Summary
Tumor heterogeneity is a major barrier to effective cancer diagnosis and treatment. We recently identified cancer-specific differentially DNA-methylated regions (cDMRs) in colon cancer, which also distinguish normal tissue types from each other, suggesting that these cDMRs might be generalized across cancer types. Here we show stochastic methylation variation of the same cDMRs, distinguishing cancer from normal, in colon, lung, breast, thyroid, and Wilms tumors, with intermediate variation in adenomas. Whole genome bisulfite sequencing shows these variable cDMRs are related to loss of sharply delimited methylation boundaries at CpG islands. Furthermore, we find hypomethylation of discrete blocks encompassing half the genome, with extreme gene expression variability. Genes associated with the cDMRs and large blocks are involved in mitosis and matrix remodeling, respectively. These data suggest a model for cancer involving loss of epigenetic stability of well-defined genomic domains that underlies increased methylation variability in cancer and could contribute to tumor heterogeneity.
Introduction
Cancer is generally viewed as over 200 separate diseases of abnormal cell growth, controlled by a series of mutations, but also involving epigenetic non-sequence changes involving the same genes1. DNA methylation at CpG dinucleotides has been studied extensively in cancer, with hypomethylation or hypermethylation reported at some genes, and global hypomethylation ascribed to normally methylated repetitive DNA elements. Until now, cancer epigenetics has focused on high-density CpG islands, gene promoters, or dispersed repetitive elements2,3.
Here we have taken a different and more general approach to cancer epigenetics. It is based on our recent observation of frequent methylation alterations in colon cancer of lower cytosine-density CpG regions near islands, termed shores; as well as the observation that these cancer-specific differentially methylated regions, or cDMRs, correspond largely to the same regions that show DNA methylation variation among normal spleen, liver, and brain, or tissue-specific DMRs (tDMRs)4. Furthermore, cDMRs are highly enriched among regions differentially methylated during stem cell reprogramming of induced pluripotent stem (iPS) cells5. We thus reasoned that the very same sites might be generalized cDMRs, since they are involved in normal tissue differentiation but show aberrant methylation in at least one cancer type (colon).
We tested this hypothesis by designing a semi-quantitative custom Illumina array for methylation analysis of 151 cDMRs consistently altered across colon cancer, and analyzed these sites in 290 samples, including matched normal and cancer from colon, breast, lung, thyroid, and Wilms’ tumor. We were surprised to discover that almost all of these cDMRs were altered across all cancers tested. Specifically, the cDMRs showed increased stochastic variation in methylation level within each tumor type, suggesting a generalized disruption of the integrity of the cancer epigenome. To investigate this idea further, we performed genome-scale bisulfite sequencing of 3 colorectal cancers, the matched normal colonic mucosa, and two adenomatous polyps. These experiments revealed a surprising loss of methylation stability in colon cancer, involving CpG islands and shores, and large (up to several megabases) blocks of hypomethylation affecting more than half of the genome, with associated stochastic variability in gene expression, which could provide an epigenetic mechanism for tumor heterogeneity.
RESULTS
Stochastic variation in DNA methylation across cancer types
We sought to increase the precision of DNA methylation measurements over our previous tiling array-based approach, termed CHARM6, analyzing 151 colon cDMRs4. We designed a custom nucleotide-specific Illumina bead array 384 probes covering 139 regions7. We studied 290 samples, including cancers from colon, lung, breast, thyroid, and Wilms’, with matched normal tissues to 111 of these 122 cancers, along with 30 colon premalignant adenomas and 27 additional normal samples (see Methods). To minimize the risk of genetic heterogeneity arising from sampling multiple clones we purified DNA from small (0.5 cm × 0.2 cm) sections verified by histopathologic examination.
Cluster analysis of the DNA methylation values revealed that the colon cancer cDMRs largely distinguished cancer from normal for each tumor type (Supplementary Fig. 1). The increased across-sample variability in methylation within the cancer samples of each tumor type compared to normal was even more striking than differences in mean methylation. We therefore computed across-sample variance within normal and cancer samples in all five tumor/normal tissue types at each CpG site. Although these CpGs sites were selected for differences in mean values in colon cancer, the great majority exhibited greater variance in cancer than normal in each tissue type (Fig. 1a–e), even accounting for differences in variability expected from mean shifts according to a binomial distribution model of methylation measurements (Supplementary Fig. 2). This increase was statistically significant (p<0.01, using an F-test) for 81%, 92%,81%, 70%, and 80% of the CpG sites in colon, lung, breast, thyroid, and Wilms tumor, respectively. Furthermore, 157 CpG sites had statistically significant increased variability in all cancer types tested. This increased stochastic variation was found in CpG islands, CpG island shores, and regions distant from islands (Fig. 1a–e). These data suggest a potential mechanism of tumor heterogeneity, namely increased stochastic variation of DNA methylation in cancers compared to normal, within each tumor type tested (see Discussion). We ruled out increased cellular heterogeneity and patient age as artifactual causes for methylation heterogeneity in cancer samples (Supplementary Figs. 3 and 4). Furthermore, there was no difference in methylation hypervariability comparing five high copy variation colon cancers to five low copy variation Wilms tumors (Supplementary Fig. 5a–b), arguing against genetic heterogeneity as a cause of methylation hypervariability. Similarly, 7 Wilms tumors without aberrant p53 expression by immunohistochemistry showed similar methylation hypervariability to 7 colon tumors with positive staining, a marker of chromosomal instability (Supplementary Fig. 6).
