Abstract
Many etiologies of fatty liver disease (FLD) are associated with hyper-activation of one of the three pathways that comprise the unfolded protein response (UPR), a harbinger of endoplasmic reticulum (ER) stress. The UPR is mediated by pathways initiated by PERK, IRE1a/XBP1and ATF6, and each of these pathways have been implicated as either protective or pathological in FLD. We use zebrafish with FLD and hepatic ER stress to explore the relationship between Atf6 and steatosis. Mutation of the foie gras (foigr) gene causes FLD and hepatic ER stress. Prolonged treatment of wild-type larvae with a dose of tunicamycin that causes chronic ER stress phenocopies foigr. In contrast, acute exposure to a high dose of tunicamycin robustly activates the UPR but is less effective at inducing steatosis. The Srebp transcription factors are not required for steatosis in any of these models. Instead, depleting larvae of active Atf6 either through mbtps1 mutation or atf6 morpholino injection protects against steatosis caused by chronic ER stress whereas it exacerbates steatosis caused by acute tunicamycin treatment.
Conclusion
ER stress causes FLD. Loss of Atf6 prevents steatosis caused by chronic ER stress but can also potentiate steatosis caused by acute ER stress. This demonstrates that Atf6 can play both protective and pathological roles in FLD.
Keywords: foie gras, unfolded protein response, steatosis, zebrafish, tunicamycin
Fatty liver disease (FLD) is emerging as a global epidemic, necessitating a comprehensive understanding of its molecular basis. Interestingly, most etiologies of FLD are associated with induction of the unfolded protein response (UPR), likely attributed to a deficit in the protein folding capacity of the endoplasmic reticulum (ER) in FLD. There is a strong, but poorly understood link between UPR activation and lipid accumulation in hepatocytes (steatosis).
UPR function is required by all cells to ensure that proteins in the secretory pathway are efficiently processed (1, 2). The three branches of the UPR are connected through the master chaperone, Bip. The proximal mediators are (i) PERK (also called EIF2AK3) which phosphorylates EIF2S1 (also called EIF2α), repressing protein synthesis and selective translation of ATF4 mRNA (ii) IRE1A (also called ERN1) which splices XBP1 mRNA to encode the XBP1s transcription factor (3) and (iii) the ATF6 transcription factor which cooperates with ATF4 and XBP1s to regulate a panel of genes that maintain ER function (1, 2). Accordingly, there is significant cooperation and crosstalk between branches. When the unfolded protein load is mitigated, homeostasis is achieved and UPR activity returns to baseline. In contrast, when the ER is overwhelmed with unfolded proteins, the UPR is chronically activated in a pathological state termed ER stress. In most cases, UPR activation protects cells by maintaining homeostasis (2). However, prolonged UPR activation in chronic ER stress results in aberrant protein secretion and apoptosis (1, 2).
Upregulation of some or all UPR branches is found in most etiologies of FLD (4–8) and that it contributes to steatosis. Obesity-related steatosis is ameliorated when Eif2s1 phosphorylation is prevented (9) and enhancing protein folding in obese mice results in a dialing down of the UPR, improving hepatic insulin resistance (10, 11). In contrast, other studies indicate that crippling the UPR causes FLD: Xbp1 heterozygosity predisposes mice to developing hepatic insulin resistance (6) and mice lacking Atf6 or Dnajc3 are unable to resolve steatosis caused by an acute block in protein glycosylation (12, 13). Intriguingly, Bip+/− mice are protected from insulin resistance due to compensatory, low-grade UPR activation (14). The theory that UPR activation plays two distinct roles – protective in the setting of acute ER stress and pathological when the UPR is chronically activated - may unify these seemingly disparate data.
The sterol regulatory binding protein (SREBP) transcription factors are the master regulators of triglyceride and cholesterol synthesis in hepatocytes, and are essential for obesity and alcohol induced steatosis (15). In some cells, UPR activation causes SREBP activation (16–18) whereas in others, SREBP expression, activation or function is suppressed by ER stress (12, 18–20). Whether activation of the UPR is coupled to SREBP driven steatosis remains to be determined.
Here, we use three means to induce steatosis in zebrafish larvae and find that each has ER stress. We demonstrate that loss of Atf6 protects against steatosis due to chronic ER stress, but increases steatosis if the insult is acute.
EXPERIMENTAL PROCEEDURES
Zebrafish
Wild-type (TAB5 and TAB14) and mutant lines (foigrhi1532b and mbtps1hi1487) were maintained in accordance with the policies of the Mount Sinai Institutional Animal Care and Use Committee. Mutants were genotyped as described (21). Tg(fabp10:RFP;ela:GFP) fish were obtained from D. Stainier (UCSF).
Morpholinos targeting the initiator ATG of atf6 (gene name si:ch211-199m3.9; 5’-ACATTAAATTCGACGACATTGTGCC-3’), or scap as previously described (22) and a non-targeting control (5’-CCTCTTACCTCAGTTACAATTTATA-3’) were ordered from Gene Tools, LLC (Philomath, OR). Morpholinos were diluted in water to a 0.5 mM stock and ~5 pmol was injected into early embryos. Tunicamycin treatment protocols are detailed in the results.
Oil Red O staining
Whole mount oil red O staining was carried out as described (22). Steatosis was scored in larvae with 3 or more lipid droplets in the liver parenchyma. A Nikon SMZ1500 equipped with a Nikon DS-2M color camera was used to acquire images that were edited using Photoshop.
The amount of oil red O staining per liver cell was quantified using Metamorph Software (Molecular Devices) on cryosections stained with oil red O and DAPI. A region outlining the liver was selected on each brightfield image and oil red O stained particles were selected by color thresholding and counted. The total area occupied by oil red O staining was measured. Each measurement was divided by the number of DAPI stained nuclei within the region. At least 5 sections per fish were measured in at least 3 fish per group.
Histology and Electron Microscopy
At least 4 wild-type and foigr mutant larvae fixed in 4% paraformaldehyde were embedded in plastic as described (23). 4 µm sections were incubated in 0.5% periodic acid, washed, stained with Schiff Reagent (5g/l basic fuchin/0.1 N HCl/0.045 K2S2O5), washed with running tap water and counterstained with hematoxylin. Images were taken on an Olympus BX41 microscope using a Nikon Ds-Ri1 color camera.
TUNEL staining was carried using the Roche In Situ Cell Death Detection Kit as described (24). Hepatocytes were stained with CY3-streptavadin (1:200; Sigma) and nuclei were labeled with DAPI. The percent of apoptotic hepatocytes was calculated for at least 15 sections representing at least 3 fish per group by dividing the number of TUNEL positive hepatocytes by the total number of nuclei on each section.
Samples from 5 days post fertilization (dpf) larvae were fixed and processed for transmission electron microscopy as described (25).
In situ hybridization
Probes were generated by PCR amplification from cDNA generated from 5 dpf RNA using primers listed in Table S1. The bip probe was generated by first creating cDNA using the zbip-3a primer. Nucleotides 1235 to 2260 of†BC063946.1 were amplified using primers bip-5b and†bip-3b. The chop probe was amplified using primers zchop-5c and†zchop-3, spanning nucleotides 248–976 of NM_001082825.1. The dnajc3 probe was amplified using primers zdnajc3-5p and zdnajc3-3p, spanning nucleotides 318–819 of NM_199610. Each fragment was cloned into PCRII (Invitrogen) and sequenced. Probes were created using digoxigenin RNA Labeling Mix (Roche). Whole mount in situ hybridizations were performed as described (24).
Blotting
Five dpf larvae were homogenized in lysis buffer (20 mM Tris pH 7.5, 150 mM NaCl, 1% NP-40, 2mM EDTA, 10% glycerol and protease inhibitors) and diluted to a final concentration of 2% SDS/5% 2-mercaptoethanol. Two embryos were loaded on a 10% polyacrylamide gel, blotted to nitrocellulose, incubated with antibodies recognizing α-tubulin (1:2000, Sigma), Bip (1:3000, Sigma) or phosphorylated Eif2s1 (1:1000, #9721 Cell Signaling) followed by anti-mouse-HRP conjugated secondary antibody (1:1500, Jackson Immunoresearch) and visualized by chemiluminescence using a FujiFilm LAS-3000. Band intensities were quantified using Quantity One software (Bio-Rad).
PCR
RNA was isolated from 5 dpf whole larvae, dissected livers or liver-less carcasses using the Qiagen RNeasy Kit. cDNA was synthesized using Superscript II Reverse Transcriptase (Invitrogen). PCR reactions were performed as described (25). Real time quantitative PCR (qPCR) was performed in triplicate using Roche Sybergreen on the Roche LightCycler 480 System. The ΔCt was calculated by 2−(Ct(target)-Ct(reference)) using ribosomal protein P0 (rpp0) as the reference. Primer specificity (Table S1) was determined by melting curve assessment; some amplicons were sequenced.
Statistics
All experiments were repeated on at least 3 clutches. When data is presented as a percent of control, we calculated either the average or the median and the standard deviation. Statistical tests used include unpaired and paired 2 tailed Student’s t-test, one sample t-test, ANOVA, Fisher’s exact test, or chi square analysis as appropriate.
RESULTS
foigr mutants develop steatosis and hepatic dysfunction
Nearly all zebrafish larvae with a homozygous mutation in the foigr gene develop hepatomegaly (Fig. 1A–B and (25)) and steatosis by 5 dpf (Fig. 1B–C). foigr mutants have other defects such as underdeveloped gut, small head and eye, yolk under-consumption and death by 7 dpf. These phenotypes are common to zebrafish mutants lacking a gene involved in basic cellular processes. However, the phenotype of steatosis in foigr mutants is unusual.
