Abstract
A complex and heterogeneous microflora performs sugar and lactic acid fermentations in food products. Depending on the fermentable food matrix (dairy, meat, vegetable etc.) as well as on the species composition of the microbiota, specific combinations of molecules are produced that confer unique flavor, texture, and taste to each product. Bacterial populations within such “fermented food microbiota” are often of environmental origin, they persist alive in foods ready for consumption, eventually reaching the gastro-intestinal tract where they can interact with the resident gut microbiota of the host. Although this interaction is mostly of transient nature, it can greatly contribute to human health, as several species within the food microbiota also display probiotic properties. Such an interplay between food and gut microbiota underlines the importance of the microbiological quality of fermented foods, as the crowded environment of the gut is also an ideal site for genetic exchanges among bacteria. Selection and spreading of antibiotic resistance genes in foodborne bacteria has gained increasing interest in the past decade, especially in light of the potential transferability of antibiotic resistance determinants to opportunistic pathogens, natural inhabitants of the human gut but capable of acquiring virulence in immunocompromised individuals. This review aims at describing major findings and future prospects in the field, especially after the use of antibiotics as growth promoters was totally banned in Europe, with special emphasis on the application of genomic technologies to improve quality and safety of fermented foods.
Keywords: Lactic acid bacteria, Fermented food, Horizontal transfer, Metagenomics
Food fermenting microflora
Fermented foods result from the metabolic activity of a complex and heterogeneous microflora, which multiplies using naturally occurring substrates within the food matrix while converting them to a broad range of molecules that confer unique composition and sensory features to the final products. The composition of such “food microbiota” represents a key factor toward obtaining final products with typical taste, texture, and nutrient profile and is specific for each food, as it depends on the fermentable substrates as well as on production technology (temperatures, pH, ripening etc.). Bacteria acting as fermentation starters in food may originate from environmental sources (i.e., pre-existing in the raw ingredients), or be added as commercial mixtures of specific strains to pasteurized matrices (milk, meat, vegetables etc.). In this latter case, the microbiological profile is more standardized, although relative proportions among the different species can be altered by technological processing (Hansen 2002). Several dairy and meat products typical of Mediterranean countries are obtained by traditional manufacturing procedures that do not employ selected industrial starters, relying on the microflora naturally present in raw ingredients (Morea et al. 1999). Fermentation in such products is therefore carried out by natural starters, whose species profile reflects local environments. As an example, a recent study reporting comparison of the typing profiles of natural Lactobacillus paracasei isolates from products manufactured in different geographical regions of Italy, has shown almost unique strain composition for each product, as expected in fermented foods employing naturally occurring starters (Comunian et al. 2009). Selective pressure exerted by the technological steps along manufacturing procedures further impacts bacterial composition in the final product, leading to a wide range of fermented foods characterized by distinct flavors, consistencies and microbiological quality, achievable only within specific geographical areas. Most of these products, which are especially abundant in countries where warm climates allow ample microbiological biodiversity in environmental sources, are identified in Europe by the Protected Designation of Origin (PDO) label, a recognition which guarantees both the specific geographical origin and the unique production process (EC Council Regulation N° 2081/92, July 14, 1992, protection of geographical indications and designations of origin for agricultural products and foodstuffs). The extent to which these products contain live bacteria mostly depends on the combination of technological and ripening processes to which each food is subjected before being consumed (Ercolini et al. 2004; Devirgiliis et al. 2008). Dairy fermented products are frequently consumed fresh, containing therefore more than other fermented fodstuffs a complex, live microbial community which enters the human body through the food chain. Lactic Acid Bacteria (LAB) represent an important component of such fermented food microbiota.