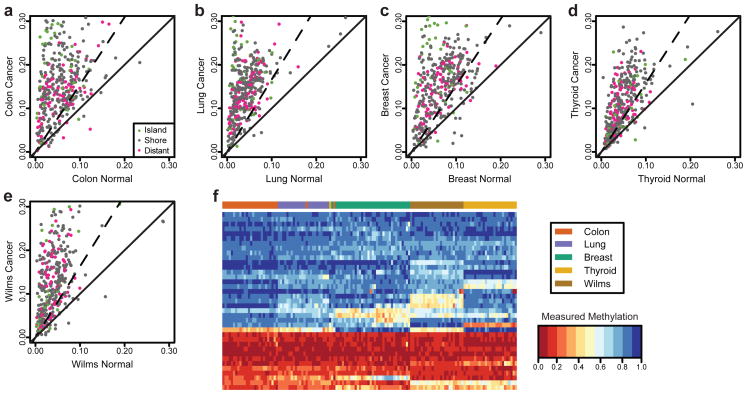
Figure 1. Increased methylation variance of common CpG sites across human cancer types.
Methylation levels measured at 384 CpG sites using a custom Illumina array exhibit an increase in across-sample variability in (a) colon, (b) lung, (c) breast, (d) thyroid, and (e) kidney (Wilms tumor) cancers. Each panel shows the across-sample standard deviation of methylation level for each CpG in normal and matched cancer samples. The solid line is the identity line; CpGs above this line have greater variability in cancer. The dashed line indicates the threshold at which differences in methylation variance become significant (F-test at 99% level). In all five tissue types, the vast majority of CpGs are above the solid line, indicating that variability is larger in cancer samples than in normal. Colors indicate the location of each CpG with respect to canonical annotated CpG islands. (f) Using the CpGs that showed the largest increase in variability we performed hierarchical clustering on the normal samples. The heatmap of the methylation values for these CpGs clearly distinguishes the tissue types, indicating that these sites of increased methylation heterogeneity in cancer are tissue-specific DMRs.
The loci where increased variability in cancer was observed are also able to distinguish the five normal tissues from each other, but this is a mean shift rather than a variation shift, apparent from cluster analysis (Supplementary Fig. 7). Interestingly, this is the case even when only using the 25 most variable sites in cancer (Fig. 1f). This result reinforces the concept of a biological relationship between normal tissue differentiation and stochastic variation in cancer DNA methylation.
To determine if the increased variability is a general property of cytosine methylation in cancer or a specific property of the CpGs selected for our custom array, we used as a control a publicly available methylation dataset comparing colorectal cancer to matched normal mucosa on the Illumina Human Methylation 27k beadchip array. In this dataset we found that only 42% of the sites showed a statistically significant increase in methylation variability, compared to 81% in the custom array (p<0.01), confirming the specificity of the cancer DMRs included in our custom array. Increased stochastic variation was more common in CpGs far from islands (57%) than in shores (44%) or islands (31%), contrasting the relative representation of these locations on the 27k array which breaks down as: distal to islands (26.4%), shores (31.6%) and islands (42%) (see Methods). This result suggested that something other than relationship to CpG islands might be defining the largest fraction of sites of altered DNA methylation in cancer.
Hypomethylation of large DNA methylation blocks in colon cancer
The methylation stochasticity described above appears to be a general property of cancer, affecting cDMRs in both island and non-island regions, in all five cancer types tested. To investigate this apparent universal loss of DNA methylation pattern integrity in cancer, and analyze lower CpG abundance regions not examined by array-based methods, we performed shotgun bisulfite genome sequencing on 3 colorectal cancers and the matched normal colonic mucosausing the ABI SOLiD platform. We wanted to obtain methylation estimates with enough precision to detect differences of 10% methylation. Because we used a local likelihood approach, which aggregated information from neighboring CpGs and combined data from 3 biological replicates, we determined that 4X coverage would suffice to estimate methylation values at this precision with a standard error of at most 3% (see Methods). We therefore obtained between 12.5 and 13.5 gigabases for each sample, providing ~5X coverage for each CpG after quality control filtering (see Methods) and alignment(Supplementary Table 1). To verify the accuracy of methylation values obtained by our approach, we performed capture bisulfite sequencing on the same 6 samples for 39,262 regions yielding 39.3k–125.6k CpG with >30× coverage (Supplementary Table 2), with correlations of 0.82–0.91 between our local likelihood approach and capture sequencing, a remarkable agreement since experiments were performed in different laboratories using different sequencing platforms and protocols. Examination of individual loci demonstrated that our methylation estimates closely track the high-coverage capture data (Supplementary Fig. 8). We also performed traditional bisulfite pyrosequencing, further confirming the accuracy of our approach (Supplementary Fig. 9).
Sequencing analysis revealed the surprising presence of large blocks of contiguous hypomethylation in cancer compared to normal (Fig. 2a–b). We identified 13,540 such regions of 5kb–10MB (Table 1, Supplementary Table 3). The across-cancer average hypomethylation throughout the blocks was 12%–23%. Remarkably, these hypomethylated blocks in cancer corresponded to more than half of the genome, even accounting for the number of CpG sites within the blocks (Table 1), and may include small hypermethylated regions. We also noted the existence of a small fraction (3%) of hypermethylated blocks in cancer (Table 1, Figs. 2a, b). A histogram of smoothed methylation values shows the shift in distribution of global DNA methylation (Fig. 2c). The predominant change in block methylation in cancer was a loss in the abundant compartment of intermediate methylation levels (mean 73%for all samples) to significantly lower levels (50–61%)(Fig. 2d).
