Abstract
Hepatitis C virus (HCV) employs various strategies to establish persistent infection that can cause chronic liver disease. Our previous study showed that both the original patient serum from which the HCV JFH-1 strain was isolated and the cell culture-generated JFH-1 virus (JFH-1cc) established infection in chimpanzees, and that infected JFH-1 strains accumulated mutations after passage through chimpanzees. The aim of this study was to compare the in vitro characteristics of JFH-1 strains emerged in each chimpanzee at early and late stages of infection, as it could provide an insight into the phenomenon of viral persistence. We generated full-genome JFH-1 constructs with the mutations detected in patient serum-infected (JFH-1/S1 and S2) and JFH1cc-infected (JFH-1/C) chimpanzees, and assessed their effect on replication, infectious virus production, and regulation of apoptosis in cell culture. The extracellular HCV core antigen secreted from JFH-1/S1-, S2-, and C-transfected HuH-7 cells was 2.5, 8.9, and 2.1 times higher than that from JFH-1 wild type (JFH-1/wt) transfected cells, respectively. Single cycle virus production assay with a CD81 negative cell line revealed that the strain JFH-1/S2, isolated from the patient serum-infected chimpanzee at later time-point of infection, showed lower replication and higher capacity to assemble infectious virus particles. This strain also showed productive infection in human hepatocyte-transplanted mice. Furthermore, the cells harboring this strain displayed lower susceptibility to the apoptosis induced by tumor necrosis factor-α or Fas ligand as compared to the cells replicating JFH-1/wt.
Conclusion
the ability of lower replication, higher virus production and less susceptibility to cytokine-induced apoptosis may be important for prolonged infection in vivo. Such control of viral functions by specific mutations may be a key strategy for establishing persistent infection.
Keywords: chimpanzee, adaptive mutation, virus assembly, cytokine
INTRODUCTION
Currently, about 200 million people are infected with Hepatitis C virus (HCV) and are at continuous risk of developing chronic liver diseases such as chronic hepatitis, liver cirrhosis, and hepatocellular carcinoma (1,2). Although acute HCV infection elicits innate and adaptive immune responses, the virus successfully evades clearance in approximately 75% of infected individuals (3,4). The mechanisms by which HCV leads to persistent infection at a high frequency are not yet fully understood. Lack of appropriate animal models, except chimpanzees, has rendered such studies difficult. Human hepatocyte-transplanted mice (5,6), a useful small animal model to study HCV infection, are unsuitable to study the mechanisms of virus persistence because of a lack of B- and T-cell-mediated immunity.
HCV is a non-cytopathic positive-stranded RNA virus of the Flaviviridae family. It primarily infects hepatocytes of humans and chimpanzees, where, thanks to error prone RNA-dependent RNA polymerase, the infected virus accumulates a high number of mutations rapidly, thus providing opportunity for selection of viruses that have the ability to escape the immune system and establish persistent infection. Deciphering the strategies employed by HCV to establish persistence can be helpful in the development of new strategies to eradicate the virus and to stop disease progression. Until recently, the lack of an HCV strain having the ability to establish infection in vivo and in vitro was a substantial hindrance in studying the molecular mechanisms of virus persistence. This problem was solved by the identification of an HCV strain, JFH-1, that was isolated from a fulminant hepatitis patient and found to be capable of replicating and assembling infectious virus particles in chimpanzees as well as in cell culture (7–10). This clone can be used to study the molecular mechanisms by which HCV evades the host immune system and causes chronic infection.
In our previous report, we inoculated patient serum from which the JFH-1 strain was originally isolated and cell culture-generated JFH-1 virus (JFH-1cc) into two different chimpanzees (11). HCV established infection in both animals within three days after inoculation. In the JFH-1cc-infected chimpanzee, genome sequence of predominant infecting virus at week 2 was identical to JFH-1 wild type (JFH-1/wt; in this study, this abbreviation was used instead of JFH-1 to distinguish it from other variant strains), and the infecting virus has 4 synonymous and 7 non-synonymous mutations at week 7. In the JFH-1 patient serum-infected chimpanzee, 19 synonymous and 6 non-synonymous mutations were observed in predominantly circulating virus at week 2 and this number increased to 35 synonymous and 17 non-synonymous mutations at the later stage of infection course (week 23) (11). From these observations, we presumed that the isolates evolved in each chimpanzee at later stages of infection might have some advantage over the viruses isolated at earlier time points for survival in the infected animals. Thus, in this study, we generated JFH-1 variants containing the mutations observed in these animals, and assessed their effect on replication and infectious virus production in cell culture. Furthermore, we examined the effects of infection of these strains to tumor necrosis factor-α (TNF-α)- or Fas ligand (FasL)- mediated apoptosis.
MATERIALS and METHODS
The complete materials and methods are provided in the Supporting Information.