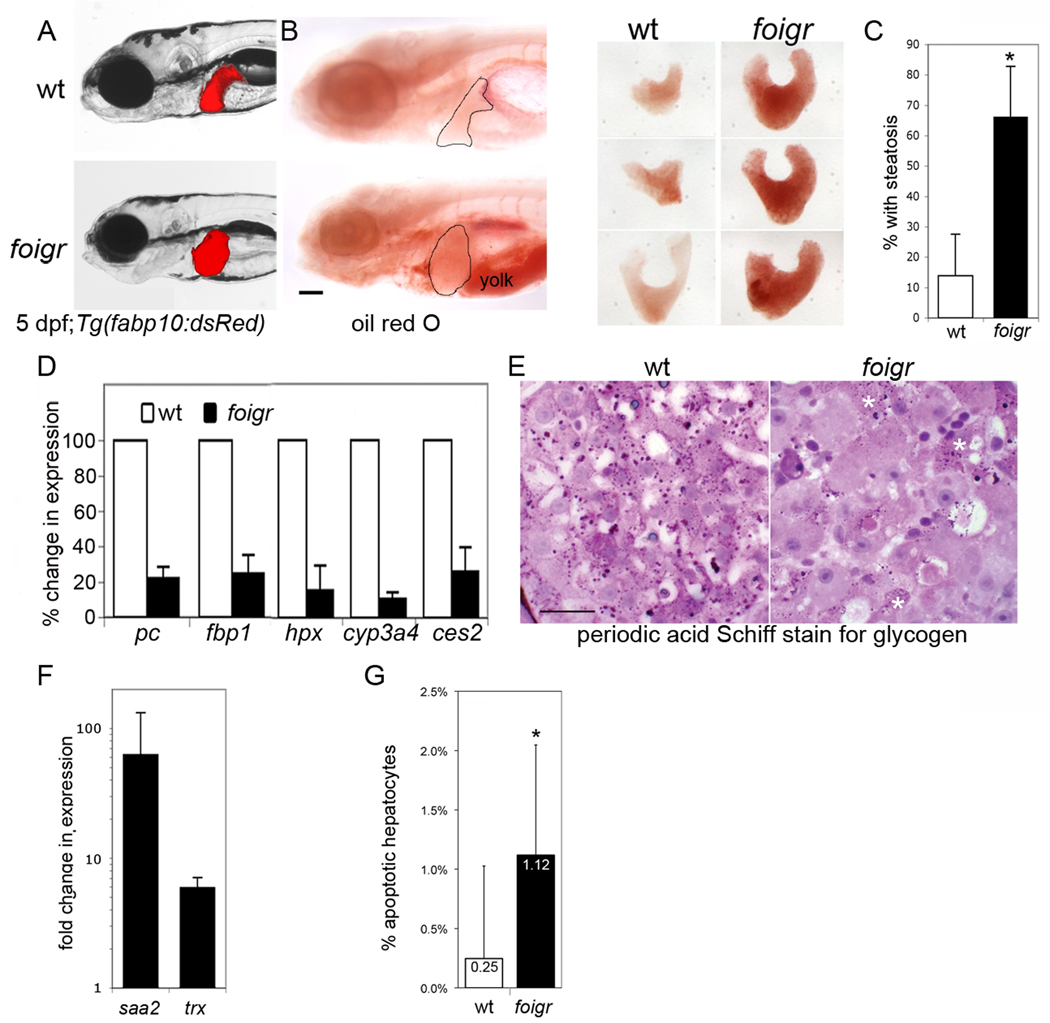
Figure 1. foigr mutants develop steatosis and liver damage by 5 dpf.
A. Live foigr larvae expressing dsRed in hepatocytes (Tg(fabp10:dsRed)) have large, round livers compared to their wild-type (wt) siblings. (left) Steatosis in foigr livers was detected by whole mount oil red O staining. The liver is circled. The incompletely consumed yolk in foigr mutants is labeled. Scale bar = 100 µm. (right). B. Livers dissected from oil red O stained 5 dpf wild-type and foigr larvae. C. Nearly all foigr mutants develop steatosis by 5 dpf. The percent of fish scored positive for steatosis by whole mount oil red O staining was averaged from 5 clutches (n=65 mutants; 70 wild-type; * indicates p<0.001 by a Student’s t-test). D. qPCR analysis of RNA samples isolated from whole 5 dpf larvae. Target gene expression was normalized to rpp0 to determine the ΔCt. The ΔCt in mutants was divided by ΔCt in wild-type siblings to obtain percent change in expression in each clutch and the average for at least 3 clutches is shown. All genes tested were significantly down regulated in mutants (p<0.01 for all samples using a 1 sample t-test). E. Glycogen was detected in sections of wild-type and foigr liver using the periodic acid Schiff stain. The scarce glycogen deposits in foigr hepatocytes are marked by white asterisks. Scale bar = 20 µm. F. Genes induced in response to hepatic damage were assessed by qPCR. The fold change for each clutch was calculated by dividing the ΔCt in mutants by the ΔCt in wild-type siblings and averaged (n>5 clutches; p<0.05 by a one tailed t-test). G. The percent of TUNEL positive hepatocytes on at least 15 sections of wild-type (n=1026) and foigr (n=1005) were averaged. * indicates p=0.0037 by a Student’s t-test. All error bars represent the standard deviation.
Impaired hepatic function, liver damage and hepatocyte death occur in FLD patients. By 5 dpf, foigr mutants have decreased expression of genes involved in key hepatocyte processes (Table S1 contains gene names), including carbohydrate metabolism (pc, fbp), iron transport (hpx) and xenobiotic metabolism (cyp3a4, ces2) (Fig. 1D). Depleted glycogen in foigr mutant hepatocytes (Fig. 1E and Fig. 2A) also suggest impaired hepatocyte function. Both saa2 and trx are significantly upregulated (Fig. 1F) and the 4-fold increase in TUNEL positive cells (Fig. 1G) in foigr mutant livers suggest hepatic damage. Together these data indicate that foigr mutants have steatosis accompanied by decreased liver function, liver damage and hepatocyte apoptosis, similar to patients with FLD. The function of the Foigr protein is unknown, although recent studies suggest a role in the secretory pathway (26–28). Regardless, the interesting phenotype of foigr mutants compelled us to investigate the mechanism of steatosis in this new FLD model.
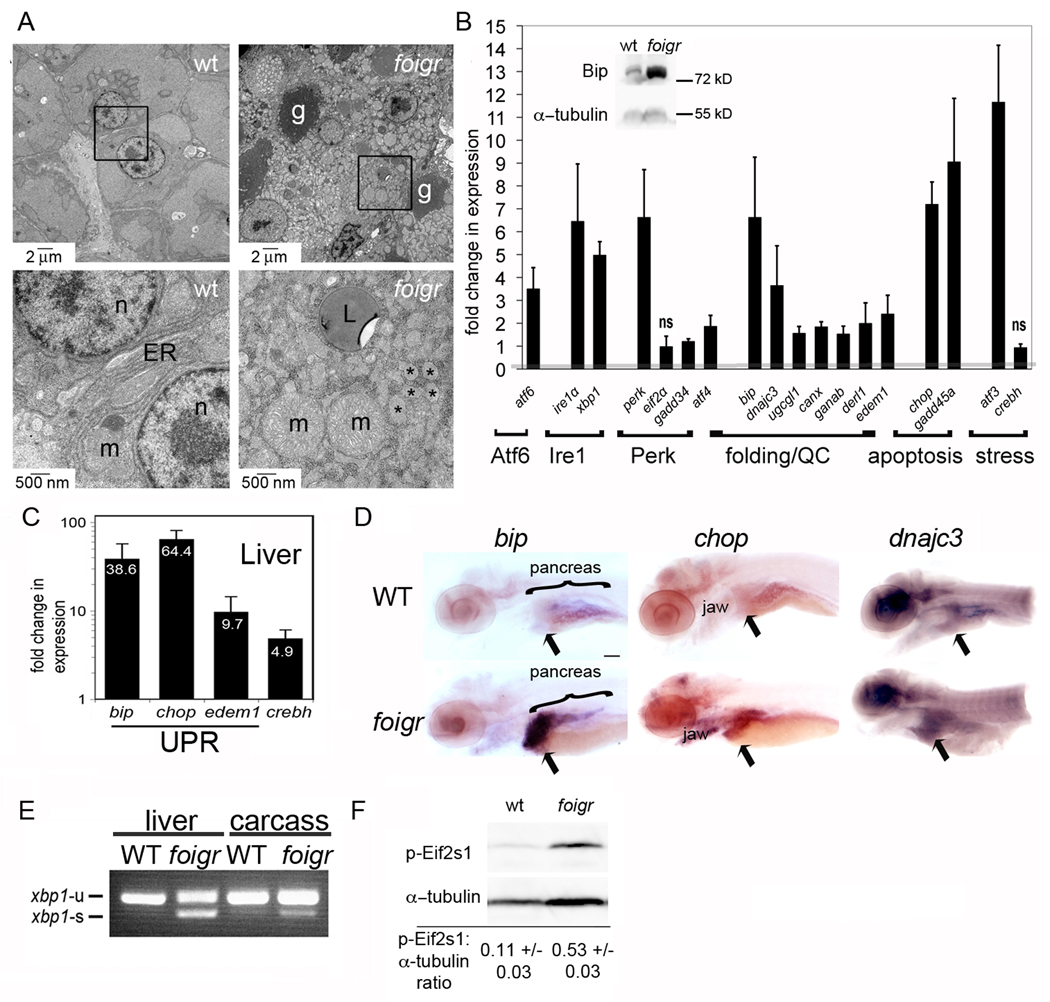
Figure 2. foigr larvae have hepatic ER stress.