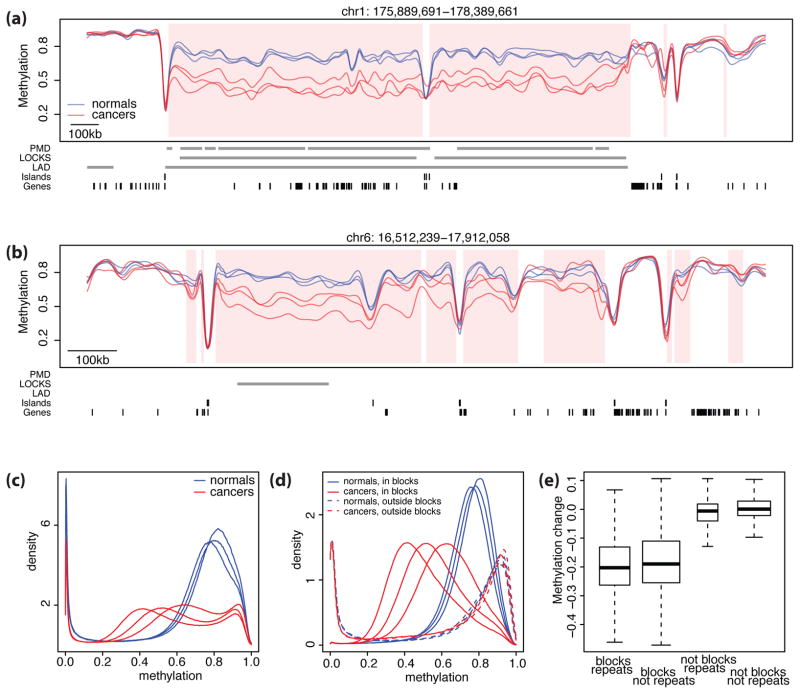
Figure 2. Large hypomethylated genomic blocks in human colon cancer.
Shown in (a) and (b) are smoothed methylation values from bisulfite sequencing data for cancer samples (red) and normal samples (blue) in two genomic regions. The hypomethylated blocks are shown with pink shading. Grey bars indicate the location of PMDs, LOCKs, LADs, CpG Islands, and gene exons. Note that the blocks coincide with the PMD, LOCKS, and LADs in panel (a) but not in (b). Also one can see small hypermethylated blocks at the right edge, which account for 3% of the blocks. (c) The distribution of high-frequency smoothed methylation values for the normal samples (blue) versus the cancer samples (red) demonstrates global hypomethylation of cancer compared to normal. (d) The distribution of methylation values in the blocks (solid lines) and outside the blocks(dashed lines) for normal samples (blue) and cancer samples (red). Note that while the normal and cancer distributions are similar outside the blocks, within the blocks methylation values for cancer exhibit a general shift. (e) Distribution of methylation differences between cancer and normal samples stratified by inclusion in repetitive DNA and blocks. Inside the blocks, the average difference was ~−20% in both in repeat and non-repeat areas. Outside the blocks, the average difference was ~0% in repeat and non-repeat areas, indicating that blocks rather than repeats account for the observed differences in DNA methylation.
Table 1.
Genomic features of Differentially Methylated Regions (DMRs) in colon cancer
| N | # CpG | Genomic size | Median size (bp) | Overlap with islands | Overlap with shores | Overlap with Ref seq mRNA TSS | |
|---|---|---|---|---|---|---|---|
| Normal genome (reference) | N/A | 28.2M | 3.10 Gb | N/A | 27.7K | 55.4K | 36,983 |
| Hypomethylated blocks | 13,540 | 16.2M | 1.95 Gb | 39,412 | 17.6% | 26.8% | 10,453 |
| Hypermethylated blocks | 2,871 | 485K | 35.8 Mb | 9,213 | 13.4% | 36.4% | 976 |
| Hypomethylated small DMRs | 4,315 | 59.5K | 2.91 Mb | 401 | 2.2% | 51.0% | 1,708 |
| Novel hypomethylated | 448 | 8.35K | 367 Kb | 658 | 2.9% | 19.9% | 30 |
| Shift of methylation boundary | 1,516 | 17.5K | 741 Kb | 261 | 2.1% | 92.8% | 1,313 |
| Other | 2,351 | 33.7K | 1.80MB | 479 | 2.1% | 29.9% | 368 |
| Hypermethylated small DMRs | 5,810 | 403K | 6.14 Mb | 820 | 67.2% | 17.0% | 3,068 |
| Loss of boundary* | 1,756 | 165K | 2.36 Mb | 1,159 | 80.9% | 3.4% | 1,091 |
| Shift of methylation boundary | 1,774 | 96.3K | 1.40 Mb | 502 | 60.3% | 33.0% | 1,027 |
| Other | 2,280 | 142K | 2.38MB | 769 | 62.2% | 15.1% | 983 |
As described in the text, loss of boundary DMRs were associated with increase of methylation in the CpG island and a decrease of methylation in the adjacent shore. We score these as a single event and classify them here since there are more CpGs in the islands than in the shores.
These blocks are common across all three cancers. An analysis of the tumors individually versus a normal profile shows consistent block boundary locations (see Fig. 2, Supplementary Fig. 10, and Methods). These blocks were not driven by copy number variation since the location of the latter was not consistent across subjects, in contrast to the consistent block boundaries (Supplementary Fig. 11a, b), and the methylation difference estimates provided by our statistical approach did not correlate with copy number values (Supplementary Fig. 11c).
Global hypomethylation in cancer8 is attributed to the presence of normally methylated repetitive elements9 and may be relevant to colon cancer as LINE-1 element hypomethylation is associated with worse prognosis in colon cancer10. We observed that in normal tissues, repetitive elements were more methylated than non-repetitive regions (76% vs. 66%). To determine whether such repetitive elements were responsible for the block hypomethylation, we compared differences in methylation levels inside and outside repeat elements (see Methods), both inside and outside blocks. Most of the global hypomethylation was due to hypomethylated blocks (Fig. 2e) and not the presence of repetitive elements. As repetitive elements are slightly enriched in blocks (odds ratio 1.4), much of the apparent repeat-associated methylation may in fact be due to blocks. This result does not exclude repeat-associated hypomethylation, since not all repeats were mappable. However, 57% of L1 elements, 94% of L2 elements, 95% of MIR sequences, and 18% of Alu elements were covered by our data (Supplementary Table 4) and did not show repeat-specific hypomethylation (Supplementary Fig. 12). Note that it is possible that Alu sequences not covered by our data are somehow more hypomethylated than covered Alu sequences and thus contribute to global hypomethylation.