RESULTS
Effects of Mutations Identified in Chimpanzees
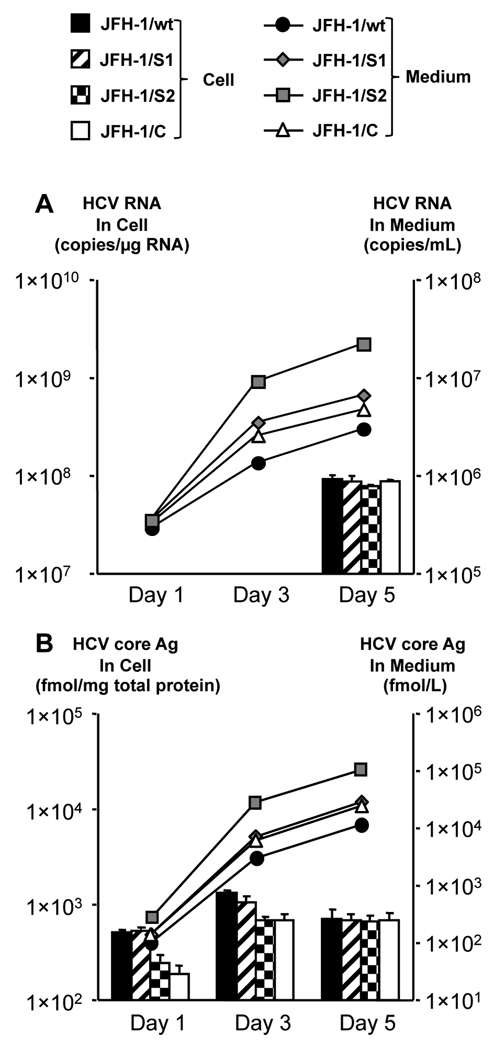
To investigate the effect of mutations on virus phenotype, we generated constructs containing the mutations observed in the JFH-1 patient serum- and JFH-1cc-infected chimpanzees at various time-points. The JFH-1 variants JFH-1/S1 and S2 contain the mutations observed in the patient serum-infected chimpanzee at week 2 and week 23, respectively, and JFH-1/C contains the mutations observed in the JFH-1cc-infected chimpanzee at week 7 (Supporting Table 1). The replication and virus production capacity of these variants in HuH-7 cells was compared with that of JFH-1/wt. After electroporation of in vitro synthesized full-genome RNA of JFH-1/wt and variant strains, extra- and intracellular HCV RNA and core antigen (Ag) were measured (Fig. 1). At day 5 post-transfection, all constructs displayed similar intracellular HCV RNA levels. However, extracellular HCV RNA level of JFH-1/C was 1.6 times higher than that of JFH-1/wt. Likewise, extracellular HCV RNA level of JFH-1/ S2 was 3.4 times higher than that of JFH-1/S1 (Fig. 1A). Intracellular HCV core Ag levels of JFH-1/S2 and C were 240.9 ± 58.2 and 189.8 ± 42.1 fmol/µg protein, respectively, and were significantly lower (p < 0.005) than that of JFH-1/S1 (526.1 ± 58.2 fmol/µg protein) and JFH-1/wt (511.7 ± 32.9 fmol/µg protein) at day 1, but reached comparable levels at day 5 post-transfection. On the other hand, extracellular HCV core Ag level of JFH-1/C was 2.2 times higher than that of JFH-1/wt, and that of JFH-1/S2 was 3.6 times higher than that of JFH-1/S1 at day 5 post-transfection (Fig. 1B). Transfection efficiency of these strains, indicated by intracellular HCV core Ag levels at 4 h post-transfection, was almost identical (data not shown).
Fig. 1. Effects of in vivo adaptive mutations on virus production in HuH-7 cells.
One million cells were transfected with 10 µg in vitro transcribed RNA of JFH-1/wt, JFH-1/S1, JFH-1/S2, and JFH-1/C. HCV RNA (A) and core Ag (B) levels in cell lysates and medium were measured at indicated time-points. Assays were performed in triplicate, and means ± standard deviation are plotted.
Single Cycle Virus Production Assay
For detailed analysis of the effects of these mutations on different stages of the virus lifecycle, we used a Huh7–25 cell line that lacks the surface expression of CD81, one of the cellular receptors for HCV entry. Three days after transfection with full-genome RNA of JFH-1/wt, S1, S2, and C, HCV RNA levels and infectivity titer were measured and the specific infectivity was calculated (Table 1). Intracellular HCV RNA levels of JFH-1/C and S2 were lower than those of JFH-1/wt and S1, suggesting lower replication efficiency of these strains. However, the intracellular infectivity titers of JFH-1/C and S2 were 2.03 and 11.0 times higher than those of JFH-1/wt and JFH-1/S1, respectively (p < 0.005). Intracellular specific infectivities (infectivity titer/ HCV RNA copy number) of JFH-1/C and S2 showed more pronounced difference from those of JFH-1/wt and JFH-1/S1 (3.92 times and 12.9 times higher, respectively; p < 0.005). The infectious virus secretion rate (extracellular infectivity titer/ intracellular infectivity titer) was not significantly different between JFH-1/wt and variant strains. These data indicate that mutations identified in chimpanzees at the later time-point of infection led to reduced viral replication and increased assembly of infectious virus particles without any effect on viral release in cell culture.
Table 1.
Infectious virus production and release of JFH-1/wt and variants in Huh7–25 cells
| Strain | Intracellular | Extracellular | Secretion Ratio (extra-/intra-) |
||
|---|---|---|---|---|---|
| HCV RNA (copies/ µg RNA) |
Infectivity Titer (ffu/well) |
Specific Infectivity (ffu/copies) |
Infectivity Titer (ffu/well) |
||
| JFH-1/wt | 7.75 × 108 ± 1.04 × 108 |
4.21 × 102 ± 4.32 × 101 |
2.09 × 10−7 ± 7.06 × 10−8 |
1.94 × 103 ± 3.76 × 101 |
4.6 ± 1.3 |
| JFH-1/S1 | 7.04 × 108 ± 8.49 × 107 |
4.72 × 102 ± 5.63 × 101 |
2.91 × 10−7 ± 6.00 × 10−8 |
3.02 × 103 ± 2.77 × 102 |
5.4 ± 2.0 |
| JFH-1/S2 | 4.16 × 108** ± 7.47 × 106 |
5.19 × 103** ± 8.24 × 101 |
3.76 × 10−6** ± 7.01 × 10−7 |
3.23 × 104** ± 3.52 × 103 |
6.2 ± 3.0 |
| JFH-1/C | 3.15 × 108* ± 5.02 × 107 |
8.59 × 102* ± 4.81 × 101 |
8.19 × 10−7* ± 5.68 × 10−8 |
3.68 × 103 ± 3.02 × 103 |
4.3 ± 1.4 |
| JFH-1/ S2-wt |
7.07 × 108 ± 8.43 × 107 |
4.40 × 103* ± 9.5 × 101 |
2.73 × 10−6* ± 2.35 × 10−7 |
3.0 × 104* ± 1.1 × 103 |
6.7 ± 0.7 |
| JFH-1/ wt-S2 |
4.21 × 108* ± 1.97 × 107 |
2.7 × 102 ± 2.9 × 101 |
2.02× 10−7 ± 4.0 × 10−8 |
1.7 × 103 ± 1.3 × 102 |
4.5 ± 0.4 |
p < 0.005 compared with JFH-1/wt
p < 0.005 compared with JFH-1/S1
Subgenomic Replicon Assay
To further confirm the replication efficiencies of strains observed in chimpanzees, we generated subgenomic replicons of JFH-1/wt, S1, S2, and C carrying the firefly luciferase reporter gene (SGR-JFH-1/Luc/wt, S1, S2, and C). In vitro transcribed RNAs of these constructs were transfected into HuH-7 cells and luciferase activity was measured to assess their replication capacity. The luciferase activities of SGR-JFH-1/Luc/C and S2 replicons were 7.30 and 7.33 times lower than those of SGR-JFH-1/Luc/wt and S1 at day 1, respectively (p < 0.00005), suggesting attenuated replication capacities of variant replicons isolated from each animal at later time-points of infection (Supporting Fig. 1A). The luciferase activity at 4 h after transfection was comparable, indicating similar levels of transfection efficiency (data not shown). Based on these data, we found that the mutations that emerged in NS3 - NS5B of JFH-1/S2 and C reduced the replication efficiency in cell culture.