A. Electron micrographs of 5 dpf wild-type (left) and foigr (right) livers. The boxed regions in the top panels are magnified in the bottom panels. g: glycogen, m: mitochondria, L: lipid; n: nucleus; * indicates characteristic dilated ER. The cytoplasm of wild-type hepatocytes is full of grey glycogen, whereas only sparse glycogen patches are visible in mutant hepatocytes (g). B. qPCR analysis of UPR gene expression in whole 5 dpf foigr larvae normalized to expression in wild-type siblings (set to 1, indicated by the grey line). The fold change in expression of each gene in mutants compared to their wild-type siblings were averaged for at least 5 clutches and found to be significant using a 1 sample t-test (p<0.01) except where noted as not significant (ns). Genes are grouped by pathway or general function. QC: protein folding quality control. The inset is a representative Western blot of Bip on 5 dpf whole foigr mutants and their wild-type siblings. C. qPCR analysis of a subset of UPR target genes in livers dissected from 5 dpf larvae. The average fold change in 5 clutches of foigr mutant livers normalized to wild-type livers is labeled on each bar. All genes are significantly increased in mutants (p value <0.05 using a 1 sample t-test). D. In situ hybridization for bip, chop and dnajc3 on 5 dpf wild-type (top) and foigr (bottom) larvae. Images are representative of at least 20 embryos from 2 clutches. Staining is observed in the liver (arrow), jaw and exocrine pancreas. Scale bar = 200 µm. E. PCR analysis of xbp1 splicing using primers to detect both unspliced (xbp1-u) and spliced (xbp1-s) xbp1 revealed robust splicing in 5 dpf foigr livers and moderate splicing in the liver-less carcass. Data are representative of 3 experiments. F. Phosphorylated Eif2s1 was detected by Western blotting. The blot was repeated with 6 batches of 5 dpf wild-type and foigr samples and relative band intensity was normalized to α-tubulin, averaged and displayed with the standard deviation; p=0.000008 determined by a t-test. A representative blot is shown. Error bars in all graphs display the standard deviation.
foigr mutation causes hepatic ER stress
ER stress is marked by UPR induction, compromised ER function and abnormal ER structure. However, moderate or partial activation of the UPR may suggest an adaptive response that is maintaining ER function. To differentiate between these possibilities, we assessed ER structure and the activation status of each UPR branch in foigr mutants.
Electron microscopy revealed that wild-type larvae have a granular cytoplasm full of glycogen, scant lipid droplets and perinuclear rough ER (Fig. 2A). In contrast, foigr mutant hepatocytes are enlarged with abundant lipid droplets and scarce glycogen patches (Fig. 2A). The most striking feature of mutant hepatocytes is the grossly dilated ER, resembling the ER in hepatocytes with ER stress due to hepatitis C infection (4) or tunicamycin injection (12).
We next assessed the degree to which each branch of the UPR is activated in foigr mutants. We found upregulation of Bip protein (Fig. 2B inset) and the RNA of major players in each UPR branch and UPR target genes including chaperones (bip, dnajc3), ER quality control (ugcgl1, canx, ganab), ER associated degradation (derl1, edem1) and apoptosis (chop, gadd45a) (Fig. 2B). Many of these genes were expressed at even higher levels in foigr mutant livers (Fig. 2C). In situ hybridization confirmed the enrichment of UPR target genes bip, chop and dnajc3 infoigr livers on 5 dpf (Fig. 2D, arrow), although moderate induction in other tissues was also found. We found robust xbp1 splicing in 5 dpf foigr livers (Fig. 2E) and, to a lesser extent, in the liver-less carcass of foigr mutants. Although Eif2s1 can be phosphorylated by kinases other than Perk, the marked increase in phospho-Eif2s1 in 5 dpf foigr mutants (Fig. 2F) suggests Perk activation. The massive upregulation of each UPR branch and disruption of ER structure unequivocally demonstrate that foigr mutation causes hepatic ER stress.
Tunicamycin causes UPR activation and steatosis
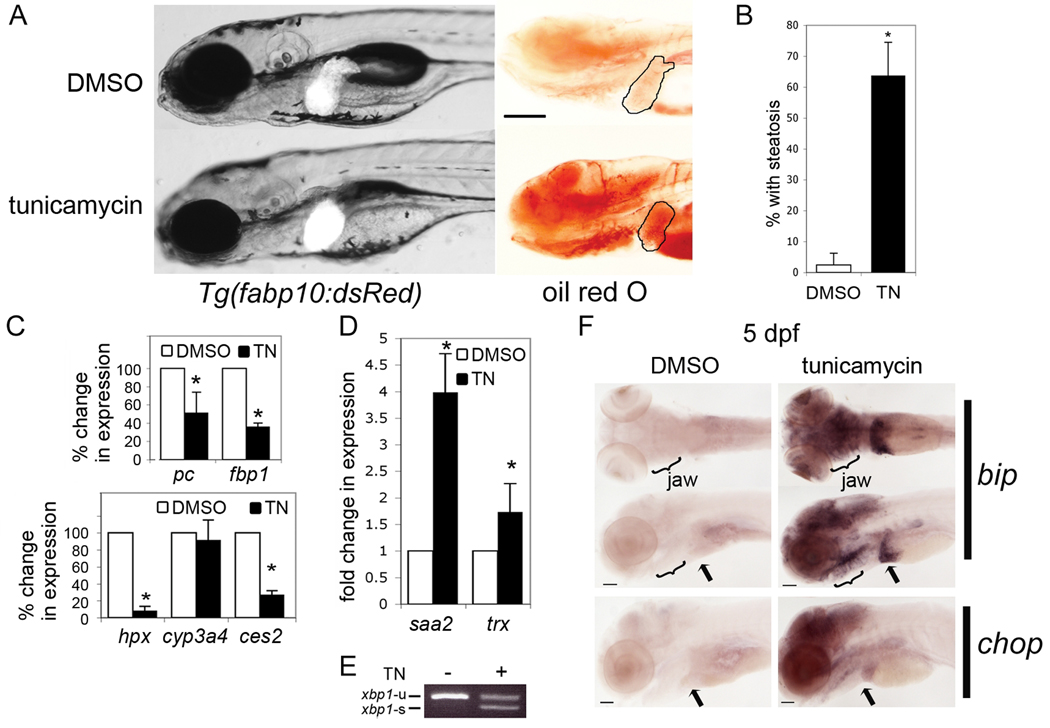
Studies in mice suggest that UPR activation can cause steatosis (6, 9, 10, 29) and acute exposure to tunicamycin, which blocks protein glycosylation and induces the UPR, causes steatosis in mice (12, 13). We used tunicamycin to ask whether ER stress causes steatosis in zebrafish. Doses exceeding 2.5 µg/ml were acutely toxic to 3 and 4 dpf larvae and 2 µg/ml was toxic when larvae were treated for longer than 12 hours. Treatment with 1 µg/ml tunicamycin from 3–5 dpf caused no mortality and moderate phenotypic abnormalities including hepatomegaly and steatosis (Fig. 3A-B). Genes required for some hepatic functions are decreased (Fig. 3C) and genes that signify hepatic damage (Fig. 3D) are increased in tunicamycin treated larvae. As expected, prolonged tunicamycin treatment induced xbp1 splicing (Fig. 3E) and UPR target genes, including bip and chop (Fig. 3F). These data demonstrate that tunicamycin causes ER stress and FLD.
Figure 3. Tunicamycin causes steatosis and liver damage.
A. Tg(fabp10:dsRed) larvae exposed from 3–5 dpf to DMSO as a control or to 1µg/ml tunicamycin (TN) were imaged on 5 dpf live (left) or after whole mount staining with oil red O (right). The liver is circled. Scale bar = 200 µm. B. Tunicamycin treatment significantly increased the average percent of larvae with steatosis (6 clutches with n>100 for each sample; * p<0.0001 by Students t-test). Error bars represent standard deviation. Expression of genes implicated in liver function (C) and damage (D) was detected by qPCR in tunicamycin-treated embryos and normalized to controls (DMSO treated). Values represent the average fold change in 3 experiments; error bars indicate standard deviation. * indicates p<0.05 by a 1 sample t-test. E. xbp1-u and xbp1-s mRNA was detected in 5 dpf larvae treated for 48 hours with DMSO (−) or tunicamycin (+). Image is representative of 5 experiments. F. In situ hybridization of 5 dpf larvae to detect bip and chop. bip stained larvae are visualized from the ventral (top) and left lateral view (bottom) to show the jaw, pancreas and liver (arrow). Scale bar = 200 µm. Images are representative of 20 larvae per sample.
Srebp activation is not required for steatosis due to tunicamycin treatment or foigr mutation
Srebps and Atf6 are activated by similar mechanisms involving the site-1 and site-2 proteases (encoded by mbtps1 and mbtps2, respectively) (see (30) and Fig. 4A). Some studies demonstrate UPR and SREBPs are activated together (16–18) whereas others report that UPR activation is accompanied by decreased SREBP activation (12, 13, 20, 31). We found that Atf6 depletion induces Srebp2 target genes (Fig. S2), consistent with the model proposed by Zeng et al. (2004) in which Atf6 suppresses Srebp2 function. Our finding that Srebp2 target genes (hmgcra, fdft1) are expressed at lower levels in foigr mutants (Fig. 4B), in which Atf6 is likely activated, support this hypothesis.