Lister et al. performed bisulfite sequencing analysis of the H1 human embryonic stem cell line compared to the IMR90 fibroblast line, identifying large regions of the genome that are less methylated in fibroblast cells than ES cells, referred to as partially methylated domains (PMDs)11. The intermediate-methylation level regions we identified above largely coincided with the PMDs, containing 85% of CpGs inside PMDs (odds ratio 6.5, P<2×10−16, Supplementary Table 5). We previously described large organized chromatin lysine (K) modifications, or LOCKs, genome-wide in normal mouse cells that are associated with both constitutive and tissue-specific gene silencing12. We mapped LOCKs in primary human cells (see Methods). Remarkably, 89% of the LOCKs were contained within the blocks (odds ratio 6.8, P<2×10−16). LOCKs are also known to overlap with nuclear lamina-associated domains or LADs12. Approximately 83% of the LADs were also contained within the blocks (odds ratio 4.9, P<2×10−16). In addition, DNase I hypersensitive sites, a structural signal for regulatory regions13 were enriched within 1 kb of block boundaries and small DMRs (p<2×10−16 for both). Thus the large hypomethylated blocks we identified in cancer correspond to a genomic organization identified in normal cells by several complementary methods. Note that although the PMDs and our hypomethylated blocks largely overlap, we demonstrate later significant differences in gene expression in cancer between non-overlapping blocks and PMDs.
We observed a relationship between the 157 CpGs that are hypervariable across all cancer types identified by our custom array and the hypomethylated blocks identified by whole genome bisulfite sequencing. We found that 63% of the hypomethylated hypervariable CpGs were within hypomethylated blocks, and 37% of the hypermethylated hypervariable CpGs were within the rare hypermethylated blocks. In contrast, hypomethylated and hypermethylated CpGs, respectively, from the control Human Methylation 27K array, that were not hypervariable in cancer were enriched only 13% and 1.5% in the hypomethylated and hypermethylated blocks, respectively, demonstrating high statistical significance for enrichment of hypervariably methylated CpGs in blocks (p<2×10−16; Supplementary Table 6).
Small DMRs in cancer involve loss of stability of DNA methylation boundaries
We developed a statistical algorithm (see Methods) for detecting DNA methylation changes in regions smaller than the blocks (≤5kb). Our analysis of biological replicates was critical as we found that regions showing across-subject variability in normal samples would be easily confused with DMRs if only one cancer-normal pair was available (Supplementary Fig. 13). Methylation measurements in these smaller regions exhibited good agreement with measurements from our previous CHARM-based microarray analysis4 (Supplementary Fig. 14). We refer to these as small DMRs to distinguish them from the large (>5 kb) differentially methylated blocks described above. The increased comprehensiveness of sequencing over CHARM and other published array-based analyses allowed us to detect more small DMRs than previously reported, 5,810 hypermethylated and 4,315 hypomethylated small DMRs (Supplementary Table 7). We also confirmed our finding4 that hypermethylated cDMRs are enriched in CpG islands while hypomethylated cDMRs are enriched in CpG island shores (Table 1). Sequencing also showed that the ratio of unmethylated to methylated islands is normally approximately 2:1, and for both types approximately 20% change methylation state in cancers (Table 2, Supplementary Table 8).
Table 2.
Methylation values* observed in CpG islands in cancer compared to normal samples
| Methylation status in normals | Total | Hypo | No change | Hyper |
|---|---|---|---|---|
| Unmethylated (<= 0.2) | 16184 | 0.1% | 83.2% | 16.7% |
| Partial methylated (>= 0.2, <=0.8) | 4796 | 17.0% | 46.7% | 36.3% |
| Methylated (>= 0.8) | 5527 | 24.0% | 75.9% | 0.1% |
Average methylation value in each island were then averaged across subject for cancer and normal samples separately
The most striking and consistent characteristic of small DMR architecture was a shift in one or both of the DNA methylation boundaries of a CpG island out of the island into the adjacent region (Fig. 3a,)or into the interior of the island (Fig. 3b). Boundary shifts into islands would appear as hypermethylated islands on array-based data, while boundary shifts out of islands would appear as hypomethylated shores.
Figure 3. Loss of methylation stability at small DMRs.
Methylation estimates plotted against genomic location for normal samples (blue) and cancer samples (red). The small DMR locations are shaded pink. Grey bars indicate the location of blocks, CpG islands, and gene exons. Tick marks along the bottom axis indicate the location of CpGs. Pictured are examples of (a) a methylation boundary shift outward, (b) a methylation boundary shift inward, (c) a loss of methylation boundary, and (d) a novel hypomethylation DMR.
The second most frequent category of small DMRs involved loss of methylation boundaries at CpG islands. For example, many hypermethylated cDMRs were defined in normal samples by unmethylated regions surrounded by highly methylated regions. In cancer, these regions exhibited stable methylation levels of approximately 40–60% throughout (Table 1, Fig. 3c). These regions with loss of methylation boundaries largely correspond to what are classified as hypermethylated islands in cancer.