Genomic Regions Responsible for Lower Replication and Higher Assembly of JFH-1/S2
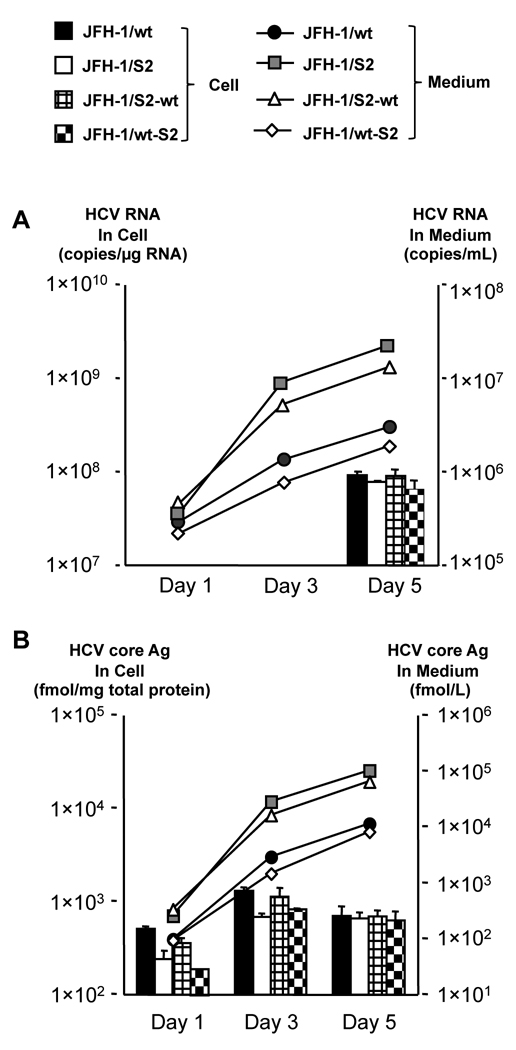
To further clarify the genomic region responsible for lower replication efficiency and higher assembly rate of JFH-1/S2, we generated the chimeric constructs JFH-1/S2-wt and wt-S2 as described in materials and methods. In vitro transcribed RNAs of JFH-1/wt, S2, S2-wt and wt-S2 were introduced into HuH-7 cells by electroporation and intra- and extracellular HCV RNA and core Ag were measured. At day 5 post-transfection, all constructs displayed comparable intracellular HCV RNA levels (Fig. 2). However, extracellular HCV RNA levels of JFH-1/S2 and S2-wt were significantly higher (p < 0.0005) than that of JFH-1/wt. On the other hand, extracellular RNA level of JFH-1/wt-S2 chimeric construct was lower than that of JFH-1/S2 and S2-wt and similar to that of JFH-1/wt. Likewise, extracellular core Ag levels of JFH-1/S2 and S2-wt were also significantly higher than that of JFH-1/wt. Intracellular HCV core Ag levels of JFH-1/S2 and wt-S2 on day 1 post-transfection were 240.9 ± 58.2 and 134.3 ± 17.1 fmol/µg protein, respectively, and were significantly lower (p < 0.005) than that of JFH-1/wt (526.1 ± 58.2 fmol/µg protein) while intracellular HCV core Ag level of JFH-1/S2-wt was comparable to that of JFH-1/wt. Transfection efficiency of these strains, indicated by intracellular HCV core Ag levels at 4 h post-transfection, was almost identical (data not shown).
Fig. 2. Virus production of JFH-1/S2 chimeric constructs in HuH-7 cells.
One million cells were transfected with 10 µg in vitro transcribed RNA of JFH-1/wt, JFH-1/S2, JFH-1/S2-wt, and JFH-1/wt-S2. HCV RNA (A) and core Ag (B) levels in cell lysates and medium were measured at indicated time-points. Assays were performed in triplicate, and means ± standard deviation are plotted.
To further elucidate, we transfected Huh7–25 cells with in vitro transcribed RNA of JFH-1/wt, S2, S2-wt and wt-S2 and measured HCV RNA, core Ag and infectivity titer in the cells and culture medium. Intracellular HCV RNA levels of JFH-1/S2 and wt-S2 were similar and lower than those of JFH-1/wt and S2-wt, suggesting mutations in NS3 - NS5B were responsible for lower replication efficiency of JFH-1/S2 (Table 1). Intracellular infectivity titer of JFH-1/S2 and S2-wt was 12.3 and 10.4 times higher than that of JFH-1/wt, respectively (p < 0.005) on day 3 post-transfection. The intracellular specific infectivities of JFH-1/S2 and S2-wt were significantly higher than that of JFH-1/wt (18 times and 13.1 times higher, respectively; p < 0.005). On the other hand, intracellular specific infectivity of JFH-1/wt-S2 was comparable to that of JFH-1/wt. The infectious virus secretion rate was not significantly different among all the constructs (Table 1). These data indicate that mutations emerged in core - NS2 region of JFH-1/S2 are responsible for the enhanced assembly of infectious virus particles as compared to JFH-1/wt.
Mapping Study for JFH-1/S2 strain
As our experiments with JFH-1/S2 subgenomic replicon and JFH-1/wt-S2 chimeric construct showed that mutations emerged in NS3 - NS5B region are responsible for reduced replication efficiency of JFH-1/S2, we performed mapping studies by generating various JFH-1 subgenomic replicons, each containing the mutations observed in individual non-structural protein. Although mutations in NS4B and NS5A were associated with attenuated replication capacity of JFH-1, the most significant decrease in replication was observed with NS5B mutations (Supporting Fig. 1B).
For detailed analysis of mutations responsible for higher assembly, in vitro transcribed RNAs of JFH-1/wt, S2, S2-wt, N397S, L752V, S2-NS2 (containing mutations G838R, A878V and V881A), G838R and A878V, were transfected into Huh7–25 cells, and intracellular specific infectivities were compared (Supporting Table 2). As reported previously, JFH1/G838R showed higher intracellular specific infectivity than that of JFH-1/wt, but could not reach to the level of JFH-1/S2 or JFH-1/S2-wt. Among the mutants, intracellular specific infectivities of JFH1/L752V, JFH1/NS2 and JFH1/G838R were 4.02, 5.42 and 3.07 times higher than that of JFH-1/wt, but of JFH1/N397S and JFH1/A878V were similar to that of JFH-1/wt. Thus the combination of mutations in P7 and NS2 was found to contribute to the higher assembly of JFH-1/S2 strain.
