pielak RM, Schnell JR, Chou JJ. Proc Natl Acad Sci U S A. 2009;106:7379–7384. doi: 10.1073/pnas.0902548106. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Sharma M, Yi M, Dong H, Qin H, Peterson E, Busath D, Zhou HX, Cross TA. Science. 2010;330:509–512. doi: 10.1126/science.1191750. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Nguyen PA, Soto CS, Polishchuk A, Caputo GA, Tatko CD, Ma C, Ohigashi Y, Pinto LH, DeGrado WF, Howard KP. Biochemistry. 2008;47:9934–9936. doi: 10.1021/bi801315m. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Luo W, Cady SD, Hong M. Biochemistry. 2009;48:6361–6368. doi: 10.1021/bi900716s. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Cady SD, Mishanina TV, Hong M. J Mol Biol. 2009;385:1127–1141. doi: 10.1016/j.jmb.2008.11.022. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Takegoshi K, Nakamura S, Terao T. Chem Phys Lett. 2001;344:631–637. [Google Scholar]
- 23.Hong M, Griffin RG. J Am Chem Soc. 1998;120:7113–7114. [Google Scholar]
- 24.Gullion T, Schaefer J. J Magn Reson. 1989;81:196–200. [Google Scholar]
- 25.Jaroniec CP, Tounge BA, Herzfeld J, Griffin RG. J Am Chem Soc. 2001;123:3507–3519. doi: 10.1021/ja003266e. [DOI] [PubMed] [Google Scholar]
- 26.Song Z, Kovacs FA, Wang J, Denny JK, Shekar SC, Quine JR, Cross TA. Biophys J. 2000;79:767–775. doi: 10.1016/S0006-3495(00)76334-X. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Cady SD, Goodman C, Tatko C, DeGrado WF, Hong M. J Am Chem Soc. 2007;129:5719–5729. doi: 10.1021/ja070305e. [DOI] [PubMed] [Google Scholar]
- 28.Cady SD, Hong M. Proc Natl Acad Sci USA. 2008;105:1483–1488. doi: 10.1073/pnas.0711500105. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Cady SD, Hong M. J Biomol NMR. 2009;45:185–196. doi: 10.1007/s10858-009-9352-9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Saffman PG, Delbruck M. Proc Natl Acad Sci USA. 1975;72:3111–3. doi: 10.1073/pnas.72.8.3111. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Hu F, Luo W, Cady SD, Hong M. Biochim Biophys Acta. 2011;1808:415–423. doi: 10.1016/j.bbamem.2010.09.014. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Yi M, Cross TA, Zhou HX. Proc Natl Acad Sci U S A. 2009;106:13311–13316. doi: 10.1073/pnas.0906553106. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Duong-Ly KC, Nanda V, DeGrado WF, Howard KP. Protein Sci. 2005;14:856–61. doi: 10.1110/ps.041185805. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Cady SD, Wang J, Wu Y, DeGrado WF, Hong M. J Am Chem Soc. 2011;133:4274–4284. doi: 10.1021/ja102581n. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Rosenberg MR, Casarotto MG. Proc Natl Acad Sci U S A. 2010;107:13866–13871. doi: 10.1073/pnas.1002051107. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Li C, Qin H, Gao FP, Cross TA. Biochim Biophys Acta. 2007;1768:3162–3170. doi: 10.1016/j.bbamem.2007.08.025. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Stouffer AL, Ma C, Cristian L, Ohigashi Y, Lamb RA, Lear JD, Pinto LH, DeGrado WF. Structure. 2008;16:1067–76. doi: 10.1016/j.str.2008.04.011. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Stouffer AL, Nanda V, Lear JD, DeGrado WF. J Mol Biol. 2005;347:169–179. doi: 10.1016/j.jmb.2005.01.023. [DOI] [PubMed] [Google Scholar]
- 39.Schroeder C, Heider H, Möncke-Buchner E, Lin TI. Eur Biophys J. 2005;34:52–66. doi: 10.1007/s00249-004-0424-1. [DOI] [PubMed] [Google Scholar]
- 40.Blough HA. J Gen Virol. 1971;12:317–320. doi: 10.1099/0022-1317-12-3-317. [DOI] [PubMed] [Google Scholar]
- 41.Klenk HD, Becht H, Rott R. Virology. 1972;47:579–591. doi: 10.1016/0042-6822(72)90547-8. [DOI] [PubMed] [Google Scholar]
- 42.Hu J, Asbury T, Achuthan S, Li C, Bertram R, Quine JR, Fu R, Cross TA. Biophys J. 2007;92:4335–4343. doi: 10.1529/biophysj.106.090183. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.