We also found hypomethylated cDMRs that arose de novo in highly methylated regions outside of blocks, which we call novel hypomethylated DMRs, usually corresponding to CpG-rich regions that were not conventional islands (Table 1). Here, regions in which normal colon tissue was 75–95% methylated dropped to lower levels (20–40%) in cancer (Fig. 3d). In summary, in addition to the hypomethylated blocks, we found 10,125 small DMRs, 5,494 of which clearly fell in three categories: shifts of methylation boundaries, loss of methylation boundaries, and novel hypomethylation. Note that not all small DMRs followed a consistent pattern across all three sample pairs and were therefore not classified (Table 1).
Methylation-based Euclidean distances show colon adenomas intermediate between normals and cancers
Using multidimensional scaling of the methylation values measured via the custom array in colon samples we noticed that normal samples clustered tightly together in contrast to dispersed cancer samples (Fig. 4a). This is consistent with the observed increase in methylation variability in cancer described earlier. We analyzed 30 colon adenomas on the custom array, and found that they were intermediate in both variability within samples and distance to the cluster of normal samples (Fig. 4a).
Figure 4. Adenomas show intermediate methylation variability.
(a) Multidimensional scaling of pairwise distances derived from methylation levels assayed on a custom Illumina array. Note that cancer samples (red) are largely far from the tight cluster of normal samples (blue), while adenoma samples (black) exhibit a range of distances: some are as close as other normal samples, others are as far as cancer samples, and many are at intermediate distances. (b) Multidimensional scaling of pairwise distances derived from average methylation values in blocks identified via bisulfite sequencing. Matching sequenced adenoma samples (labeled 1 and 2) appear in the same locations relative to the cluster of normal samples in both (a) and (b). (c) Methylation values for normal (blue), cancer (red) and two adenoma samples (black). Adenoma 1, which appeared closer to normal samples in the multidimensional scaling analysis (a), follows a similar methylation pattern to the normal samples. However, in some regions (shaded with pink) differences between Adenoma 1 and the normal samples are observed. Adenoma 2 shows a similar pattern to cancers.
We subsequently performed whole genome bisulfite sequencing on two of these adenomas, a premalignant colon adenoma with relatively small methylation-based distance to the normal colons and an adenoma with a large methylation-based distance to the normal colons, similar to the cancer samples. We computed average methylation levels over each block from each sequenced sample and computed pairwise Euclidean distances between samples using these values. These measurements from hypomethylated blocks confirm the characteristic observed the array data: genome-wide increased variability in cancers compared to normals with adenomas exhibiting intermediate values (Fig. 4).
Expression of cell cycle genes associated with hypomethylated shores in cancer
Whole genome analysis has demonstrated an inverse relationship between gene expression and methylation, especially at transcriptional start sites14. To study this relationship in small DMRs, we obtained public microarray gene expression data from cancer and normal colon samples (see Methods) and compared to results fromour sequencing data. We mapped 6,869 genes to DMRs within 2 kb of the gene’s transcription start site and observed the expected inverse relationship between DNA methylation and gene expression (r = −0.27, p< 2×10−16, Supplementary Fig. 15).
We examined the inverse relationship between methylation and gene expression for each category of small DMRs separately and noticed that the strongest relationship for hypomethylated shores is due to methylation boundary shifts (Supplementary Table 9). We performed gene ontology enrichment analysis15 for differentially expressed genes (FDR<0.05), comparing those associated with hypomethylated boundary shifts to the other categories. Categories (Supplementary Table 10) were strongly enriched for mitosis and cell-cycle related genes CEP55, CCNB1, CDCA2, PRC1, CDC2, FBXO5, AURKA, CDK1, CDKN3, CDK7, and CDC20B, among others (Supplementary Table 11).
Increased variation in gene expression in hypomethylated blocks and DMRs
We compared across-subject methylation variability levels between cancer and normal, within the blocks, and found a striking similarity to the cancer methylation hypervariability found with the custom array (Fig. 1a–e compared to Supplementary Fig. 16). To study the relationship to gene expression in colon cancer, we obtained public gene expression data from cancer and normal samples (see Methods). Genes in the blocks were generally silenced (80% genes silenced in all samples) both in normal and cancer samples. Of the genes consistently transcribed in normal tissue, albeit at low levels, 36% are silenced in blocks in cancers, compared to 15% expected by chance. This is consistent with other reports in the literature, e.g. Frigola et al16.
More striking than subtle differences in gene silencing, we found substantial enrichment of genes exhibiting increased expression variability in cancer compared to normal samples in the hypomethylated blocks. First, we ruled out that this observed increased variability was due to the potential high cellular heterogeneity of cancer (Supplementary Fig. 17a). Then, we noticed a clear and statistically significant association between increased variability in expression of a gene and its location within a hypomethylated block (Supplementary Fig. 17b). For example, 26 of the 50 genes exhibiting the largest increase in expression variability were inside the blocks; 52% compared to the 17% expected by chance (p = 3×10−9). Expression levels for 25 of these exhibited an interesting pattern: while never expressed in normal samples, they exhibited stochastic expression in cancer (Fig. 5 and Supplementary Fig. 18). For example the genes MMP3, MMP7, MMP10, SIM2, CHI3L1, STC1, and WISP (described in the Discussion) were expressed in 96%, 100%, 67%, 8%, 79%, 50%, and 17% of the cancer samples, respectively, but never expressed in normal samples (Supplementary Table 12).