Human Hepatocyte-transplanted Mice Assay
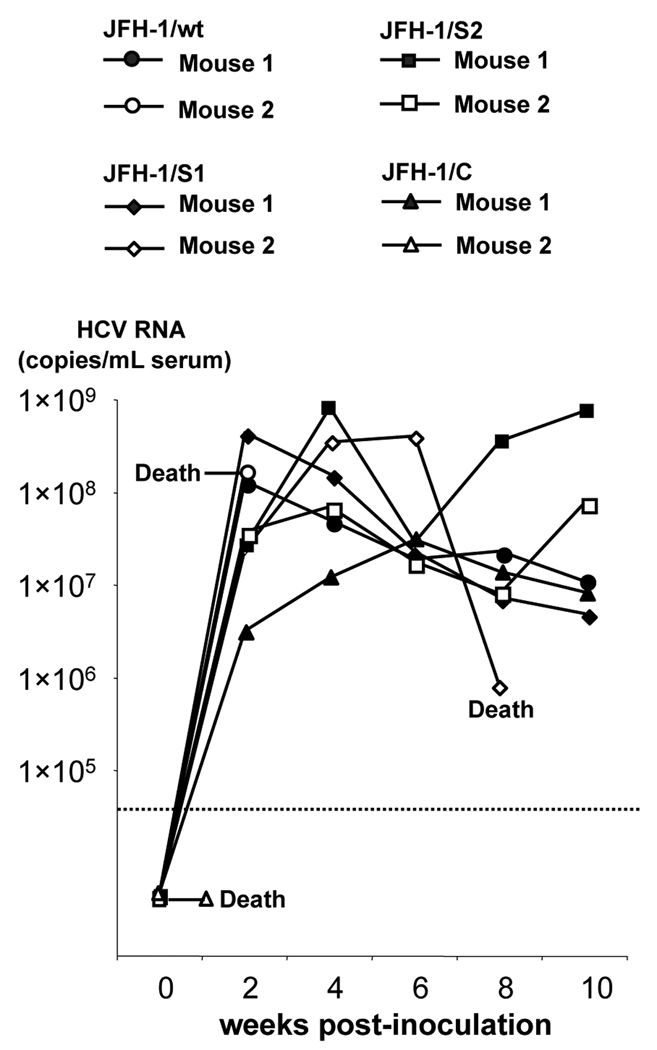
To assess the in vivo infectivity of these strains, we inoculated culture medium containing 107 copies (HCV RNA titer measured by RTD-PCR) of JFH-1/wt, S1, S2, and C viruses into human hepatocyte-transplanted mice. Two mice were used for each virus. Two weeks after intravascular inoculation, all mice but one became HCV RNA positive (Fig. 3). Two mice died 3 weeks after inoculation; one was inoculated with JFH-1/wt and had developed infection, while the other was inoculated with JFH-1/C and died without developing infection. HCV RNA levels in infected mice fluctuated, ranging from 106 to 109 copies/mL. We could not observe much difference of infected HCV RNA titer among these inoculated mice. Sequence analyses of the complete ORFs revealed that infecting JFH-1/wt virus and variant strains had no non-synonymous mutations at the time of development of infection. From these data, we concluded that not only JFH-1/wt virus but also JFH-1/S1, S2 and C viruses were able to establish productive infection in human hepatocyte-transplanted mice.
Fig. 3. In vivo infection study of JFH-1/wt and its variants in human hepatocyte-transplanted mice.
Cell culture medium containing 1 × 107 HCV RNA copies of JFH-1/wt, S1, S2, and C were inoculated into human hepatocyte-transplanted mice, and HCV RNA levels in mice serum were monitored.
Apoptosis Induction Assay
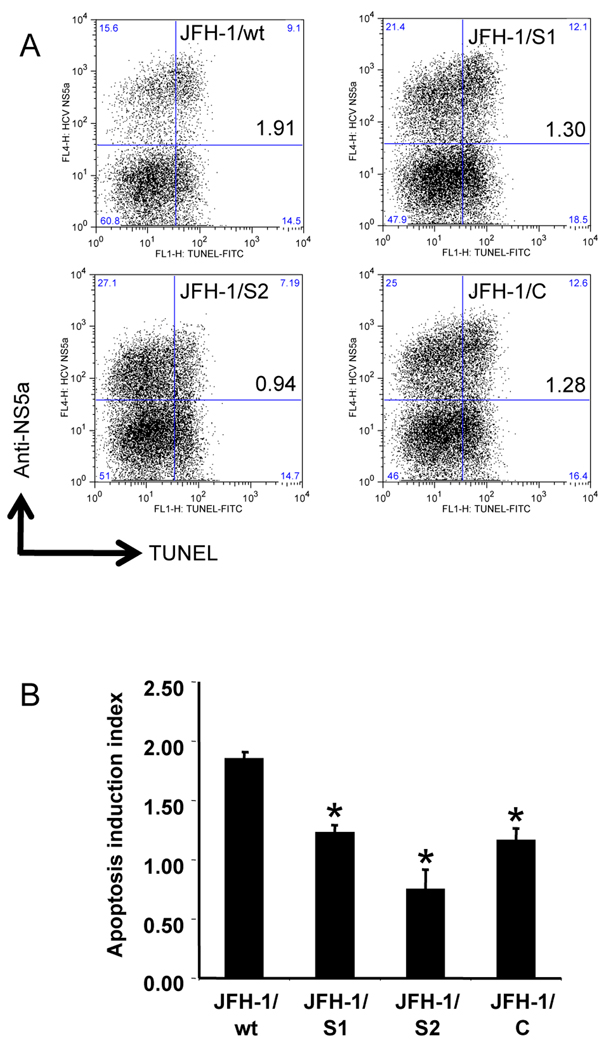
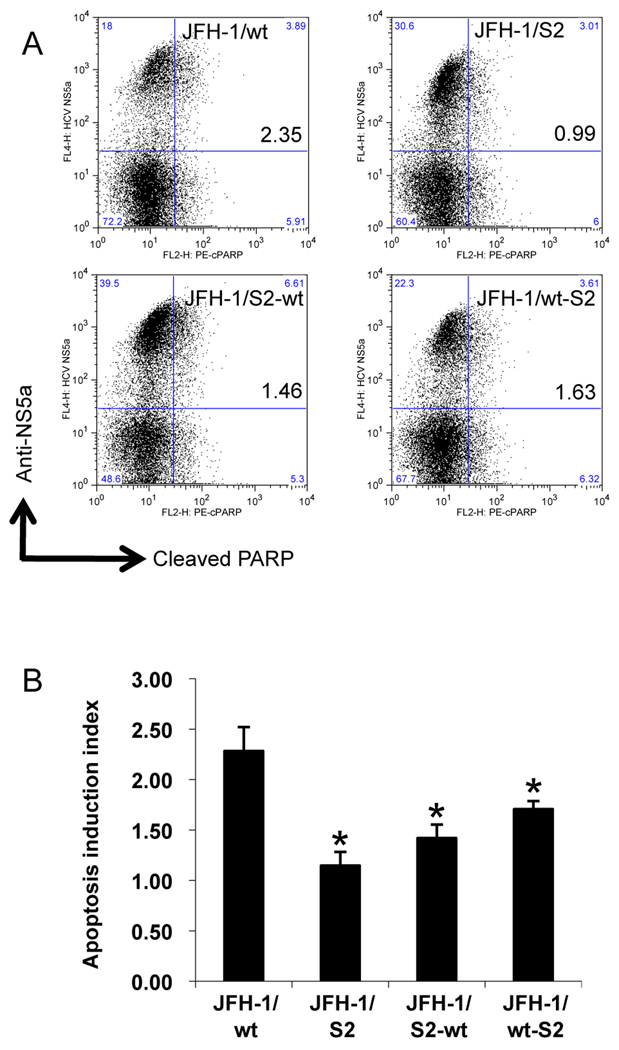
To investigate the survival strategy against the host defense system, we examined the susceptibility of JFH-1/wt and variant strains to TNF-α-mediated apoptosis induction. After transfection with in vitro transcribed RNA of JFH-1/wt, S1, S2, and C, Huh-7.5.1 cells were exposed to TNF-α plus Actinomycin D (Act D). Without exposure, apoptosis was observed in a limited number of HCV positive cells (Supporting Fig. 2A). Forty-eight hours later, cells were harvested, fixed, and subjected to terminal deoxynucleotidyltrasferase mediated dUTP nick end-labeling (TUNEL) assay and anti-HCV NS5a staining. The effects of JFH-1/wt, S1, S2, and C transfection on apoptosis induction were determined by calculating the ratio of apoptosis between HCV-positive and -negative populations and expressed as an apoptosis induction index. After treatment of JFH-1/wt-transfected cells with TNF-α, apoptosis was observed in 36.8% of the HCV-positive population and in 19.3% of the HCV-negative population, and the apoptosis induction index was 1.85 ± 0.06 (Fig. 4). The apoptosis induction indexes of JFH-1/S1- and C- transfected cells were 1.23 ± 0.06 and 1.16 ± 0.10, respectively, suggesting lower susceptibility to apoptosis induction compared to JFH-1/wt. On the other hand, the apoptosis induction index of JFH-1/S2 was 0.74 ± 0.17, which was substantially lower than that of JFH-1/wt demonstrating the more reduced apoptosis in the cells harboring this strain. Similar results were obtained by treatment with FasL plus Act D (Supporting Fig. 2B). To confirm the lower susceptibility of JFH-1/S2 transfected cells, apoptosis was also detected by staining with anti-cleaved poly [ADP-Ribose] polymerase (PARP) antibody. The apoptosis induction indexes of JFH-1/wt and S2 transfected cells were 2.28 ± 0.24 