Lactic acid bacteria (LAB) in food fermentations
LAB represent a heterogeneous group of Gram-positive, catalase-negative, non-spore-forming, low G + C content, facultative anaerobic bacteria able to ferment sugars primarily into lactic acid via homo- or hetero-fermentative pathways (Bernardeau et al. 2008). Several LAB species are involved in fermentation of a wide range of foods and beverages, such as dairy, meat, fish, vegetable, sourdough, wine, and cider (Liu 2003). In dairy technologies in particular, they play different roles: starter LAB (SLAB) initiate the fermentation process, rapidly converting lactose to high concentrations of lactic acid whereas non-starter LAB (NSLAB) are responsible for ripening, which occurs through biological modifications leading to cheese maturation, greatly contributing to the quality of finished products. SLAB mainly include the mesophilic species Lactococcus lactis and Leuconostoc spp., as well as the thermophilic species Streptococcus thermophilus, Lactobacillus delbrueckii, and L. helveticus. On the contrary, the group of NSLAB is very heterogeneous, with Lactobacillus as the most represented genus (Settanni and Moschetti 2010), followed by other genera including Enterococcus, Lactococcus, and Streptococcus (Liu 2003). Lactobacilli can colonize raw foods (e.g., milk and meat) and multiply during fermentation. Environmental species of lactobacilli also display probiotic features, they can survive food processing and persist in finished products, constituting a large portion of the natural, live microflora in most fermented dairy and meat foodstuffs (Turpin et al. 2010). This aspect is extremely important, as they are also highly represented within the intestinal microbiota of humans and animals (Bernardeau et al. 2008). Among lactobacilli, L. paracasei is the most common foodborne isolate from raw milk, as well as from dairy products such as typical mediterranean cheeses (Abriouel et al. 2008; Jokovic et al. 2008; Van Hoorde et al. 2008). Together with the closely related species L. casei, L. rhamnosus, and L. zeae, it is also one of the most common bacterial species employed as probiotic additives in the food industry (Andriulli et al. 2008; Chouraqui et al. 2008). Enterococcus species represent a substantial proportion of foodborne NSLAB. They perform important functions during cheese ripening and in the development of specific flavors in traditional cheeses and sausages, although they have also been reported to spoil processed meats (Franz et al. 2003). Their use as human probiotics remains controversial, in light of the capability of most Enterococcus species to turn into opportunistic pathogens in immunocompromised individuals, leading to nosocomial infections such as bacteraemia or endocarditis (Foulquie Moreno et al. 2006; Ogier and Serror 2008). The most relevant bacterial functions in dairy fermentations are encoded by genes involved in sugar metabolism, lipid metabolism, and in the proteolytic system. Proteolysis plays a key role in cheese ripening, especially toward the achievement of final texture and flavors. Availability of the complete genome sequence of L. helveticus DPC4571, a strain isolated from cheese whey, has enabled to identify specific genes of technological interest. Noteworthy, the genome of this foodborne strain is very similar to those of intestinal lactobacilli, although originating from considerably different environments. Bioinformatic analysis demonstrated that 65–75% of the genes were conserved between dairy and commensal lactobacilli, suggesting that a relatively small, though highly specific gene set accounts for the difference between dairy and gut species (Slattery et al. 2010).
Fermented food microbiota
The relevance of food microbiological biodiversity for human health becomes evident when considering the contribution it provides to the resident microbiota of the human gut. The human digestive tract harbors trillions of bacteria, many of which establish lifetime, symbiotic relationships with their hosts (Bik 2009). The gut microbiota provides the host with enhanced metabolic capabilities (i.e., enzymes that break down hard-to-digest polysaccharides in plant foods), protection against pathogens, development of the immune system and other yet poorly investigated regulatory functions, overall playing an essential role in the balance between health and disease (Martins dos Santos et al. 2010; Sekirov et al. 2010). Changes in microbiota composition have often been associated with health disorders, such as IBS (Frank et al. 2007), Crohn’s Disease (Manichanh et al. 2006), obesity and weight loss (Backhed et al. 2007; Zhang et al. 2009; Tsukumo et al. 2009). LAB and bifidobacterial populations have been detected as part of the complex intestinal ecosystem in the majority of humans, even though species composition is highly subject-specific (Turnbaugh and Gordon 2009). These unique autochthonous or indigenous populations can be supplemented with LAB from dairy or probiotic products that are generally allochthonous, due to their transient passage through the GI tract (Vaughan et al. 2002). Increasing evidence points to the ability of diet to shape the human gut microbiota, acting on the balance between Bacteroidetes and Firmicutes, the two major phyla of Gram-positive commensal bacteria residing in the gastro-intestinal tract. An important metabolic difference between these two phyla is the efficiency of energy extraction from undigestible dietary carbohydrates. Therefore, alterations in their relative ratio can result in increased energy accumulation, which is of special concern in certain pathological conditions. For this reason, several studies have focused in recent years on the composition of human gut microbiota in obese subjects under different dietary regimens (Turnbaugh and Gordon 2009; Backhed et al. 2007; Bajzer and Seeley 2006; Tsai and Coyle 2009; Neyrinck and Delzenne 2010 and references therein). Nevertheless, the cause-effect relationship between microbial composition and this pathology is not yet entirely clear. Other studies investigated diet-gut microbiota interaction in non-pathological conditions: for example, a polysaccharide-rich diet was recently reported to lead to significant enrichment in Bacteroidetes and depletion in Firmicutes in the fecal microbiota of children (De Filippo et al. 2010). An interesting example of the interaction between food and gut microbiota is provided by a study on Japanese individuals, whose diet includes regular consumption of sushi. Their gut microbiota was shown to harbor resident bacteria that had acquired genetic material from a marine species, enabling to digest the components of the seaweed used to wrap sushi (Hehemann et al. 2010; Sonnenburg 2010). Such an interplay between food and gut microbiota underlines the importance of food microbial composition in terms of both quality (e.g., presence of probiotic species) and safety (e.g., ratio of probiotics/pathogens). From the food safety viewpoint, selection and spreading of antibiotic resistance genes in foodborne bacteria has gained increasing interest in the past decade. Environmental bacteria are inevitably found in milk and unprocessed meat, even when milking and slaughtering occur under proper hygenic conditions. When antibiotic resistant (AbR) strains persist alive in fermented products, they eventually reach the human GI tract through food consumption (Teuber and Perreten 2000) thereby increasing the risk of AbR gene transfer to human pathogens which might be present within the intestinal microbiota (reviewed in: Kazimierczak and Scott 2007).
