Abstract
The ratio of genetic diversity on chromosome X to that on the autosomes is sensitive to both natural selection and demography. Based on whole-genome sequences of 69 females, we report that while this ratio increases with genetic distance from genes across populations, it is lower in Europeans than in West Africans independent of proximity to genes. This relative reduction is most parsimoniously explained by differences in demographic history without the need to invoke natural selection.
The genetic diversity of chromosome X is expected, under equilibrium assumptions, to be that of the autosomes in a population where the two sexes have an identical distribution of offspring numbers. However, deviations from this ratio can result from at least four forces known to have been prevalent in human history: (i) sex-biased demographic events leading to different effective population sizes of males and females, (ii) changes in population size over time (since chromosome X is proportionally more sensitive to recent epochs, owing to its reduced effective population size1), (iii) natural selection, which also affects chromosome X differently, and (iv) differences in mutation rates between sexes or between chromosome X and the autosomes. The possible effect of these forces on human genetic variation has received recent attention: Hammer et al. reported that nucleotide diversity is higher than expected on chromosome X, with a mean X-to-Autosome diversity ratio (X/A) of 0.9 across six populations and with no significant differences between populations.2,3 Another recent study reported a significantly reduced X/A in non-African populations relative to West Africans, beyond the reduction expected from known historical changes in population size4, with similar conclusions having been drawn from analyses of inter-population allele frequency differences and the distribution of allele frequencies within populations4–7.
Estimates of the absolute X/A ratio are sensitive to details of the methods used to obtain them, including the normalization by divergence from an outgroup8 and differences in SNP ascertainment biases between chromosome X and the autosomes. To eliminate factors of this kind, we examine here the relative X/A ratio between different populations. To compare the diversity of chromosome X and the autosomes in different populations, we considered intergenic SNPs from whole-genome sequences of 36 West African (YRI) and 33 European (CEU) females from the 1000 Genomes Project9, following rigorous quality control (Supplementary Methods). We normalized estimates of nucleotide diversity by divergence from a primate outgroup to correct for differences in mutation rates. Genome-wide X/A estimates are 0.73±0.016 in YRI and 0.61±0.018 in CEU (Supplementary Table 1; normalization by divergence from rhesus macaque), which are consistent with previous estimates4 and support a reduced ratio in non-Africans relative to Africans.
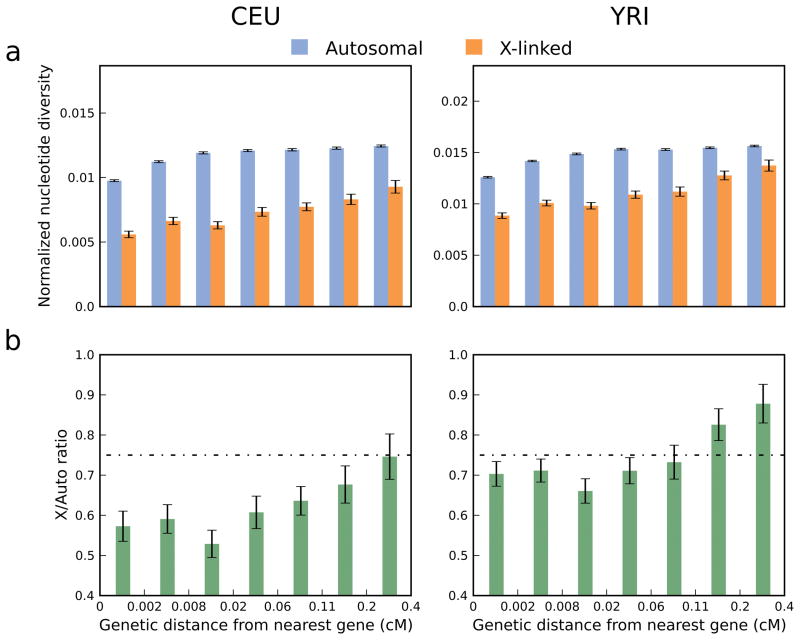
To examine the effect of natural selection, we partitioned the data by genetic distance from the nearest gene. Both X-linked and autosomal diversity increase with distance from genes (Figure 1a; P=0.002 and P=0.077 for CEU, P=0.0008 and P=0.070 for YRI). This increase in diversity with distance to genes closely matches predictions of the model of McVicker et al.10 for both the autosomes and chromosome X (Supplementary Figure 1; P<0.01 for all four cases), consistent with a diversity-reducing effect of selection on linked sites, either through purifying selection (background selection), positive selection (genetic hitchhiking), or both. We also observed a skew of the site frequency spectrum towards lower frequency alleles closer to genes, as expected from the action of natural selection (Supplementary Note). Importantly, the observed increase in diversity with distance from genes is greater for chromosome X than for the autosomes (Figure 1a), suggesting that diversity reduction due to selection at linked sites has been a more powerful force on chromosome X. As a result, X/A increases with distance from genes (Figure 1b; P<0.001 for both CEU and YRI), consistent with recent results of Hammer et al. based on 6 individuals of European descent3, as well as in line with the observation that the increase in inter-population allele frequency differentiation as recombination rate decreases is greater for chromosome X than for the autosomes11. The high X/A observed in the loci sequenced by Hammer et al.2,3 is in accordance with the large distance from genes and high local recombination rate of these loci (Figure 1b).