and 1.15 ± 0.14, respectively, and were consistent with TUNEL assay (Fig. 5). Although the HCV NS5a-positive rate in JFH-1/S2 transfected cells was higher than that in JFH-1/wt, the mean fluorescence intensity (MFI) of the NS5a-positive population in JFH-1/S2 transfected cells was significantly lower (185.0 ± 8.7) than that in JFH-1/wt transfected cells (395.0 ± 98.0), corresponding to the observed phenotype of the JFH-1/S2 strain in the single cycle virus production assay; i.e., lower replication efficiency and rapid spread to surrounding cells.
Fig. 4. Apoptosis induction in Huh-7.5.1 cells transfected with JFH-1/wt and its variants.
(A) Three million cells were transfected with 3 µg in vitro transcribed full-genome RNA of JFH-1/wt, S1, S2, and C. Forty-eight hours later, apoptosis was induced by exposing cells to 20 ng/mL TNF-α plus 50 ng/mL Act D. Cells were harvested after 48 h of treatment, and subjected to TUNEL and anti-HCV NS5a staining. Dot-plots show HCV replication and apoptosis at the single cell level. Quadrant gates were determined using unstained- and a TdT-untreated-control in each culture condition. The clone names and apoptosis induction indexes are indicated in the upper right box. (B) Apoptosis induction indexes of JFH-1/wt-, S1-, S2-, and C- transfected cells. Means ± standard deviations of 3 independent experiments are shown. *p < 0.005 compared to JFH-1/wt.
Fig. 5. Apoptosis induction in Huh-7.5.1 cells transfected with JFH-1/wt, S2, and their chimeric constructs.
(A) Three million cells were transfected with 3 µg in vitro transcribed full-genome RNA of JFH-1/wt, S2, S2-wt, and wt-S2. Apoptosis was induced by exposing cells to 20 ng/mL TNF-α plus 50 ng/mL Act D and detected by anti-cleaved PARP staining. The clone names and apoptosis induction indexes are indicated in the upper right box. (B) Apoptosis induction indexes of JFH-1/wt-, S2-, S2-wt-, and wt-S2- transfected cells. Means ± standard deviations of 3 independent experiments are shown. *p < 0.05 compared to JFH-1/wt.
To clarify the genomic region responsible for lower susceptibility of JFH-1/S2 to cytokine-induced apoptosis, we examined the effect of TNF-α on the cells carrying subgenomic reporter replicons. The apoptosis induction index of SGR-JFH1/Luc/S2 transfected cells was lower than that of SGR-JFH1/Luc/wt transfected cells (Supporting Fig. 2C), however the difference was not as pronounced as with full-genome constructs, indicating that mutations in NS3 - NS5B region contribute to lower susceptibility of JFH-1/S2 to cytokine-induced apoptosis but they are not sufficient to explain the difference between JFH-1/wt and JFH-1/S2. We confirmed these results by use of chimeric constructs JFH-1/S2-wt and wt-S2. The apoptosis induction indexes of JFH-1/S2-wt- and wt-S2- transfected cells were 1.42 ± 0.13 and 1.71 ± 0.08, respectively (Fig. 5). These data indicated that both structural and non-structural regions of JFH-1/S2 were associated with lower susceptibility to cytokine-induced apoptosis, although mutations in core-NS2 seemed to have higher contribution towards this phenotype. Altogether, these results indicate that the JFH-1/S2 strain, which was selected after passage in the patient serum-infected chimpanzee, acquired less susceptibility to the cytokine-induced apoptosis.