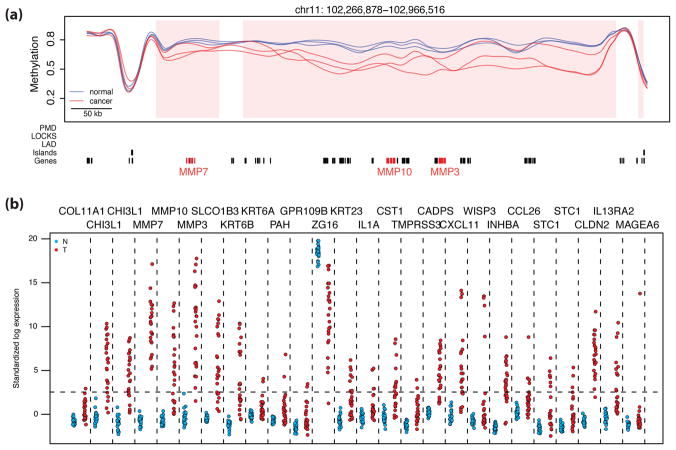
Figure 5. High variability of gene expression associated with blocks.
(a) An example of hypervariably expressed genes contained within a block; note genes MMP7, MMP10, and MMP3 highlighted in red. Methylation values for cancer samples (red) and normal samples (blue) with hypomethylated block locations highlighted (pink shading) are plotted against genomic location. Grey bars are as in Fig. 2. (b) Standardized log expression values for 26 hypervariable genes in cancer located within hypomethylated block regions (normal samples in blue, cancer samples in red). Standardization was performed using the gene expression barcode. Genes with standardized expression values below 2.54, or the 99.5th percentile of a normal distribution (horizontal dashed line) are determined to be silenced by the barcode method26. Vertical dashed lines separate the values for the different genes. Note there is consistent expression silencing in normal samples compared to hypervariable expression in cancer samples. A similar plot drawn from an alternative GEO dataset is shown in Supplementary Figure 18.
Functional differences between hypomethylated blocks and PMDs
As noted above, the hypomethylated blocks we observed substantially overlapped PMDs reported in a fibroblast cell line by Lister et al.11. We examined the genomic regions of no overlap between blocks and PMDs to identify potential functional differences between them. We grouped them into two sets: 1) regions within the hypomethylated blocks but not in the PMDs (B+P−) and 2) regions within the PMDs but not in the hypomethylated blocks (B−P+). We obtained microarray gene expression data from fibroblast samples (see Methods) and, as expected, the genes in the fibroblast PMDs were relatively silenced in the fibroblast samples (p<2×10−16). Furthermore, genes that were silenced in fibroblast samples and consistently expressed in normal colon were enriched in the B-P+ regions (odds ratio of 3.2, p<2×10−16), while genes consistently silenced in colon and consistently expressed in fibroblast samples were enriched in the B+P-regions (odds ratio 2.8, p = 0.0004). Finally, the 50 hypervariable genes described above were markedly enriched in the B+P− regions (p=0.00013), yet showed no enrichment in the B−P+ regions. These results suggest that hypervariable gene expression in colon cancer may be related to their presence in hypomethylated blocks.
DISCUSSION
In summary, we show that colon cancer cDMRs are generally involved in the common solid tumors of adulthood, lung, breast, thyroid, and colon cancer, and the most common solid tumor of childhood, Wilms tumor, with tight clustering of methylation levels in normal tissues, and marked stochastic variation in cancers. Efforts to exploit DNA methylation for cancer screening focus on identifying narrowly defined cancer-specific profiles17. Our data suggests future efforts might instead be directed at defining the cancer epigenome as the departure from a narrowly defined normal profile.
Surprisingly, two-thirds of all methylation changes in colon cancer involve hypomethylation of large blocks, with consistent locations across samples, comprising more than half of the genome. The functional relevance is supported by the fact that genes in colon blocks not in fibroblast blocks tend to be silenced in colon and not in fibroblasts and vice-versa.
The most variably expressed genes in cancer are enriched in the blocks, and involve genes associated with tumor heterogeneity and progression, including three matrix metalloproteinase genes, MMP3, MMP7, and MMP1018, and a fourth, SIM2, which acts through metalloproteinases to promote tumor invasion19. Another, STC1, helps mediate the Warburg effect of reprogramming tumor metabolism20. CHI3L1 encodes a secreted glycoprotein associated with inflammatory responses and poor prognosis in multiple tumor types including colon21. WISP genes are targets of Wnt-1 thought to contribute to tissue invasion in breast and colon cancer22. Our gene ontology enrichment analysis15 of genes associated with hypervariable expression in blocks (FDR<0.05)showed enrichment for categories including extracellular matrix remodeling genes (Supplementary Table 13). One cautionary note raised by these findings is that treatment of cancer patients with nonspecific DNA methylation inhibitors could have unintended consequences in the activation of tumor-promoting genes in hypomethylated blocks. It is also important to note that while previous studies23,24 have shown large-region hypermethylation or no regional methylation change, this study is based on whole-genome bisulfite sequencing. Nevertheless, future studies are needed to show whether block hypomethylation is a feature of cancer epigenomes in general.
Small DMRs, while representing a relatively small fraction of the genome (0.3%), are numerous (10,125), and frequently involve loss of boundaries of DNA methylation at the edge of CpG islands, shifting of DNA methylation boundaries, or the creation of novel hypomethylated regions in CG-dense regions that are not canonical islands. These data underscore the importance of hypomethylated CpG island shores in cancer since shores associated with hypomethylation and gene overexpression in cancer are enriched for cell cycle related genes, suggesting a role in the unregulated growth that characterizes cancer.
We propose a model relating tissue-specific DMRs to the sites of methylation hypervariability in cancer. Normal pluripotency might require stochastic gene expression at some loci, allowing for differentiation along alternative pathways in response to external stimuli or even intrinsically. The epigenome could collaborate to create a permissive state by changing its physical configuration to relax the stringency of epigenetic marks, since variance increases away from the extremes, and a similar process may occur in cancer. One way is by altering LOCKs/LADs/blocks, which could involve a change in the chromatin packing density or proximity to the nuclear lamina. Similarly, subtle shifts in DNA methylation boundaries near CpG islands may drive normal chromatin organization and tissue-specific gene expression. Given the importance of boundary regions for both small DMRs and large blocks identified in this study, it will be important to focus future epigenetic investigations on the boundaries of blocks and CpG islands (shores), and on genetic or epigenetic changes in genes encoding factors that interact with them.