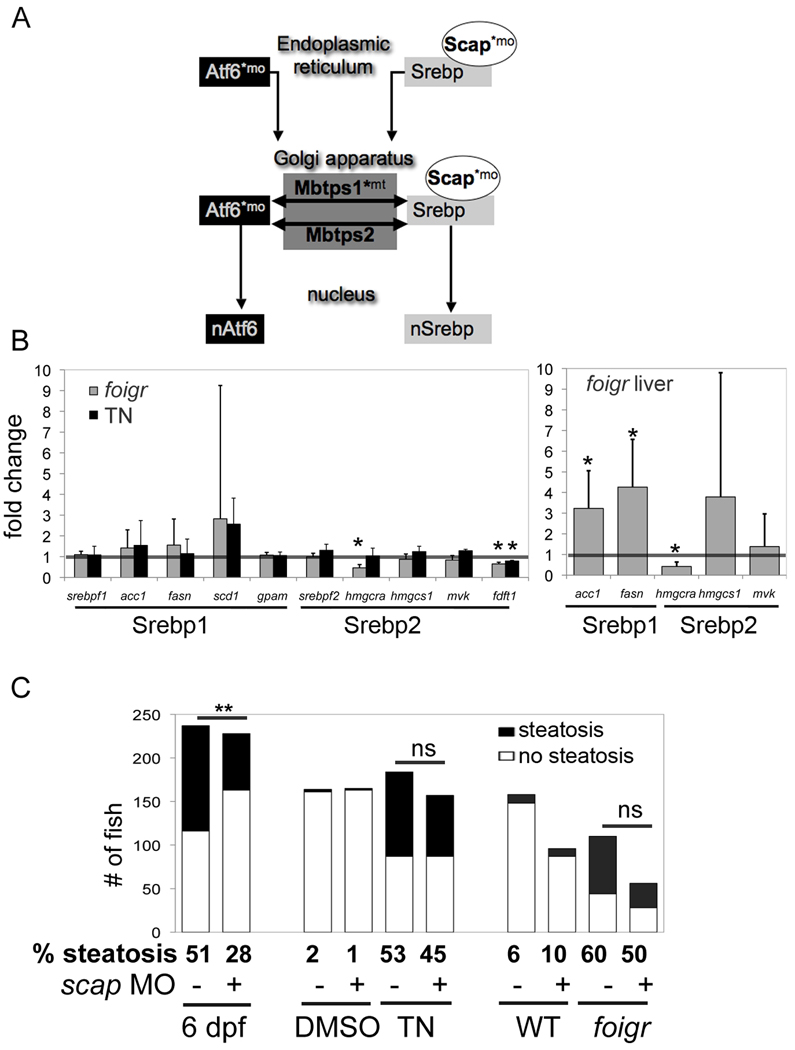
Figure 4. Steatosis in response to chronic ER stress does not require Srebp activation.
A. The activation pathways for the nSrebp and nAtf6 transcription factors utilize common components. * indicates components targeted in our studies using either a morpholino (mo) or mutant (mt). B. Srebp1 and Srebp2 target genes were assessed in at least 3 clutches of RNA isolated from whole larvae (left) and dissected livers (right). Expression in foigr mutants was normalized to wild-type siblings (grey bars) and expression in tunicamycin treated larvae was normalized to DMSO treated siblings (black bars). The level of expression in the respective controls was set to 1 (horizontal bar). * indicates p<0.01 by a 1 sampled t-test. C. The effect of scap morpholino injection on steatosis in three models (fasting, chronic tunicamycin and foigr mutation) was assessed. First, scap morphants and uninjected siblings were fasted until 6 dpf (left) or, second, they were treated with tunicamycin or DMSO from 3–5 dpf (center). Third, scap morpholino was injected into foigr mutants and wild-type siblings (right). All larvae were collected on 5 dpf, whole mount stained with oil red O and scored for steatosis. The number larvae scored positive larvae (black) and negative (white) are plotted. The percent of fish with steatosis is indicated below each bar. ** indicates a p <0.001 by Fisher’s exact test, ns: not significant.
While the genes encoding Srebps or their target genes were mostly unchanged in whole tunicamycin treated larvae, foigr mutants and mutant livers (Fig. 4B), acc1 and fasn were upregulated in foigr livers. We thus explored the possibility that Srebp activation contributes to steatosis due to foigr mutation and tunicamycin treatment using two genetic tools to block Srebp activation (indicated by * in Fig. 4A). Scap is specific for Srebp processing (32) while Mbtps1 and Mbtps2 also cleave other substrates (30, 33). Both are highly effective at blocking steatosis due to other causes and Mbtps1 mutants have significant reduction of Srebp target gene expression (22). A morpholino blocking scap translation was injected into either wild-type fish that were treated with tunicamycin from 3–5 dpf or into foigr mutants and their phenotypically wild-type siblings. Larvae were collected on 5 dpf, stained with oil red O and scored for steatosis. Uninjected siblings or those injected with a non-targeting control morpholino were used interchangeably as controls, since we found no difference in viability, gross appearance, liver size, steatosis or expression of UPR and Srebp target gene (Fig. S1A–C) between these two. scap morpholino efficacy was demonstrated resistance to steatosis caused by fasting (Fig. 4C) and alcohol (22). However, scap morphants are not protected from steatosis caused by tunicamycin or foigr mutation (Fig. 4C). Thus, steatosis due to ER stress is independent of Srebp activation.
mbtps1hi1487 mutants have defects in jaw, brain and liver development, do not develop steatosis unperturbed or in response to alcohol (Fig. 5A and (34)). We found no difference in the expression of Srebp target genes in mbtps1hi1487 mutants in response to tunicamycin (Fig. 5B). This supports the hypothesis that Srebps are neither induced by ER stress nor required for steatosis. The mechanism by which the Srebp-1c target genes acc1 and fasn are induced in foigr mutant livers is unclear.
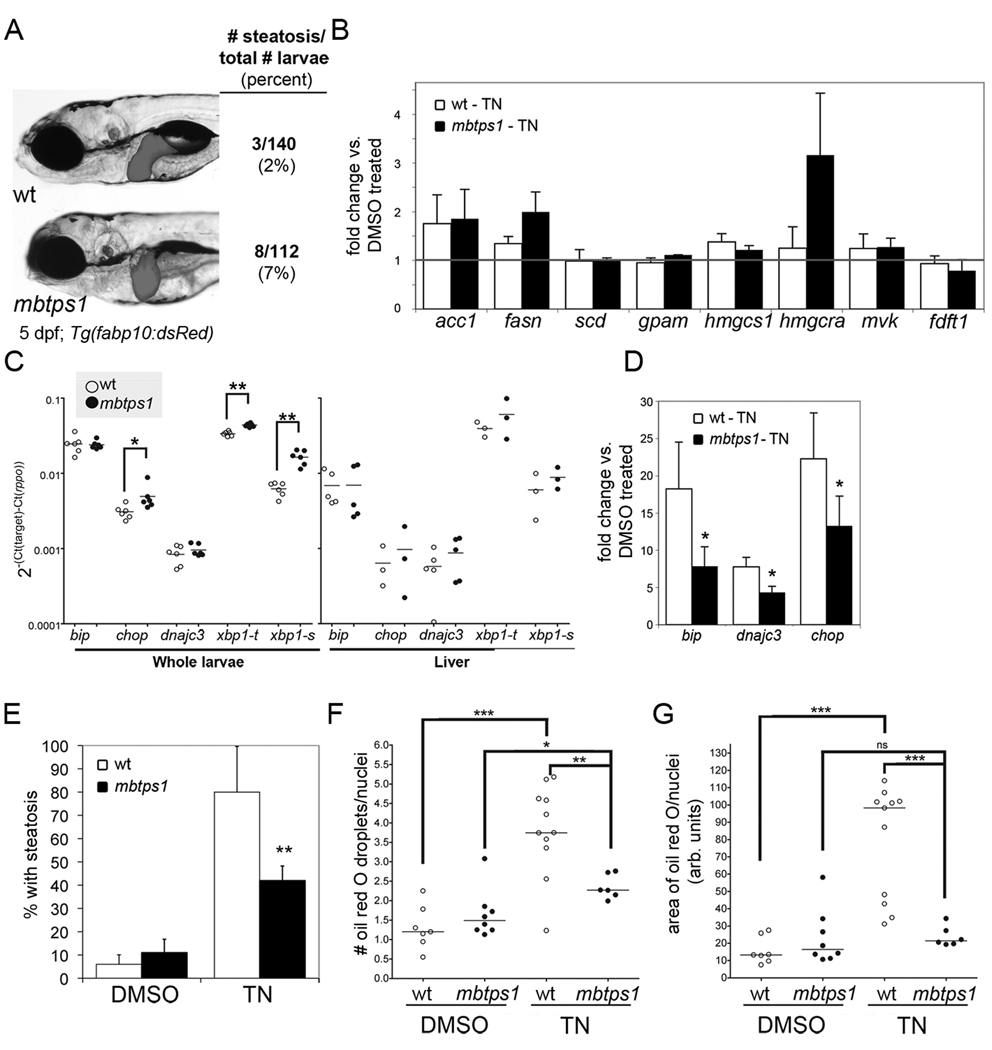
Figure 5. mbtps1hi1487 mutants are protected from steatosis caused by chronic tunicamycin treatment.