DISCUSSION
HCV develops chronic infection in the vast majority of infected patients (1), however, the mechanisms of its persistence are still under investigation. Many viruses have evolved different strategies to cope with host immune systems, thus causing the development of persistent infection. For example, some viruses interfere with the major histocompatibility complex (MHC) class I presentation of viral antigens, while others modulate lymphocyte and macrophage functions, including cytokine production (12–16). In our previous study, we detected increasing number of mutations in HCV genome isolated from JFH-1 patient serum-infected chimpanzees. Thus we reasoned that these detected mutations might have imparted some advantage to this virus for long-time survival. To examine this hypothesis, we compared the phenotypes of JFH-1 variant strains emerged at early and late stages of infection in JFH-1 patient serum- and JFH-1cc- infected chimpanzees, and found that the JFH-1/S2 strain isolated from the patient serum-infected chimpanzee at a later time-point of infection replicated slowly, produced more infectious viruses, and displayed reduced susceptibility to cytokine-induced apoptosis.
The JFH-1 variant strains JFH-1/C, which contains 7 non-synonymous mutations identified in JFH-1cc infected chimpanzee at week 7, showed comparatively slower replication kinetics and slightly enhanced infectious virus production in cell culture. The intracellular specific infectivity of this strain in Huh7–25 cells was 3.9 times higher than that of JFH-1/wt (Table 2). These characteristics might have imparted some advantage to this strain for establishing productive infection in the chimpanzee. The other JFH-1 variant strains, JFH-1/S1 and S2, contain 6 and 17 non-synonymous mutations identified in JFH-1 patient serum infected chimpanzee at weeks 2 and 23 post-infection, respectively. Replication kinetics and infectious virus production of the JFH-1/S1 strain were comparable to that of JFH-1/wt in cultured cells (Fig. 1, Table 2). In contrast, the JFH-1/S2 strain showed lower replication efficiency. Although the intracellular HCV RNA level of this strain in Huh7–25 cells was lower than that of JFH-1/wt and S1, and almost the same as that of JFH-1/C (Table 2), intracellular specific infectivity was 18.0 and 12.9 times higher than that of JFH-1/wt and S1, respectively, suggesting a significant increase in the assembly of infectious virus particles (p < 0.005, Table 2). The enhanced capacity of this strain to assemble infectious virus particles resulted in a higher extracellular infectivity titer that contributed to the rapid spread of virus to surrounding cells. Flow cytometry analyses of cells transfected with JFH-1/wt and variant strains revealed that the percentage of the HCV NS5a-positive population in JFH-1/S2 transfected cells was higher, but the MFI of the anti-NS5a signal was lower than that in JFH-1/wt transfected cells, thus confirming higher spread and lower replication of this strain. Taken together, both JFH-1/C and JFH-1/S2 exhibited a tendency towards decreased replication and increased infectious virus production. However, the extent of enhanced virus production was substantially lower in JFH-1/C than JFH-1/S2 strain, which might have led to the earlier elimination of infection in the JFH-1cc-infected chimpanzee. In other words, the potency of infectious virus production and spread seems to correspond to the duration of infection in infected animals.
The association between a lower replication efficiency and persistent infection is still unclear. It has been reported that an escape mutant with an amino acid substitution at the CTL epitope in the NS3 region exhibits lower NS3/4 protease activity and replication capacity in vitro (17,18). The JFH-1/S2 strain contains the T1077A mutation in the NS3 region (Table 1), and this mutation is located close to mutations reported to be associated with immune evasion and lower replication (17). Thus, the lower replication efficiency of the JFH-1/S2 strain may be a result of an immune escape mutation at the expense of viral fitness. Meanwhile, we cannot deny the advantage of lower replication in establishing persistent infection. Lower replication may contribute to the avoidance of MHC class I mediated antigen presentation and to escape from the host immune system. Either way, by acquiring the ability to produce more viral particles, the JFH-1/S2 strain could rapidly spread to surrounding cells, irrespective of its lower replication efficiency. Importantly, these emerged mutations did not attenuate in vivo infectivity, unlike cell culture adaptive mutations reported to cause attenuated infection in vivo (19). Upon inoculation into human hepatocyte-transplanted mice, JFH-1/S1, S2 and C strains could establish infection without any mutations, produced levels of viremia similar to JFH-1/wt, and persisted for a similar observed period of infection (Fig. 2). This observation is different from that in chimpanzees, where JFH-1/wt and JFH-1/C strains were eliminated earlier than JFH-1/S2. In contrast to chimpanzees, human hepatocyte-transplanted mice lack cytotoxic T lymphocytes (CTL) and the natural killer (NK) cell-mediated immune system, which could be responsible for this difference (6). Taken together, our results suggest that along with efficient infectious virus production, the JFH-1/S2 strain might have acquired an advantage that helps it evade the CTL and NK cell-mediated immune system.
Apoptosis of virus-infected cells by the immune system is crucial as a general mechanism of clearing infections (20, 21). The J6/JFH-1 chimeric virus has been reported to exhibit pro-apoptotic characteristics in cell culture (22). However, because HCV needs to escape the host immune system in order to establish chronic infection, immune cell-mediated apoptosis may be inhibited in infected hepatocytes. In the liver, HCV-infected hepatocytes are eliminated by targeted apoptosis induced by NK cells, macrophages, and CTL with ligand- and receptor-mediated signals such as TNF-α, FasL, and TNF-related apoptosis inducing ligand (TRAIL) (23–26). Thus, we used TNF-α to mimic natural immune-mediated apoptosis, and found that the JFH-1/S2 strain replicating cells have lower susceptibility to the apoptosis induced by these cytokines. In JFH-1/S2-transfected cells, TNF-α-induced apoptosis detected by TUNEL assay was substantially lower than that of JFH-1/wt transfected cells (Fig. 4). We confirmed it by staining with anti cleaved PARP. In complete agreement with the results produced by TUNEL assay, number of anti cleaved PARP stained cells among JFH-1/S2-infected cells was significantly lower than that among JFH-1/wt infected cells (Fig. 5). In our previous study, we reported that HCV-specific immune responses with T-cell proliferation and interferon gamma production were maintained until the disappearance of viremia in the patient serum-infected chimpanzee (11). This indicates that continuous selection pressure in the infected chimpanzee might have contributed to the emergence of a clone with an ability to escape the cytokine-induced apoptosis. We are not sure whether this phenotype of JFH-1/S2 is due to its lower replication efficiency and thus lower production of HCV proteins. The accumulation of viral proteins might predispose cells to the apoptosis induced by TNF-α. To answer this question, it will be necessary to investigate the genomic region(s) of JFH-1/S2 and cellular host factors responsible for the ability of this strain to escape the apoptosis.