The increased methylation and expression variability in each cancer type is consistent with the potential selective value of increased epigenetic plasticity in a varying environment first suggested for evolution but applicable to the strong but variable selective forces under which a cancer grows, such as varying oxygen tension or metastasis to a distant site25. Thus, increased epigenetic heterogeneity in cancer at cDMRs (which we show are also tDMRs) could underlie the ability of cancer cells to adapt rapidly to changing environments, such as increased oxygen with neovascularization, then decreased oxygen with necrosis; or metastasis to a new intercellular milieu.
Supplementary Material
Acknowledgments
We thank Applied Biosystems, Inc. for supplying reagents for the sequencing experiments, Bert Vogelstein and Martha Zeiger for tumor samples, and Marvin Newhouse for computer assistance. This work was supported by NIH Grants R37CA054358, R01HG005220, 5P50HG003233, F32CA138111, 5R01GM083084, andR01DA025779 (KZ).
Footnotes
DATA ACCESSION
Whole genome bisulfite sequencing data, capture bisulfite sequencing data, custom GoldenGate microarray data, ChIP-chip LOCK data, Wilms’ tumor copy number microarray data are submitted, pending assignment of accession numbers.
Author contributions: K.D.H. and R.A.I wrote the DMR finder and smoothing algorithms; W.T. performed and analyzed the arrays with H.C.B. who wrote new software for this purpose; S.S. made the libraries and performed validation; B.L. wrote new methylation sequence alignment software; O.G.M. performed histopathologic analysis; B.W. and H.W. performed LOCK experiments; Y.L performed copy number experiments; D.D. and K.Z. performed bisulfite capture; E.B. performed the sequencing; R.A.I. and A.P.F. conceived and led the experiments and wrote the paper with the predominant assistance of K.D.H., W.T., H.C.B. and B.L.
References
- 1.Jones PA, Baylin SB. The fundamental role of epigenetic events in cancer. Nat Rev Genet. 2002;3:415–28. doi: 10.1038/nrg816. [DOI] [PubMed] [Google Scholar]
- 2.Feinberg AP, Tycko B. The history of cancer epigenetics. Nat Rev Cancer. 2004;4:143–53. doi: 10.1038/nrc1279. [DOI] [PubMed] [Google Scholar]
- 3.Esteller M. Epigenetics in cancer. N Engl J Med. 2008;358:1148–59. doi: 10.1056/NEJMra072067. [DOI] [PubMed] [Google Scholar]
- 4.Irizarry RA, et al. The human colon cancer methylome shows similar hypo-and hypermethylation at conserved tissue-specific CpG island shores. Nat Genet. 2009;41:178–86. doi: 10.1038/ng.298. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Doi A, et al. Differential methylation of tissue-and cancer-specific CpG island shores distinguishes human induced pluripotent stem cells, embryonic stem cells and fibroblasts. Nat Genet. 2009;41:1350–3. doi: 10.1038/ng.471. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Irizarry RA, et al. Comprehensive high-throughput arrays for relative methylation (CHARM) Genome Res. 2008;18:780–90. doi: 10.1101/gr.7301508. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Bibikova M, et al. High-throughput DNA methylation profiling using universal bead arrays. Genome Res. 2006;16:383–93. doi: 10.1101/gr.4410706. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Feinberg AP, Gehrke CW, Kuo KC, Ehrlich M. Reduced genomic 5-methylcytosine content in human colonic neoplasia. Cancer Res. 1988;48:1159–61. [PubMed] [Google Scholar]
- 9.Ehrlich M. DNA methylation in cancer: too much, but also too little. Oncogene. 2002;21:5400–13. doi: 10.1038/sj.onc.1205651. [DOI] [PubMed] [Google Scholar]
- 10.Ogino S, et al. A cohort study of tumoral LINE-1 hypomethylation and prognosis in colon cancer. J Natl Cancer Inst. 2008;100:1734–8. doi: 10.1093/jnci/djn359. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Lister R, et al. Human DNA methylomes at base resolution show widespread epigenomic differences. Nature. 2009;462:315–22. doi: 10.1038/nature08514. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Wen B, Wu H, Shinkai Y, Irizarry RA, Feinberg AP. Large histone H3 lysine 9 dimethylated chromatin blocks distinguish differentiated from embryonic stem cells. Nat Genet. 2009;41:246–50. doi: 10.1038/ng.297. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Hesselberth JR, et al. Global mapping of protein-DNA interactions in vivo by digital genomic footprinting. Nat Methods. 2009;6:283–9. doi: 10.1038/nmeth.1313. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Li Y, et al. The DNA Methylome of Human Peripheral Blood Mononuclear Cells. PLoS Biol. 2010;8:e1000533. doi: 10.1371/journal.pbio.1000533. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Falcon S, Gentleman R. Using GOstats to test gene lists for GO term association. Bioinformatics. 2007;23:257–8. doi: 10.1093/bioinformatics/btl567. [DOI] [PubMed] [Google Scholar]
- 16.Frigola J, et al. Epigenetic remodeling in colorectal cancer results in coordinate gene suppression across an entire chromosome band. Nat Genet. 2006;38:540–9. doi: 10.1038/ng1781. [DOI] [PubMed] [Google Scholar]
- 17.Gal-Yam EN, Saito Y, Egger G, Jones PA. Cancer epigenetics: modifications, screening, and therapy. Annu Rev Med. 2008;59:267–80. doi: 10.1146/annurev.med.59.061606.095816. [DOI] [PubMed] [Google Scholar]
- 18.Yu AE, Hewitt RE, Connor EW, Stetler-Stevenson WG. Matrix metalloproteinases. Novel targets for directed cancer therapy. Drugs Aging. 1997;11:229–44. doi: 10.2165/00002512-199711030-00006. [DOI] [PubMed] [Google Scholar]