A. mbtps1hi1487 mutant larvae have a round liver, but do not develop more steatosis than their wild-type siblings. Tg(fabp10:dsRed) wild-type (top) and mbtps1hi1487 (bottom) larvae were imaged on 5 dpf and scored for steatosis by whole mount oil red O staining. B. Expression of Srebp target genes in 5 dpf wild-type andmbtps1hi1487 larvae treated with tunicamycin was analyzed by qPCR. The expression in tunicamycin treated larvae was normalized to their DMSO treated fish of the same genotype to obtain the fold change for each gene. The average fold change in wild-type and mutants from 3 clutches is plotted with error bars showing standard deviation. C. mbtps1 mutation induces UPR target genes. cDNA from 5 dpf wild-type (white dots) or mbtps1hi1487 (black dots) whole larvae (left) or dissected livers (right) was analyzed by qPCR. The ΔCt obtained for each gene is plotted as a single point for each individual clutch analyzed, with the average indicated by a horizontal bar. * indicates p<0.05, ** p<0.005 by a Student’s t-test. D. DMSO or tunicamycin treated WT and mbtps1hi1487 larvae were analyzed by qPCR for Atf6 target genes. The ΔCt in the wild-type and mutant tunicamycin treated larvae was normalized to DMSO treated wild-type larvae to obtain the fold change. The average fold change from 5 clutches is plotted with error bars indicating the standard deviation. * indicates p<0.05 by a one sampled t-test. E. mbtps1 mutation protects 5 dpf larvae from steatosis caused by chronic tunicamycin treatment. Wild-type and mbtps1hi1487 larvae treated from 3–5 dpf with 1 µg/ml tunicamycin or DMSO were stained with oil red O and scored for steatosis. Steatosis was scored in at least 8 clutches and the median percent of steatosis is plotted with bars indicating the standard deviation. ** indicates p<0.001 by Student’s t-test. In addition to whole mount oil red O staining, a subset of the samples described above were sectioned, stained with oil red O and quantified for the number of oil red O droplets (F.) and area of oil red O (G.) per liver cell. The median value for each fish is plotted as a dot, with a horizontal line to indicate the mean. * indicates p<0.05, ** indicates p<0.005 and *** indicates p<0.0005 by ANOVA. ns: not significant.
We predicted that Atf6 target genes would be expressed at lower levels in mbtps1hi1487 mutants compared to wild-type fish. Surprisingly, chop, xbp1-u and xbp1-s expression was increased in mbtps1hi1487 mutants (Fig. 5C). This suggests that Xbp1s is induced to compensate for Atf6 loss; a similar response occurs in atf6 morphants (Fig. 6A–B). Despite the increase in Xbp1s, however, mbtps1hi1487 mutants did not fully activate some Atf6 target genes when challenged with tunicamycin (Fig. 5D).
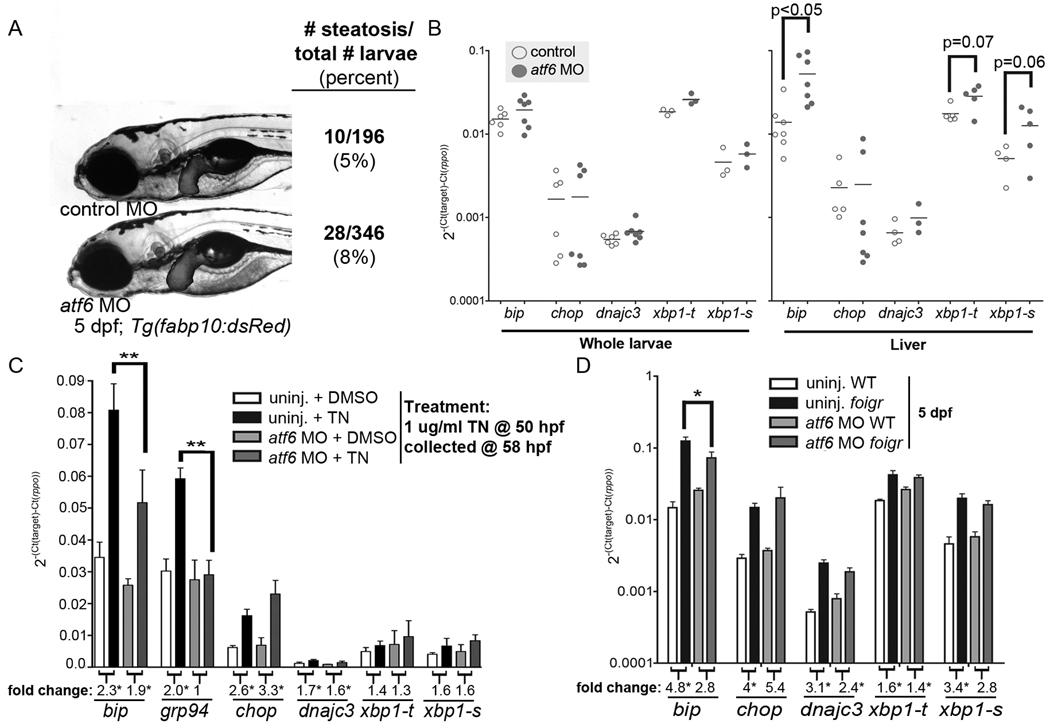
Figure 6. Atf6 knockdown affects UPR activation.
A. atf6 morpholino injected into Tg(fabp10:dsRed) embryos does not affect development or steatosis based on whole mount oil red O staining. B. cDNA prepared from whole (left) or dissected livers (right) from 5 dpf larvae injected with the standard control (white) or atf6 morpholino (grey) was analyzed by qPCR. Each gene was normalized to rpp0 and the ΔCt from each cDNA sample is plotted as a dot with the mean indicated as a bar. p values are indicated and * indicates p<0.05. C. Uninjected embryos and atf6 morphants were treated from 24–48 hpf with 1 µg/ml tunicamycin or DMSO, collected at 48 hpf and Atf6 target gene expression was examined by qPCR. The average ΔCt value of 4 clutches is plotted with error bars indicating the standard deviation. ** indicates p<0.001 by ANOVA. The fold change for each gene is labeled below each column, and numbers with an * represent a significant difference in expression in tunicamycin treated fish compared to DMSO treated fish of the same genotype. D. atf6 morpholino was injected into embryos generated by in-crossing foigr heterozygotes. On 5 dpf, foigr mutants and phenotypically wild-type siblings with and without the atf6 morpholino were collected for qPCR analysis. The ΔCt normalized to rpp0 was calculated from 3 clutches, averaged and plotted with error bars indicating the standard deviation. * indicates p<0.01 by ANOVA. The fold change for each gene is labeled below each column, and numbers with an * represent a p<0.05 comparing expression in mutants compared to wild-type fish as by ANOVA.
Unexpectedly, both the number of fish and degree of steatosis caused by tunicamycin is significantly reduced in mbtps1hi1487 mutants. Only 40% of mutants develop steatosis following tunicamycin treatment (Fig. 5E). Moreover, wild-type larvae treated with tunicamycin have 3 times more lipid droplets per liver cell (white dots; Fig. 5F) and 7 times greater area occupied by oil red O staining in the liver (white dots; Fig. 5G). Both measures of steatosis were significantly reduced in mbtps1hi1487 mutants challenged with tunicamycin (black dots; Fig. 5F–G). Since Atf6 target genes (Fig. 5D) but not Srebp targets (Fig. 5B), are decreased in mbtps1hi1487 mutants following tunicamycin treatment, we predict that loss of Atf6 activity, not Srebps, account for the protection of these mutants from steatosis caused by ER stress.
Atf6 depletion prevents steatosis in foigr mutants
To determine whether loss of Atf6 protects fish from steatosis due to prolonged UPR activation, we injected foigr mutants with a morpholino to block atf6 translation and assessed the effects on UPR target genes and steatosis. As in mice (12, 13), loss of atf6 does not affect viability, development, or the size, shape or lipid accumulation in the liver (Fig. 6A). Similar to mbtps1hi1487 mutants, the Ire1a/Xbp1 branch was induced in atf6 morphants (Fig. 6B), yet they were impaired in their ability to fully induce expression of Atf6 target genes in response to tunicamycin (Fig. 6C) or foigr mutation (Fig. 6D).