Origin and spread of antibiotic resistance in foodborne bacteria
Antimicrobial agents are commonly used in animal husbandry to cure or prevent the onset of bacterial infections. However, their use at sub-therapeutic doses as growth promoters has led over decades of use to selection of AbR strains within the intestinal microflora of treated livestock, and as a consequence AbR commensal bacteria have been detected in fermented foods derived from meat and milk. The cause-effect relationship between spread of antibiotic resistance in commensal bacteria and antimicrobial use in animal farming started to be seriously considered in the early 80s (reviewed in: Wegener 2003), but a growing body of literature reporting identification of AbR genes in foodborne bacteria appeared after several European countries started monitoring their fermented foods for presence of AbR strains (Witte 2000; Teuber 2001; Hammerum et al. 2007; Klare et al. 2007; Maietti et al. 2007; Ammor et al. 2008; Ouoba et al. 2008; Zonenschain et al. 2009; Nawaz et al. 2010; Toomey et al. 2010). As an example of such a tight link, comparative analysis of 121 strains of L. paracasei isolated from Italian dairy and meat products manufactured in different geographical regions was recently reported. Phenotypic and molecular characterization demonstrated the presence of genetic determinants of tetracycline [tet(M), tet(W)] and/or erythromycin resistance [erm(B)] (Table 1) in the different AbR isolates. However, the majority of L. paracasei strains susceptible to these two drugs originated from cheeses produced in a region where livestock are traditionally pastured and no systematic use of antibiotics as growth promoters has been carried out over the years. Conversely, the highest number of resistant strains was detected in fermented meat and dairy foods produced in areas where more intensive practices have been applied in animal husbandry (Comunian et al. 2009).
Table 1.
Antibiotic resistance genes most commonly found in LAB
| Antibiotic (pharmacological class) | Resistance gene(s) | Mechanism of action | References |
|---|---|---|---|
|
Tetracycline (Tetracyclines) |
tet(M), (S), (O), (W), (36) | Ribosomal protection | (Roberts 2005); (Thaker et al. 2010) |
| tet(L) | Efflux | (Roberts 2005); (Thaker et al. 2010) | |
| tet(K) | Efflux | (Roberts 2005); (Thaker et al. 2010) | |
|
Erythromycin (Macrolides) |
erm(A), (B), (C), (F), (G) | rRNA Methylases | (Roberts 2008); (Mayrhofer et al. 2010) |
| msr(A/B), (D) | Efflux | (Roberts 2008); (Toomey et al. 2010) | |
| isa(A) | Efflux | (Roberts 2008) | |
| vga(B) | Efflux | (Roberts 2008) | |
|
Vancomycina (Glycopepdtides) |
van(A), (B), (D), (E), (G), (L)b | Peptidoglycan modification | (Werner et al. 2008) |
| van(C)c | Peptidoglycan modification | (Werner et al. 2008) |
aSome LAB genera (Lactobacillus, Pediococcus, Leuconostoc) are intrinsically resistant to Vancomycin
bAcquired
cIntrinsic
Antibiotics for human use belong to the same pharmacological classes as those employed in veterinary medicine (mainly tetracyclines, macrolides, glycopeptides, and aminoglycosides), even though represented by different molecules. Selection of antibiotic resistance by exposure to a specific drug therefore leads to cross-resistance against all antimicrobials belonging to the same class, through inhibition of common chemical features or action mechanisms (Wright 2003). Simultaneous resistance to multiple unrelated antibiotics belonging to different pharmacological classes has also been reported to arise in bacterial populations, following exposure to a single drug. The most common mechanism proposed for this phenomenon is the genetic linkage of different AbR genes within gene clusters, followed by acquisition of such resistance islands by susceptible bacteria through horizontal gene transfer under selective pressure (Gilmore and Ferretti 2003). This mechanism is supported by genomic identification of AbR islands flanked by mobile elements (Alekshun and Levy 2007). A novel alternative mechanism was recently demonstrated, which occurs via ROS-induced mutagenesis stimulated by sublethal concentrations of bactericidal, DNA damaging antibiotics such as β-lactams, quinolones, and aminoglycosides. Increased mutation rates were associated in Escherichia coli with heterogeneous increase in the minimum inhibitory concentration (MIC) for several antibiotics, due to selection of multidrug resistance proteins with wider selectivity toward antimicrobials (Kohanski et al. 2010).