Figure 1. Autosomal, X-linked, and absolute X/A diversity increase with genetic distance from the nearest gene.
(a) Nucleotide diversity normalized by genetic divergence from rhesus macaque for a partition of the genome by distance from the nearest gene (Supplementary Methods). Note the different scale of the y-axis for the two populations (CEU and YRI), which is proportional to autosomal normalized diversity. (b) X/A ratios corresponding to the estimates from panel a (horizontal line represents the expectation of ¾). In all panels, x-axis labels represent the boundaries between partitions, which were selected such that each partition encompasses an equal fraction of chromosome X (Supplementary Figure 2). Error bars denote ± one standard error estimated by a block bootstrap approach (Supplementary Methods). Similar results were obtained when divergence from orangutan was used for normalization (Supplementary Figure 3) and similar trends were also observed when considering only levels of human nucleotide diversity, without any normalization by divergence (Supplementary Figure 4).
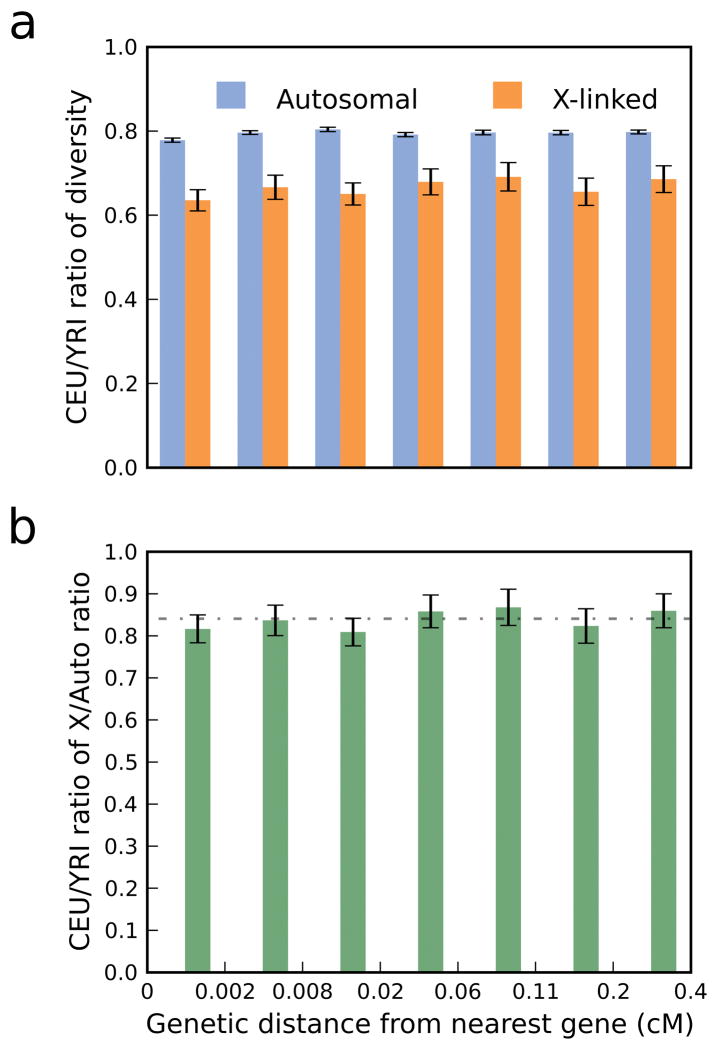
So far, we have shown that the absolute X/A ratio is likely to have been strongly influenced by natural selection. To test whether the observed differences between Africans and non-Africans are also due to differential selective forces, we studied the relative levels of diversity between populations, considering the CEU-to-YRI ratio of nucleotide diversity (relative diversity), and the CEU-to-YRI ratio of the X/A ratio (relative X/A). Interestingly, neither X-linked nor autosomal relative diversity is sensitive to distance from genes (Figure 2a; P=0.28 and P=0.53 in a test of correlation), and the levels of relative diversity are consistently lower for chromosome X than for the autosomes (Figure 2a). As a consequence, the relative X/A remains nearly constant across all distances from genes (Figure 2b; P=0.42 in a test of correlation), and is always consistent with the genome-wide estimate of 0.84±0.03, despite the pronounced dependence of selective effects on proximity to genes. Notably, Keinan et al. also observed no clear relationship between relative X/A and distance to genes4, and the improved methodology and much richer data set used here enable us to more definitively establish that relative X/A is indeed not sensitive to proximity to genes.