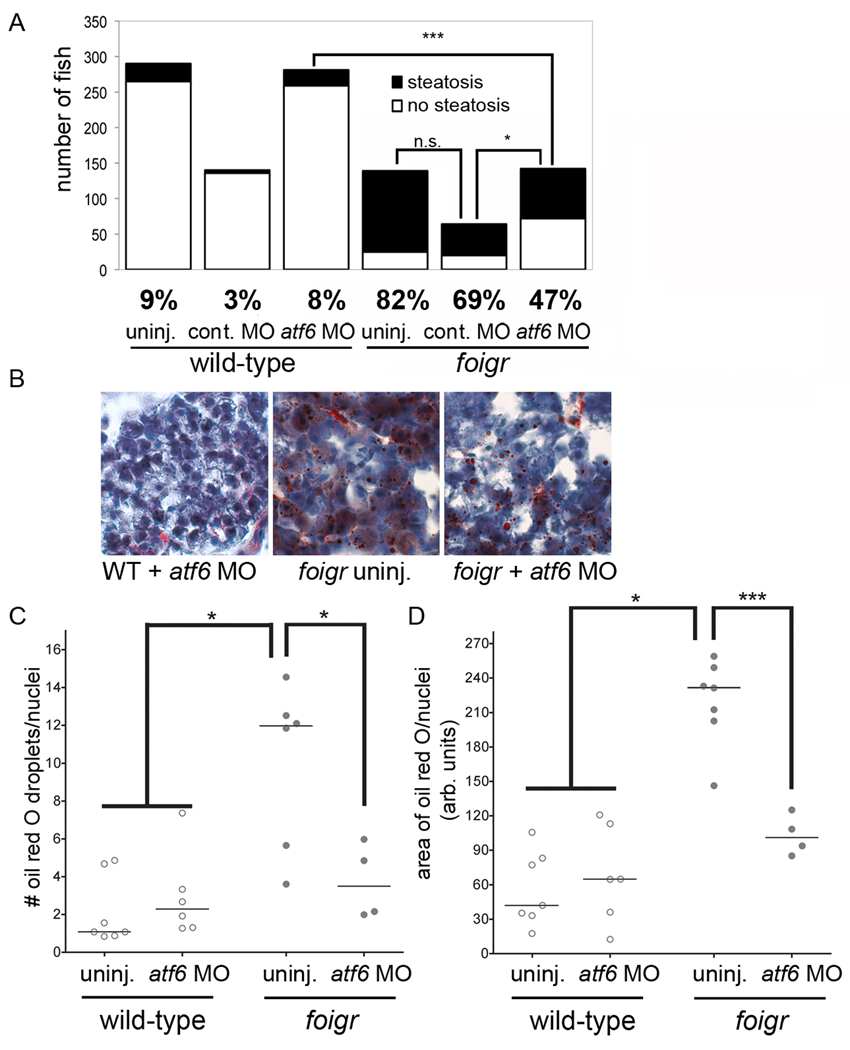
atf6 morpholino injection into foigr mutants reduced the number of mutants with steatosis to 47%, compared to 82% in uninjected mutants and 69% in mutants injected with the control morpholino (Fig. 7A). This finding was confirmed using a splice-blocking atf6 morpholino: less than 30% of mutants injected with the atf6 splice blocking morpholino developed steatosis compared to 70% of their uninjected mutant sibilings (not shown).
Figure 7. atf6 depletion alleviates steatosis in foigr mutants.
A. atf6 morpholino decreases the number of 5 dpf foigr that develop steatosis. Embryos generated by crossing foigr heterozygotes were uninjected (15 clutches) or injected with the standard control (9 clutches) or atf6 morpholino (15 clutches). Larvae were separated based on phenotype on 5 dpf, stained with oil red O, and scored for steatosis. The total number of embryos in each category is plotted and the percent fish with steatosis is labeled under each graph. * indicates p=0.01, *** indicates p< 0.0001 by chi square. B. atf6 morpholino injection decreases the amount of oil red O staining in foigr hepatocytes. Cryosections of 5 dpf either uninjected or atf6 morpholino injected wild-type and foigr mutant embryos were stained with oil red O and hematoxylin. C–D. The number of oil red O droplets per liver cell (C) and the area of each liver cell that is stained with oil red O (D) was quantified. The median value from at least 3 sections for each fish is plotted as an individual dot with the horizontal bar indicating the mean for all fish in each sample. The foigr mutants injected with atf6 morpholino are not significantly different from the wild-type controls. * indicates p<0.05, *** indicates p<0.005 by ANOVA.
Steatosis was less severe in foigr mutants injected with the atf6 morpholino (Fig. 7B). Control, uninjected and atf6 morpholino injected wild-type larvae had a median number of lipid droplets per cell that ranged from 0.8–4 with an overall median of 2 droplets/cell (Fig. 7C, left) whereas the were over 12 droplets per cell in foigr mutant livers. Similarly, the area of each cell stained with oil red O was more than 5 times greater in foigr mutants compared to wild-types (Fig. 7D). Both these measures of hepatic lipid accumulation were significantly reduced in foigr mutants by injection of the atf6 morpholino (Fig. 7D). Collectively, these data demonstrate that loss of Atf6 protects against steatosis caused by ER stress due to foigr mutation or prolonged tunicamycin treatment.
Atf6 depletion enhances steatosis due to acute tunicamycin treatment
Acute ER stress induced by intraperitoneal injection of tunicamycin causes steatosis that resolves within 3 days in wild-type mice but does not resolve in mice lacking Atf6α (12, 13). This is in contrast to our findings that loss of Atf6 protects against steatosis due to prolonged ER stress. We hypothesized that the difference is attributed to the acute ER stress experienced by mice injected with tunicamycin compared to the chronic ER stress that occurs in foigr mutations and in larvae bathed in tunicamycin for 48 hours.
We tested this by developing a protocol to induce acute ER stress in zebrafish larvae. Larvae were exposed to 2 µg/ml tunicamycin for 12 hour intervals during 4–5 dpf as outlined in Figure 8A. In protocols B and C, larvae were collected immediately following exposure. In protocol D, tunicamycin was washed out following exposure from 4–4.5 dpf and larvae were collected at 5 dpf. We compared acute and prolonged (i.e. chronic; protocol A, Fig. 8A) tunicamycin and DMSO treatment on UPR activation and steatosis.
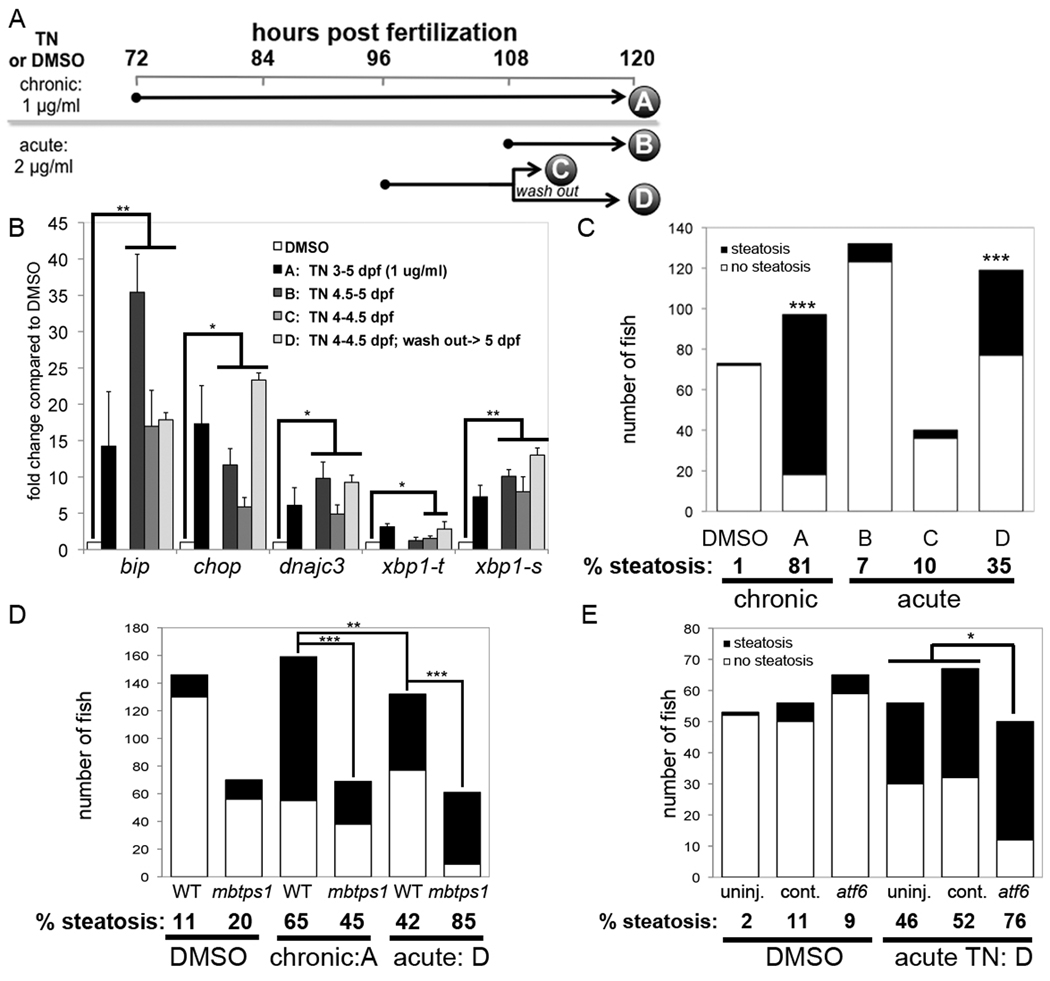
Figure 8. Acute tunicamycin treatment causes UPR activation and steatosis that is augmented by atf6 depletion.
A. Diagram of tunicamycin exposure protocols. Larvae treated with DMSO or tunicamycin for 48 (chronic; 1 µg/ml) or 12 (acute; 2 µg/ml) hours. Samples were collected at the times indicated by each letter for qPCR or oil red O staining. For protocol D, larvae were treated as in C, the tunicamycin was washed out and larvae were incubated for an additional 12 hours. B. UPR target genes were induced by all 4 protocols. The fold change for each gene in was calculated relative to DMSO (set as 1) at least 3 clutches and averaged. Error bars indicate the standard deviation, * indicates p<0.05, ** indicates p<0.005 by a ANOVA. C. Tunicamycin was administered to zebrafish according to the chronic and acute protocols outlined in panel A. The number of fish with steatosis was scored in whole mount oil red O stained larvae. The percent of steatosis in 3–5 clutches in each sample is labeled below each bar. Control fish were treated with DMSO from 3–5 dpf. The difference between tunicamycin treated and DMSO treated fish was significant for protocols A and D; *** indicates p<0.0001 by Fisher’s exact test. D. mbtps1hi1487 mutants are predisposed to steatosis caused by acute tunicamycin exposure. The number of fish with steatosis was counted in 6 clutches of mbtps1hi1487 mutants and their wild-type siblings that were treated with DMSO from 3–5 dpf or with chronic (protocol A) or acute (protocol D) tunicamycin. ** indicates p<0.002 and **** indicates p<0.0001 by Fisher’s exact test. E. atf6 morphants develop more steatosis in response to acute tunicamycin treatment. atf6 morphants, standard control morphants and uninjected embryos were treated with tunicamycin of DMSO according to protocol D. The total number of 5 dpf fish in 3 clutches that were scored for steatosis based on whole mount oil red O staining is plotted. The percent steatosis for each sample is labeled below each bar. * indicates p<0.02 by Fisher’s exact test.