The main threat associated with AbR in non-pathogenic, commensal bacteria is the risk of horizontal transfer of resistance determinants to pathogens, thus impairing successful antibiotic treatment of common microbial infections. Over 1,000 species are represented within the intestinal microbiota of humans and animals (Bik 2009), including opportunistic pathogens such as enterococci, natural members of the gut microbiota but capable of acquiring virulence genes (Ogier and Serror 2008). Up to now, published reports on selection and dissemination of antibiotic resistance genes within the complex bacterial community of the human gut were focused mainly on clinically relevant species (Franz et al. 2003). However, it is now becoming clear that foodborne LAB may act as reservoir of AbR genes potentially transferable to human pathogens (Mathur and Singh 2005). The crowded environment of the gut in humans and animals represents an ideal site for genetic exchanges between the resident and transient components of the microbiota. On the basis of the precautionary principle and to prevent potential risks to human health, the use of antimicrobials as growth promoters in animal farming was progressively banned in European countries over the past 20 years (Casewell et al. 2003; Phillips 2007). Since then, several studies have shown steadily decreasing counts of antibiotic resistant bacteria in the feces of livestock (Wegener 2003). However, low yet consistent levels of AbR species are still reported to occur in the environment, including food products (Mathur and Singh 2005; Ammor et al. 2007). Clinical investigations have documented persistence of AbR strains in the human gut even in the absence of selective pressure (reviewed in Jernberg et al. 2010), indicating that drug exposure induces long-term alterations within complex microbial communities.
Genetic transferability of AbR in fermented food microbiota
Mobile elements are key players in the horizontal spread of resistance genes in bacteria. Plasmids and transposons represent highly organized mobile elements, whose presence in bacterial genomes indicates potential intra- and inter-species transfer of genetic material, including AbR genes (Alekshun and Levy 2007). Other than plasmids and transposons, simple mobile elements such as insertion sequences (IS), capable of autonomous transposition, have been identified in association with AbR genes. IS are small segments of DNA flanked by short repeated sequences required for transposition, and encoding only few functions involved in their own mobility (Mahillon and Chandler 1998). Like transposons, IS elements have been found on the chromosome, on plasmids or on both, but their horizontal transfer occurs only when they are associated with conjugative elements. Noteworthy, genome sequencing of the vancomycin resistant V583 strain of E. faecalis has shown the presence of 38 IS which, together with other mobile elements and exogenously acquired DNA, represent more than 25% of the entire genome (Paulsen et al. 2003). It is therefore crucial to investigate the presence of mobile elements in the genomic context of AbR genes. To date, several works have reported such associations in LAB, mostly focusing on tetracycline and erythromycin resistance, as these antimicrobials are widely used in both human and animal therapy (Beaber et al. 2002; Pembroke et al. 2002; Teuber et al. 2003; Foulquie Moreno et al. 2006; Boguslawska et al. 2009 and references therein). Moreover, an increasing body of literature describes the presence of tetracycline resistance genes in the fermenting microflora derived from different food sources (reviewed in: Ammor et al. 2007). Horizontal transmission of AbR through conjugal transfer, however, has not always been demonstrated, indicating that it does not depend solely upon association with mobile elements. The most frequent AbR determinants reported to occur in LAB are listed in Table 1: tet genes, especially those encoding ribosomal protection proteins (RPP), followed by erm genes which encode methyl-transferases conferring resistance to macrolides (Bernardeau et al. 2008). The Tn916-1545 transposon family includes Tn916, a 18 kb element carrying the tet(M) gene which was the first conjugative transposon to be identified. It displays broad host-range, comprising both Gram-positive and Gram-negative bacteria (Clewell et al. 1995). A variety of antibiotic resistance determinants associated with this family of transposons have been described in foodborne strains of E. faecalis and E. faecium, although conjugative transfer of resistance genes could not be demonstrated in all cases (Hummel et al. 2007). In other reports, Tet resistant enterococci strains, originating from meat products and harboring Tn916-1545, were able to transfer tetracycline resistance to E. faecalis or Listeria innocua recipients not only in filter-mating assays, but also in meat food matrices (Rizzotti et al. 2009). In our studies, a group of tetracycline-resistant L. paracasei isolates from water buffalo raw milk and whey was shown to harbor Tn916-encoded tet(M) which could be transferred in filter-mating assays to E. faecalis via a circular intermediate containing a novel coupling sequence that confers low frequency-conjugation phenotype (Devirgiliis et al. 2009).