By mapping analysis for JFH-1/S2 strain, we could determine responsible regions; NS5B was for lower replication efficiency (Supporting Fig. 1B), and P7 and NS2 were for enhanced viral particle assembly (Supporting Table 2). For the evasion of apoptosis, we could not specify the responsible region, because both chimeric constructs, JFH-1/S2-wt and wt-S2, showed less susceptibility to cytokine-induced apoptosis to a certain extent. These data indicated that both structural and non-structural regions might have contributed to the acquisition of this phenotype. Previously, a potent anti-apoptotic effect of the HCV NS5a protein was described (27). NS5a interacts with Bin1, which is a nucleocytoplasmic c-Myc-interacting protein with tumor suppressor and apoptotic properties, thus inhibiting Bin1-associated apoptosis. As JFH-1/S2 contains several mutations in the NS5a region (Supporting Table 1), one or more mutations in this protein may be associated with anti-apoptotic effects.
In conclusion, we demonstrated that the JHF-1/S2 strain acquired phenotypes of lower replication, higher virus production, and less susceptibility to cytokine-induced apoptosis. These phenotypes were associated with mutations that emerged 23 weeks after infection in a chimpanzee, and might have contributed to long-term infection in vivo. Such control of viral functions by specific mutations may be a key viral strategy to establish persistent infection.
Supplementary Material
ACKNOWLEDGEMENT
The authors are grateful to Francis V. Chisari for providing Huh-7.5.1 cell line and Nao Sugiyama for technical assistance.
Financial Support:
This work was supported by a grant-in-aid from the Japan Society for the Promotion of Science, from the Ministry of Health, Labor and Welfare of Japan, from the Ministry of Education, Culture, Sports, Science and Technology, and by the Research on Health Sciences Focusing on Drug Innovation from the Japan Health Sciences Foundation.
List of Abbreviations
- Act D
Actinomycin D
- Ag
antigen
- CTL
cytotoxic T lymphocytes
- DMEM
Dulbecco’s modified Eagle’s medium
- FasL
Fas ligand
- FFU
focus-forming units
- HCV
Hepatitis C virus
- JFH-1cc
cell culture-generated JFH-1 virus
- LU
light units
- MFI
mean fluorescence intensity
- MHC
major histocompatibility complex
- NK
natural killer
- NS
non-structural
- SCID
severe combined immunodeficiency
- TdT
terminal deoxynucleotidyltrasferase
- TNF-α
tumor necrosis factor-α
- TRAIL
TNF-related apoptosis inducing ligand
- TUNEL
TdT mediated dUTP nick end-labeling
- uPA
urokinase-type plasminogen activator
- wt
wild type
Footnotes
Disclosures:
All authors have nothing to disclose.
Author Contributions:
K.K, T.J.L, T.W and T.K. generated the idea and planned the project;
M. Siina, T.S., H.W., K.C., T.W. and T.K. designed the experiments;
M. Saeed, T.D., D.A., N.W., A.M., Y.C. and T.K. performed the experiments and collected the data;
N.H., M.I. and K.C. contributed in experiment of human hepatocyte-transplanted mice;
M. Saeed and T.K. analyzed the data and wrote the manuscript;
All authors discussed the results and commented on the manuscript.
REFERENCES
- 1.Liang TJ, Rehermann B, Seeff LB, Hoofnagle JH. Pathogenesis, natural history, treatment, and prevention of hepatitis C. Ann Intern Med. 2000;132:296–305. doi: 10.7326/0003-4819-132-4-200002150-00008. [DOI] [PubMed] [Google Scholar]
- 2.Feld JJ, Liang TJ. Hepatitis C -- identifying patients with progressive liver injury. Hepatology. 2006;43:S194–S206. doi: 10.1002/hep.21065. [DOI] [PubMed] [Google Scholar]
- 3.Thimme R, Oldach D, Chang KM, Steiger C, Ray SC, Chisari FV. Determinants of viral clearance and persistence during acute hepatitis C virus infection. J Exp Med. 2001;194:1395–1406. doi: 10.1084/jem.194.10.1395. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Thimme R, Bukh J, Spangenberg HC, Wieland S, Pemberton J, Steiger C, Govindarajan S, Purcell RH, Chisari FV. Viral and immunological determinants of hepatitis C virus clearance, persistence, and disease. Proc Natl Acad Sci U S A. 2002;99:15661–15668. doi: 10.1073/pnas.202608299. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Mercer DF, Schiller DE, Elliott JF, Douglas DN, Hao C, Rinfret A, Addison WR, Fischer KP, Churchill TA, Lakey JR, Tyrrell DL, Kneteman NM. Hepatitis C virus replication in mice with chimeric human livers. Nat Med. 2001;7:927–933. doi: 10.1038/90968. [DOI] [PubMed] [Google Scholar]
- 6.Tateno C, Yoshizane Y, Saito N, Kataoka M, Utoh R, Yamasaki C, Tachibana A, Soeno Y, Asahina K, Hino H, Asahara T, Yokoi T, Furukawa T, Yoshizato K. Near completely humanized liver in mice shows human-type metabolic responses to drugs. Am J Pathol. 2004;165:901–912. doi: 10.1016/S0002-9440(10)63352-4. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Kato T, Furusaka A, Miyamoto M, Date T, Yasui K, Hiramoto J, Nagayama K, Tanaka T, Wakita T. Sequence analysis of hepatitis C virus isolated from a fulminant hepatitis patient. J Med Virol. 2001;64:334–339. doi: 10.1002/jmv.1055. [DOI] [PubMed] [Google Scholar]