Both the acute and chronic exposure protocols had equivalent effects in inducing UPR target gene expression (Fig. 8B). Steatosis occurred in 81% of fish treated with the chronic protocol, but not following short exposure (protocols B and C). However, when the tunicamycin is washed out (protocol D), 35% of fish developed steatosis (Fig. 8C).
We then tested whether depleting Atf6 affected steatosis caused by acute tunicamycin treatment protocol D. The percent of fish with steatosis significantly reduced in mbtps11487 mutants (45%) compared to wild-type larvae (65%) challenged with chronic tunicamycin, but increased in response to acute tunicamycin treatment (85%) compared to wild-type siblings (42%; Fig. 8D). Similar results were obtained in atf6 morphants: 76% developed steatosis following acute tunicamycin treatment, compared to 46% and 52% in uninjected and control injected larvae (Fig. 8D). Thus depleting Atf6 potentiates steatosis caused by acute ER stress in both zebrafish and mice (12, 13).
DISCUSSION
We use zebrafish as a novel tool to understand the complex relationship between UPR activation and steatosis. Our data demonstrate that both acute and chronic ER stress can lead to steatosis and illustrates the opposing roles that Atf6 plays in these different scenarios. We found that Atf6 depletion protects fish from steatosis due to chronic ER stress induced either by foigr mutation or by prolonged exposure to tunicamycin, but can accentuate steatosis caused by acute tunicamycin treatment. This is an important distinction, as most FLD etiologies are likely associated with chronic UPR activation, if not frank ER stress. In these cases, attempts to improve protein folding and reduce UPR signaling are predicted to be therapeutic. Exciting data from mouse models suggests the efficacy of this approach (10, 11, 14, 18).
How does chronic UPR activation affect lipid metabolism in the liver? One possibility is that components of the UPR may directly modulate lipid metabolism. While some studies implicate lipid synthesis directed by Xbp1 (35) or Srebps (17, 18, 36, 37) as a factor in steatosis associated with ER stress, we do not believe that lipid synthesis is a major contributing factor to steatosis in our models. We hypothesize that foigr mutation and tunicamycin treatment induces Atf6 and that this, in turn, may suppress Srebp2 activity, consistent with data from mammalian cells (20). Although depletion of Atf6 causes slight upregulation of Srebp2 target genes, this is insufficient to cause steatosis (see Figs. 7A, 8A, 8C and 8D). On the contrary, atf6 morphants were protected from steatosis induced by foigr mutation. Together, our data suggest that triglyceride and cholesterol synthesis are unlikely to significantly contribute to steatosis caused by chronic ER stress.
It is likely that disruption of the secretory pathway prevents lipoprotein secretion. This is supported by the finding of decreased ApoB levels in hepatocytes of mice injected with tunicamycin (13). In foigr mutant hepatocytes, we observed some lipid droplets within what appears to be dilated ER, perhaps reflecting lipoprotein retention. As ApoB secretion is impaired in treatments that cause prolonged ER stress (38), it is feasible lipoprotein retention in hepatocytes can contribute to steatosis. It is not known whether Atf6 impacts lipoprotein secretion or other lipid metabolic pathways in hepatocytes, such as β-oxidation.
A complex mechanism likely accounts for our finding that Atf6 depletion both prevents and accentuates steatosis. We found that Atf6 loss results in upregulation of other UPR branches. This may be due to direct cross-talk between branches or as a response to a transient increased unfolded protein load due to depletion of Atf6. Regardless of the mechanism, the result is that the cells are adapted so that they are better equipped to handle a gradual increase in unfolded proteins that likely occurs in foigr or chronic tunicamycin treated larvae. Paradoxically, Atf6 depletion effectively reduces the amount of ER stress caused by these two insults, similar to what has been reported in Bip+/− mice (14). We speculate that the reduction of ER stress in turn reduces the amount of steatosis. In contrast, an acute onslaught of unfolded proteins in the ER caused by a short exposure to a high does of tunicamycin requires a robust UPR which cannot be achieved when Atf6 is depleted. In this acute scenario, the absence of Atf6 exacerbates ER stress and disrupts lipid metabolism via a mechanism that remains elusive.
Foigr is highly conserved, yet its function remains elusive. Recent data suggests that Foigr functions in the secretory pathway (26–28), consistent with our findings of ER dysfunction in foigr mutants. If foigr mutation causes a defect in the Golgi apparatus (28), the backup of secretory pathway cargo may cause ER stress. If this were the case, then treating zebrafish with brefeldin A to disrupt the Golgi apparatus should cause ER stress and phenocopy foigr. Our preliminary studies to test this are not compelling (not shown). In contrast, the similarities between chronic tunicamycin treatment and foigr mutants lead us to speculate that loss of foigr induces a defect in protein glycosylation. It will be important to define the mechanism by which foigr mutation leads to UPR activation and to understand the function of Foigr.
Supplementary Material
ACKNOWLEDGEMENTS
We are indebted to Deanna Howarth, Mike Passeri, and Chris Monson for technical assistance. Drs. Friedman, Krauss and Burdine provided helpful comments on the manuscript. The American Gastrological Association, March of Dimes and NIAAA (p20AA017067-01 and 1RO1AA18886-01) provided support to KCS. The Medical Scientist Training Program (T32GM007280) and Training Program in Cellular and Molecular Biology (NIGMS/T32GM08633) partially supported DI.
List of abbreviations
Full gene names and corresponding abbreviations for all genes examined by PCR are listed in Table S1.
- ATF6
activating transcription factor 6
- dpf
days post-fertilization
- EIF2S1
eukaryotic translation initiation factor 2, subunit 1 alpha
- ER
endoplasmic reticulum
- FLD
fatty liver disease
- foigr
foie gras
- hpf
hours post-fertilization
- IRE1
inositol-requiring 1
- mbtps1
membrane-bound transcription factor peptidase, site 1
- PBS
phosphate buffered saline
- PCR
polymerase chain reaction
- PERK
PRKR-like endoplasmic reticulum kinase (also called eukaryotic translation initiation factor 2-alpha kinase 3)
- qPCR
real time quantitative PCR
- SCAP
sterol regulatory element binding protein cleavage-activating protein
- SREBP
sterol regulatory element binding protein
- SDS
Sodium dodecyl sulfate
- TUNEL
Terminal deoxynucleotidyl transferase dUTP nick end labeling
- UPR
unfolded protein response
- XBP1
X box binding protein-1
Contributor Information
Ayca Cinaroglu, Department of Medicine/Division of Liver Diseases and Department of Developmental and Regenerative Biology, Gustave L. Levy Place Box 1020 New York, NY 10029.
Chuan Gao, Department of Medicine/Division of Liver Diseases and Department of Developmental and Regenerative Biology, Gustave L. Levy Place Box 1020 New York, NY 10029.
Dru Imrie, Department of Medicine/Division of Liver Diseases and Department of Developmental and Regenerative Biology, Gustave L. Levy Place Box 1020 New York, NY 10029.
Kirsten C. Sadler, Mount Sinai School of Medicine, Gustave L. Levy Place Box 1020 New York, NY 10029.