- 19.Aleman MJ, et al. Inhibition of Single Minded 2 gene expression mediates tumor-selective apoptosis and differentiation in human colon cancer cells. Proc Natl Acad Sci U S A. 2005;102:12765–70. doi: 10.1073/pnas.0505484102. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Yeung HY, et al. Hypoxia-inducible factor-1-mediated activation of stanniocalcin-1 in human cancer cells. Endocrinology. 2005;146:4951–60. doi: 10.1210/en.2005-0365. [DOI] [PubMed] [Google Scholar]
- 21.Eurich K, Segawa M, Toei-Shimizu S, Mizoguchi E. Potential role of chitinase 3-like-1 in inflammation-associated carcinogenic changes of epithelial cells. World J Gastroenterol. 2009;15:5249–59. doi: 10.3748/wjg.15.5249. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Fischer H, et al. COL11A1 in FAP polyps and in sporadic colorectal tumors. BMC Cancer. 2001;1:17. doi: 10.1186/1471-2407-1-17. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Clark SJ. Action at a distance: epigenetic silencing of large chromosomal regions in carcinogenesis. Human Molecular Genetics. 2007;16:R88–R95. doi: 10.1093/hmg/ddm051. [DOI] [PubMed] [Google Scholar]
- 24.Feber A, et al. Comparative methylome analysis of benign and malignant peripheral nerve sheath tumours. Genome Research. 2011 doi: 10.1101/gr.109678.110. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Feinberg A, Irizarry R. Stochastic epigenetic variation as a driving force of development, evolutionary adaptation, and disease. Proceedings of the National Academy of Sciences. 2010;107:1757. doi: 10.1073/pnas.0906183107. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Zilliox MJ, Irizarry RA. A gene expression bar code for microarray data. Nat Methods. 2007;4:911–3. doi: 10.1038/nmeth1102. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Bolstad BM, Irizarry RA, Astrand M, Speed TP. A comparison of normalization methods for high density oligonucleotide array data based on variance and bias. Bioinformatics. 2003;19:185–93. doi: 10.1093/bioinformatics/19.2.185. [DOI] [PubMed] [Google Scholar]
- 28.Leek JT, et al. Tackling the widespread and critical impact of batch effects in high-throughput data. Nat Rev Genet. 2010;11:733–9. doi: 10.1038/nrg2825. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Aryee MJ, et al. Accurate genome-scale percentage DNA methylation estimates from microarray data. Biostatistics. 2011;12:197–210. doi: 10.1093/biostatistics/kxq055. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Bormann Chung CA, et al. Whole methylome analysis by ultra-deep sequencing using two-base encoding. PLoS One. 2010;5:e9320. doi: 10.1371/journal.pone.0009320. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Deng J, et al. Targeted bisulfite sequencing reveals changes in DNA methylation associated with nuclear reprogramming. Nat Biotechnol. 2009;27:353–60. doi: 10.1038/nbt.1530. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Xi Y, Li W. BSMAP: whole genome bisulfite sequence MAPping program. BMC Bioinformatics. 2009;10:232. doi: 10.1186/1471-2105-10-232. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Eckhardt F, et al. DNA methylation profiling of human chromosomes 6, 20 and 22. Nat Genet. 2006;38:1378–85. doi: 10.1038/ng1909. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Loader C. Local regression and likelihood. Springer Verlag; 1999. [Google Scholar]
- 35.Jurka J. Repbase update: a database and an electronic journal of repetitive elements. Trends Genet. 2000;16:418–20. doi: 10.1016/s0168-9525(00)02093-x. [DOI] [PubMed] [Google Scholar]
- 36.Olshen AB, Venkatraman ES, Lucito R, Wigler M. Circular binary segmentation for the analysis of array-based DNA copy number data. Biostatistics. 2004;5:557–72. doi: 10.1093/biostatistics/kxh008. [DOI] [PubMed] [Google Scholar]
- 37.Sabates-Bellver J, et al. Transcriptome profile of human colorectal adenomas. Mol Cancer Res. 2007;5:1263–75. doi: 10.1158/1541-7786.MCR-07-0267. [DOI] [PubMed] [Google Scholar]
- 38.Gyorffy B, Molnar B, Lage H, Szallasi Z, Eklund AC. Evaluation of microarray preprocessing algorithms based on concordance with RT-PCR in clinical samples. PLoS One. 2009;4:e5645. doi: 10.1371/journal.pone.0005645. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Galamb O, et al. Reversal of gene expression changes in the colorectal normal-adenoma pathway by NS398 selective COX2 inhibitor. Br J Cancer. 2010;102:765–73. doi: 10.1038/sj.bjc.6605515. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Smith JC, Boone BE, Opalenik SR, Williams SM, Russell SB. Gene profiling of keloid fibroblasts shows altered expression in multiple fibrosis-associated pathways. J Invest Dermatol. 2008;128:1298–310. doi: 10.1038/sj.jid.5701149. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Chen Y, et al. Developing and applying a gene functional association network for anti-angiogenic kinase inhibitor activity assessment in an angiogenesis co-culture model. BMC Genomics. 2008;9:264. doi: 10.1186/1471-2164-9-264. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Duarte TL, Cooke MS, Jones GD. Gene expression profiling reveals new protective roles for vitamin C in human skin cells. Free Radic Biol Med. 2009;46:78–87. doi: 10.1016/j.freeradbiomed.2008.09.028. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.