Several reports clearly indicate that bacterial genome plasticity leads to a great variety of genomic contexts which play a key role in the efficiency of genetic transfer in-between different species and genera. Few examples will better clarify this aspect. A strain of E. italicus isolated from robiola cheese was shown to harbor the tet(M) gene within a non-conjugative plasmid, which could not be transferred to a recipient E. faecalis (Borgo et al. 2009). On the contrary, a plasmid borne tet(S) gene identified in the E. italicus strain LMG 22195 isolated from fermented milk was shown to transfer to E. faecalis JH2-2 recipient by conjugation (Zago et al. 2010). We have characterized tetracycline-resistant E. faecalis, Streptococcus bovis, and Lc. lactis harboring the tet(M) gene, showing that only in Lc. lactis the gene was plasmid borne and could be transferred with high efficiency to E. faecalis in in vitro conjugation assays. Independent erythromycin and tetracycline doubly resistant isolates of Lc. lactis were shown to harbor the erythromicin resistance methylase gene erm(B), and the RPP gene tet(S), genetically linked and plasmid borne, although they could not be transferred to E. faecalis recipients (Devirgiliis et al. 2010). In Lc. lactis strains isolated from a Spanish, starter-free cheese and harboring the tet(M) gene within a functional Tn916 transposon inserted into a resident plasmid, the gene could be transferred by conjugation to other lactococci and enterococci (Florez et al. 2008). A newly characterized erythromycin resistance plasmid (pLFE1) was described in the L. plantarum strain M345, isolated from raw-milk cheese. This plasmid was transferred with high frequency to E. faecalis in a gnotobiotic rat model (Feld et al. 2008). In filter-mating experiments pLFE1 displayed broad host-range, yielding transconjugants when mated to L. rhamnosus, Lc lactis, Listeria innocua, E. faecalis, as well as to the pathogen Listeria monocytogenes (Feld et al. 2009). Overall, these studies indicate that no exhaustive picture has yet emerged regarding all factors affecting transferability of AbR genes in foodborne LAB, although association with mobile elements as well as favorable genomic context appear to be essential requirements.
Metagenomic approaches to the study of antibiotic resistance in food microbiota
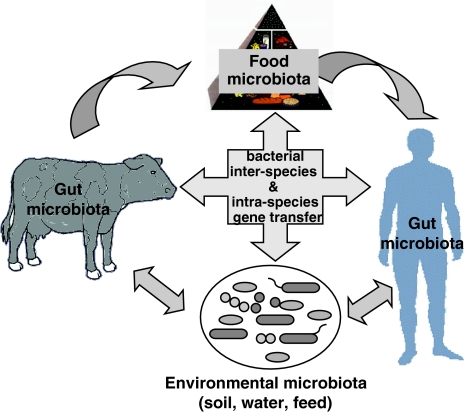
Metagenomics is a promising emerging field that allows to study complex microbial ecosystems through culture-independent approaches. This latter feature is particularly relevant, since several species of environmental bacteria are unculturable in the standard media commonly used in the laboratory. It was estimated that 106–108 separate genospecies of prokaryotic cells live on earth, and the majority of them is still uncharacterized (Simon and Daniel 2009). Metagenomic approaches are usually applied to study the microbial community as a single dynamic entity (reviewed in: Streit and Schmitz 2004; Green and Keller 2006; Simon and Daniel 2009). Widely applied to soil, water (marine environments), oral and gut microbial communities (Tuohy et al. 2009), metagenomics has still limited applications in food ecosystems. The food microbiota is extremely complex, partly specific for each fermented food and in constant interplay with the environmental bacterial community, including the gut microbiota of animals and humans (Fig. 1). Just like the gut microbiota, it is constituted by a large portion of unculturable species. The emergence of culture-independent methods can therefore greatly contribute to deeper understanding of the diversity and functionality of such a complex ecosystem, with the added value of providing also functional information on genes and pathways that could be exploited to improve microbiological composition and health-promoting functions of fermented foods.