- 8.Wakita T, Pietschmann T, Kato T, Date T, Miyamoto M, Zhao Z, Murthy K, Habermann A, Krausslich HG, Mizokami M, Bartenschlager R, Liang TJ. Production of infectious hepatitis C virus in tissue culture from a cloned viral genome. Nat Med. 2005;11:791–796. doi: 10.1038/nm1268. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Zhong J, Gastaminza P, Cheng G, Kapadia S, Kato T, Burton DR, Wieland SF, Uprichard SL, Wakita T, Chisari FV. Robust hepatitis C virus infection in vitro. Proc Natl Acad Sci U S A. 2005;102:9294–9299. doi: 10.1073/pnas.0503596102. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Lindenbach BD, Evans MJ, Syder AJ, Wolk B, Tellinghuisen TL, Liu CC, Maruyama T, Hynes RO, Burton DR, McKeating JA, Rice CM. Complete replication of hepatitis C virus in cell culture. Science. 2005;309:623–626. doi: 10.1126/science.1114016. [DOI] [PubMed] [Google Scholar]
- 11.Kato T, Choi Y, Elmowalid G, Sapp RK, Barth H, Furusaka A, Mishiro S, Wakita T, Krawczynski K, Liang TJ. Hepatitis C virus JFH-1 strain infection in chimpanzees is associated with low pathogenicity and emergence of an adaptive mutation. Hepatology. 2008;48:732–740. doi: 10.1002/hep.22422. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Johannessen I, Crawford DH. In vivo models for Epstein-Barr virus (EBV)-associated B cell lymphoproliferative disease (BLPD) Rev Med Virol. 1999;9:263–277. doi: 10.1002/(sici)1099-1654(199910/12)9:4<263::aid-rmv256>3.0.co;2-d. [DOI] [PubMed] [Google Scholar]
- 13.Oglesbee MJ, Pratt M, Carsillo T. Role for heat shock proteins in the immune response to measles virus infection. Viral Immunol. 2002;15:399–416. doi: 10.1089/088282402760312296. [DOI] [PubMed] [Google Scholar]
- 14.Stevenson PG, Boname JM, de Lima B, Efstathiou S. A battle for survival: immune control and immune evasion in murine gamma-herpesvirus-68 infection. Microbes Infect. 2002;4:1177–1182. doi: 10.1016/s1286-4579(02)01643-x. [DOI] [PubMed] [Google Scholar]
- 15.Alcami A. Viral mimicry of cytokines, chemokines and their receptors. Nat Rev Immunol. 2003;3:36–50. doi: 10.1038/nri980. [DOI] [PubMed] [Google Scholar]
- 16.Wilkinson GW, Tomasec P, Stanton RJ, Armstrong M, Prod'homme V, Aicheler R, McSharry BP, Rickards CR, Cochrane D, Llewellyn-Lacey S, Wang EC, Griffin CA, Davison AJ. Modulation of natural killer cells by human cytomegalovirus. J Clin Virol. 2008;41:206–212. doi: 10.1016/j.jcv.2007.10.027. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Soderholm J, Ahlen G, Kaul A, Frelin L, Alheim M, Barnfield C, Liljestrom P, Weiland O, Milich DR, Bartenschlager R, Sallberg M. Relation between viral fitness and immune escape within the hepatitis C virus protease. Gut. 2006;55:266–274. doi: 10.1136/gut.2005.072231. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Uebelhoer L, Han JH, Callendret B, Mateu G, Shoukry NH, Hanson HL, Rice CM, Walker CM, Grakoui A. Stable cytotoxic T cell escape mutation in hepatitis C virus is linked to maintenance of viral fitness. PLoS Pathog. 2008;4:e1000143. doi: 10.1371/journal.ppat.1000143. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Bukh J, Pietschmann T, Lohmann V, Krieger N, Faulk K, Engle RE, Govindarajan S, Shapiro M, St Claire M, Bartenschlager R. Mutations that permit efficient replication of hepatitis C virus RNA in Huh-7 cells prevent productive replication in chimpanzees. Proc Natl Acad Sci U S A. 2002;99:14416–14421. doi: 10.1073/pnas.212532699. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Kagi D, Seiler P, Pavlovic J, Ledermann B, Burki K, Zinkernagel RM, Hengartner H. The roles of perforin- and Fas-dependent cytotoxicity in protection against cytopathic and noncytopathic viruses. Eur J Immunol. 1995;25:3256–3262. doi: 10.1002/eji.1830251209. [DOI] [PubMed] [Google Scholar]
- 21.Kagi D, Vignaux F, Ledermann B, Burki K, Depraetere V, Nagata S, Hengartner H, Golstein P. Fas and perforin pathways as major mechanisms of T cell-mediated cytotoxicity. Science. 1994;265:528–530. doi: 10.1126/science.7518614. [DOI] [PubMed] [Google Scholar]
- 22.Deng L, Adachi T, Kitayama K, Bungyoku Y, Kitazawa S, Ishido S, Shoji I, Hotta H. Hepatitis C virus infection induces apoptosis through a Bax-triggered, mitochondrion-mediated, caspase 3-dependent pathway. J Virol. 2008;82:10375–10385. doi: 10.1128/JVI.00395-08. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Kafrouni MI, Brown GR, Thiele DL. Virally infected hepatocytes are resistant to perforin-dependent CTL effector mechanisms. J Immunol. 2001;167:1566–1574. doi: 10.4049/jimmunol.167.3.1566. [DOI] [PubMed] [Google Scholar]
- 24.Guicciardi ME, Gores GJ. Apoptosis: a mechanism of acute and chronic liver injury. Gut. 2005;54:1024–1033. doi: 10.1136/gut.2004.053850. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Fischer R, Baumert T, Blum HE. Hepatitis C virus infection and apoptosis. World J Gastroenterol. 2007;13:4865–4872. doi: 10.3748/wjg.v13.i36.4865. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Stegmann KA, Bjorkstrom NK, Veber H, Ciesek S, Riese P, Wiegand J, Hadem J, Suneetha PV, Jaroszewicz J, Wang C, Schlaphoff V, Fytili P, Cornberg M, Manns MP, Geffers R, Pietschmann T, Guzman CA, Ljunggren HG, Wedemeyer H. Interferon-alpha-induced TRAIL on natural killer cells is associated with control of hepatitis C virus infection. Gastroenterology. 2010;138:1885–1897. doi: 10.1053/j.gastro.2010.01.051. [DOI] [PubMed] [Google Scholar]
- 27.Nanda SK, Herion D, Liang TJ. The SH3 binding motif of HCV NS5A protein interacts with Bin1 and is important for apoptosis and infectivity. Gastroenterology. 2006;130:794–809. doi: 10.1053/j.gastro.2005.12.030. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.