Figure 2. Relative autosomal, X-linked, and X/A diversity are not correlated with genetic distance from the nearest gene.
For a partition of the genome as in Figure 1, (a) X-linked and autosomal nucleotide diversity in CEU divided by the corresponding in YRI, and (b) X/A in CEU divided by X/A in YRI. Estimates of less than 1 in panel a reflect the reduced diversity in non-Africans, most notably due to the out-of-Africa population bottleneck. Estimates of less than 1 in panel b indicate a reduction in X-linked diversity compared to autosomal diversity that is specific to non-Africans (the horizontal line denotes the estimate based on pooled, genome-wide intergenic data). In all panels, error bars denote ± one standard error estimated by a block bootstrap approach (Supplementary Methods). These results are independent of normalization by divergence since normalizing diversity in both populations by the same divergence estimates would have canceled out in the CEU-to-YRI ratio.
The lack of correlation between relative X/A and distance from genes strongly suggests that the difference in X/A between populations cannot be attributed to the effects of diversity-reducing selection acting on genes. On the other hand, several plausible demographic explanations have been offered for the observed differences between populations, including the increased impact of recent history on chromosome X1,4 and sex-biased demographic events7,12. One such sex-biased event has been highlighted in a recent simulation study: waves of primarily male migration during the dispersal of modern humans out of Africa12. Another recent modeling study supports that for a demographic event to explain observed differences, it would have to coincide with the time of the out-of-Africa event7. The results presented here indicate that the difference in X/A between African and non-African populations primarily derives from demographic forces such as those explored in these studies. It would require a very specific, consistent, and highly improbable form of population-specific natural selection to drive the observed pattern.
In principle, our results could be influenced by ascertainment biases stemming from differences in sequencing coverage and in the number of chromosomes sampled on chromosome X and the autosomes. However, three features of our analysis minimize the impact of such biases. First, to equalize sample size and coverage, we considered only females in all analyses. Second, differential ascertainment biases are not likely to correlate with genetic distance from genes. Third, and most important, such biases are not likely to affect estimates of relative diversity and relative X/A since ascertainment is similar for the two population samples we compared.
In conclusion, we have demonstrated a positive correlation between X/A and distance from genes, indicating that diversity on chromosome X has been shaped by selection at linked sites more than has diversity on the autosomes, probably in large part due to X-linked recessive variants being exposed in males13–15. More importantly, we have shown that the reduced X/A in non-Africans relative to Africans remains essentially constant across a wide range of genetic distances from genes. Hammer et al. stressed that demographic history is best studied by focusing on “neutral” loci that are located as far as possible from known functional elements3. The results of the current study lead us to propose a complementary approach of analyzing ratios of diversity between different populations, which is not sensitive to the effects of natural selection if these are similar on a genome-wide average across populations. Contrasting populations allows focusing—with increased resolution—on events occurring after their split, excluding their shared history. This is much in the same spirit as studying X/A based on inter-population allele frequency differentiation4–7, which considers changes in allele frequencies accumulated after the populations have split. In contrast to considering putatively neutral regions in a single population, the approach of contrasting statistics between populations is also not sensitive to (i) unannotated functional elements confounding the inference of “neutral” loci, (ii) normalization by genetic divergence (Figure 2), and (iii) differential ascertainment biases between X and autosomes. Finally, our approach allows the inclusion of orders of magnitude more data, thereby providing increased statistical power. Here, analysis of whole-genome sequences, in conjunction with an approach focusing on more recent epochs, revealed a non-African reduction in X/A that likely results from demographic events associated with the human dispersal out of Africa.