REFERENCES
- 1.Bernales S, Papa FR, Walter P. Intracellular signaling by the unfolded protein response. Annu Rev Cell Dev Biol. 2006;22:487–508. doi: 10.1146/annurev.cellbio.21.122303.120200. [DOI] [PubMed] [Google Scholar]
- 2.Rutkowski DT, Hegde RS. Regulation of basal cellular physiology by the homeostatic unfolded protein response. J Cell Biol. 2010;189:783–794. doi: 10.1083/jcb.201003138. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Yoshida H, Matsui T, Yamamoto A, Okada T, Mori K. XBP1 mRNA is induced by ATF6 and spliced by IRE1 in response to ER stress to produce a highly active transcription factor. Cell. 2001;107:881–891. doi: 10.1016/s0092-8674(01)00611-0. [DOI] [PubMed] [Google Scholar]
- 4.Asselah T, Bieche I, Mansouri A, Laurendeau I, Cazals-Hatem D, Feldmann G, Bedossa P, et al. In vivo hepatic endoplasmic reticulum stress in patients with chronic hepatitis C. J Pathol. 2010;221:264–274. doi: 10.1002/path.2703. [DOI] [PubMed] [Google Scholar]
- 5.Hotamisligil GS. Endoplasmic reticulum stress and the inflammatory basis of metabolic disease. Cell. 2010;140:900–917. doi: 10.1016/j.cell.2010.02.034. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Ozcan U, Cao Q, Yilmaz E, Lee AH, Iwakoshi NN, Ozdelen E, Tuncman G, et al. Endoplasmic reticulum stress links obesity, insulin action, and type 2 diabetes. Science. 2004;306:457–461. doi: 10.1126/science.1103160. [DOI] [PubMed] [Google Scholar]
- 7.Ji C, Kaplowitz N. Betaine decreases hyperhomocysteinemia, endoplasmic reticulum stress, and liver injury in alcohol-fed mice. Gastroenterology. 2003;124:1488–1499. doi: 10.1016/s0016-5085(03)00276-2. [DOI] [PubMed] [Google Scholar]
- 8.Puri P, Mirshahi F, Cheung O, Natarajan R, Maher JW, Kellum JM, Sanyal AJ. Activation and dysregulation of the unfolded protein response in nonalcoholic fatty liver disease. Gastroenterology. 2008;134:568–576. doi: 10.1053/j.gastro.2007.10.039. [DOI] [PubMed] [Google Scholar]
- 9.Oyadomari S, Harding HP, Zhang Y, Oyadomari M, Ron D. Dephosphorylation of translation initiation factor 2alpha enhances glucose tolerance and attenuates hepatosteatosis in mice. Cell Metab. 2008;7:520–532. doi: 10.1016/j.cmet.2008.04.011. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Nakatani Y, Kaneto H, Kawamori D, Yoshiuchi K, Hatazaki M, Matsuoka TA, Ozawa K, et al. Involvement of endoplasmic reticulum stress in insulin resistance and diabetes. J Biol Chem. 2005;280:847–851. doi: 10.1074/jbc.M411860200. [DOI] [PubMed] [Google Scholar]
- 11.Ozcan U, Yilmaz E, Ozcan L, Furuhashi M, Vaillancourt E, Smith RO, Gorgun CZ, et al. Chemical Chaperones Reduce ER Stress and Restore Glucose Homeostasis in a Mouse Model of Type 2 Diabetes. Science. 2006;313:1137–1140. doi: 10.1126/science.1128294. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Rutkowski DT, Wu J, Back SH, Callaghan MU, Ferris SP, Iqbal J, Clark R, et al. UPR pathways combine to prevent hepatic steatosis caused by ER stress-mediated suppression of transcriptional master regulators. Dev Cell. 2008;15:829–840. doi: 10.1016/j.devcel.2008.10.015. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Yamamoto K, Takahara K, Oyadomari S, Okada T, Sato T, Harada A, Mori K. Induction of liver steatosis and lipid droplet formation in ATF6alpha-knockout mice burdened with pharmacological endoplasmic reticulum stress. Mol Biol Cell. 2010;21:2975–2986. doi: 10.1091/mbc.E09-02-0133. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Ye R, Jung DY, Jun JY, Li J, Luo S, Ko HJ, Kim JK, et al. Grp78 heterozygosity promotes adaptive unfolded protein response and attenuates diet-induced obesity and insulin resistance. Diabetes. 2010;59:6–16. doi: 10.2337/db09-0755. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Horton JD, Goldstein JL, Brown MS. SREBPs: activators of the complete program of cholesterol and fatty acid synthesis in the liver. J Clin Invest. 2002;109:1125–1131. doi: 10.1172/JCI15593. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Colgan SM, Tang D, Werstuck GH, Austin RC. Endoplasmic reticulum stress causes the activation of sterol regulatory element binding protein-2. Int J Biochem Cell Biol. 2007;39:1843–1851. doi: 10.1016/j.biocel.2007.05.002. [DOI] [PubMed] [Google Scholar]
- 17.Lee JN, Ye J. Proteolytic activation of sterol regulatory element-binding protein induced by cellular stress through depletion of Insig-1. J Biol Chem. 2004;279:45257–45265. doi: 10.1074/jbc.M408235200. [DOI] [PubMed] [Google Scholar]
- 18.Kammoun HL, Chabanon H, Hainault I, Luquet S, Magnan C, Koike T, Ferre P, et al. GRP78 expression inhibits insulin and ER stress-induced SREBP-1c activation and reduces hepatic steatosis in mice. J Clin Invest. 2009 doi: 10.1172/JCI37007. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Harding HP, Zhang Y, Khersonsky S, Marciniak S, Scheuner D, Kaufman RJ, Javitt N, et al. Bioactive small molecules reveal antagonism between the integrated stress response and sterol-regulated gene expression. Cell Metab. 2005;2:361–371. doi: 10.1016/j.cmet.2005.11.005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Zeng L, Lu M, Mori K, Luo S, Lee AS, Zhu Y, Shyy JY. ATF6 modulates SREBP2-mediated lipogenesis. Embo J. 2004;23:950–958. doi: 10.1038/sj.emboj.7600106. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Amsterdam A, Nissen RM, Sun Z, Swindell EC, Farrington S, Hopkins N. Identification of 315 genes essential for early zebrafish development. Proc Natl Acad Sci U S A. 2004;101:12792–12797. doi: 10.1073/pnas.0403929101. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Passeri MJ, Cinaroglu A, Gao C, Sadler KC. Hepatic steatosis in response to acute alcohol exposure in zebrafish requires sterol regulatory element binding protein activation. Hepatology. 2009;49:443–452. doi: 10.1002/hep.22667. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Ho SY, Lorent K, Pack M, Farber SA. Zebrafish fat-free is required for intestinal lipid absorption and Golgi apparatus structure. Cell Metab. 2006;3:289–300. doi: 10.1016/j.cmet.2006.03.001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Sadler KC, Krahn KN, Gaur NA, Ukomadu C. Liver growth in the embryo and during liver regeneration in zebrafish requires the cell cycle regulator, uhrf1. Proc Natl Acad Sci U S A. 2007;104:1570–1575. doi: 10.1073/pnas.0610774104. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Sadler KC, Amsterdam A, Soroka C, Boyer J, Hopkins N. A genetic screen in zebrafish identifies the mutants vps18, nf2 and foie gras as models of liver disease. Development. 2005;132:3561–3572. doi: 10.1242/dev.01918. [DOI] [PubMed] [Google Scholar]
- 26.Behrends C, Sowa ME, Gygi SP, Harper JW. Network organization of the human autophagy system. Nature. 2010;466:68–76. doi: 10.1038/nature09204. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Gavin AC, Bosche M, Krause R, Grandi P, Marzioch M, Bauer A, Schultz J, et al. Functional organization of the yeast proteome by systematic analysis of protein complexes. Nature. 2002;415:141–147. doi: 10.1038/415141a. [DOI] [PubMed] [Google Scholar]
- 28.Wendler F, Gillingham AK, Sinka R, Rosa-Ferreira C, Gordon DE, Franch-Marro X, Peden AA, et al. A genome-wide RNA interference screen identifies two novel components of the metazoan secretory pathway. EMBO J. 2010;29:304–314. doi: 10.1038/emboj.2009.350. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Ji C, Mehrian-Shai R, Chan C, Hsu YH, Kaplowitz N. Role of CHOP in hepatic apoptosis in the murine model of intragastric ethanol feeding. Alcohol Clin Exp Res. 2005;29:1496–1503. doi: 10.1097/01.alc.0000174691.03751.11. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Ye J, Rawson RB, Komuro R, Chen X, Dave UP, Prywes R, Brown MS, et al. ER stress induces cleavage of membrane-bound ATF6 by the same proteases that process SREBPs. Mol Cell. 2000;6:1355–1364. doi: 10.1016/s1097-2765(00)00133-7. [DOI] [PubMed] [Google Scholar]
- 31.Kovacs WJ, Tape KN, Shackelford JE, Wikander TM, Richards MJ, Fliesler SJ, Krisans SK, et al. Peroxisome deficiency causes a complex phenotype because of hepatic SREBP/Insig dysregulation associated with endoplasmic reticulum stress. J Biol Chem. 2009;284:7232–7245. doi: 10.1074/jbc.M809064200. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Nohturfft A, DeBose-Boyd RA, Scheek S, Goldstein JL, Brown MS. Sterols regulate cycling of SREBP cleavage-activating protein (SCAP) between endoplasmic reticulum and Golgi. Proc Natl Acad Sci U S A. 1999;96:11235–11240. doi: 10.1073/pnas.96.20.11235. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Zhang K, Shen X, Wu J, Sakaki K, Saunders T, Rutkowski DT, Back SH, et al. Endoplasmic reticulum stress activates cleavage of CREBH to induce a systemic inflammatory response. Cell. 2006;124:587–599. doi: 10.1016/j.cell.2005.11.040. [DOI] [PubMed] [Google Scholar]
- 34.Schlombs K, Wagner T, Scheel J. Site-1 protease is required for cartilage development in zebrafish. Proc Natl Acad Sci U S A. 2003;100:14024–14029. doi: 10.1073/pnas.2331794100. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Lee AH, Scapa EF, Cohen DE, Glimcher LH. Regulation of hepatic lipogenesis by the transcription factor XBP1. Science. 2008;320:1492–1496. doi: 10.1126/science.1158042. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Wang H, Kouri G, Wollheim CB. ER stress and SREBP-1 activation are implicated in beta-cell glucolipotoxicity. J Cell Sci. 2005;118:3905–3915. doi: 10.1242/jcs.02513. [DOI] [PubMed] [Google Scholar]
- 37.Werstuck GH, Lentz SR, Dayal S, Hossain GS, Sood SK, Shi YY, Zhou J, et al. Homocysteine-induced endoplasmic reticulum stress causes dysregulation of the cholesterol and triglyceride biosynthetic pathways. J Clin Invest. 2001;107:1263–1273. doi: 10.1172/JCI11596. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Ota T, Gayet C, Ginsberg HN. Inhibition of apolipoprotein B100 secretion by lipid-induced hepatic endoplasmic reticulum stress in rodents. J Clin Invest. 2008;118:316–332. doi: 10.1172/JCI32752. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.