Supplementary Material
Acknowledgments
We thank Rasmus Nielsen and Thorfinn Korneliussen for sharing their software (Supplementary Note), David Reich and Elaine Zhong for advice about this project, Diana Chang, Elodie Gazave, and three anonymous reviewers for comments on earlier versions of this manuscript, and the 1000 Genomes Project. This work was supported in part by NIH grant U01-HG005715.
Footnotes
Author contributions
A.K. conceived and designed the study. S.G. and L.A. performed the experiments (contributed equally). S.G., L.A. and A.K. analysed the results and performed statistical analysis. A.S., A.G.C. and A.K. contributed analysis tools. A.K. wrote the paper, with review and contributions by all authors.
References
- 1.Pool JE, Nielsen R. Population size changes reshape genomic patterns of diversity. Evolution Int J Org Evolution. 2007;61:3001–6. doi: 10.1111/j.1558-5646.2007.00238.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Hammer MF, Mendez FL, Cox MP, Woerner AE, Wall JD. Sex-biased evolutionary forces shape genomic patterns of human diversity. PLoS Genet. 2008;4:e1000202. doi: 10.1371/journal.pgen.1000202. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Hammer MF, et al. The ratio of human Xchromosome to autosome diversity is positively correlated with genetic distance from genes. Nat Genet. 2010;42:830–1. doi: 10.1038/ng.651. [DOI] [PubMed] [Google Scholar]
- 4.Keinan A, Mullikin JC, Patterson N, Reich D. Accelerated genetic drift on chromosome X during the human dispersal out of Africa. Nat Genet. 2008;41:66–70. doi: 10.1038/ng.303. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Amato R, et al. Genome-wide scan for signatures of human population differentiation and their relationship with natural selection, functional pathways and diseases. PLoS One. 2009;4:e7927. doi: 10.1371/journal.pone.0007927. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Casto AM, et al. Characterization of X-linked SNP genotypic variation in globally distributed human populations. Genome Biol. 2010;11:R10. doi: 10.1186/gb-2010-11-1-r10. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Emery LS, Felsenstein J, Akey JM. Estimators of the human effective sex ratio detect sex biases on different timescales. Am J Hum Genet. 2010;87:848–56. doi: 10.1016/j.ajhg.2010.10.021. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Bustamante CD, Ramachandran S. Evaluating signatures of sex-specific processes in the human genome. Nat Genet. 2009;41:8–10. doi: 10.1038/ng0109-8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Durbin RM, et al. A map of human genome variation from population-scale sequencing. Nature. 2010;467:1061–73. doi: 10.1038/nature09534. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.McVicker G, Gordon D, Davis C, Green P. Widespread genomic signatures of natural selection in hominid evolution. PLoS Genet. 2009;5:e1000471. doi: 10.1371/journal.pgen.1000471. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Keinan A, Reich D. Human population differentiation is strongly correlated with local recombination rate. PLoS Genet. 2010;6:e1000886. doi: 10.1371/journal.pgen.1000886. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Keinan A, Reich D. Can a gender-biased human demography account for the reduced effective population size of chromosome X in non-Africans? Mol Biol Evol. 2010;27:2312–21. doi: 10.1093/molbev/msq117. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Charlesworth B, Morgan MT, Charlesworth D. The effect of deleterious mutations on neutral molecular variation. Genetics. 1993;134:1289–303. doi: 10.1093/genetics/134.4.1289. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Vicoso B, Charlesworth B. Effective population size and the faster-X effect: an extended model. Evolution. 2009;63:2413–26. doi: 10.1111/j.1558-5646.2009.00719.x. [DOI] [PubMed] [Google Scholar]
- 15.Betancourt AJ, Kim Y, Orr HA. A pseudohitchhiking model of X vs. autosomal diversity. Genetics. 2004;168:2261–9. doi: 10.1534/genetics.104.030999. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.