Fig. 1.
Graphic representation of the interplay among environmental, food, and gut microbiota of humans and animals. Foods derived from animal sources represent the key factors in vehiculating environmental bacteria to the human gut. Genetic exchanges can occur at any step, but they are greatly favored within the crowded environment of the gut
One of the advantages of metagenomic libraries is their stability, which allows long-term storage and subsequent screening with any specific probe for additional genes of interest. In particular, fosmid libraries have the advantage of bearing large DNA inserts (30–40 Kb), thus allowing to characterize the genomic context surrounding specific genes and to unambiguosly identify the bacterial species from which they originate. In the field of antibiotic resistance, metagenomics has contributed to the characterization of known AbR genes and of their association with mobile elements, as well as to the identification of novel ones, thus leading to the definition of “resistomes” (Kazimierczak et al. 2009; Thaker et al. 2010). For example, investigation of human oral and fecal metagenomes from healthy volunteers identified novel as well as known tetracycline and erythromycin resistance genes. Among them tet(M) was the prominent AbR determinant in oral samples and tet(W) in the feces (Seville et al. 2009). Sequencing of the Group A Streptococcus (GAS) metagenome led to the identification of exogenous integrated conjugative elements (ICEs), carrying multiple genes encoding resistance to antimicrobials, including tetracycline and erythromycin (Beres and Musser 2007). Moreover, PCR analysis of a plasmid metagenome obtained from wastewater treatment plant bacteria revealed the presence of several clinical relevant AbR genes (Szczepanowski et al. 2009). Functional screening of metagenomic expression libraries on plates containing different antibiotics has been successfully applied in the case of oral (Diaz-Torres et al. 2006), activated sludge (Mori et al. 2008) or soil (Donato et al. 2010) microbial communities. It is important to consider that possible limitations of function-based screenings might reside in the inability of a foreign host (E. coli in most cases) to properly express heterologous genes and to produce functional gene products. However, it was demonstrated that even evolutionary distant AbR genes (i.e., beta-lactamases isolated from a remote Alaskan soil bacterium) could confer AbR to E. coli without the need to manipulate the gene expression machinery of the host (Allen et al. 2009).
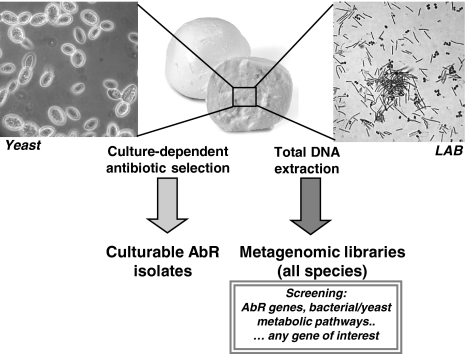
Application of metagenomics to the analysis of food microbiota requires evaluation of the specific questions to be addressed experimentally. The majority of genome sequences within the food microbiome are still unknown. Metagenomic approaches would be extremely helpful not only to expand taxonomic analysis to both culturable and unculturable species but also to identify metabolic functions and/or pathways which play essential roles in human health as well as in the quality and safety of fermented foods. Within this context, the approach based on cloning amplified 16S ribosomal DNA fragments poses some limitations (Ventura et al. 2009). Cloning large genomic fragments, on the other hand, presently appears the best strategy to obtain the necessary sequence information from flanking regions, allowing species identification as well as determination of the genomic context of target genes (Streit and Schmitz 2004). Our laboratory is involved in the isolation and characterization of AbR genes in foodborne LAB, and we have recently extended our work to the analysis of the food microbiome. To this aim, we have constructed a fosmid metagenomic library containing large fragments of total DNA extracted from water buffalo mozzarella, a traditional fermented dairy food widely consumed in Italy (unpublished results) (Fig. 2). Extraction of total bacterial DNA directly from food is more easily applicable to dairy products because, unlike fermented meats, they contain almost exclusively microbial DNA (from bacteria and yeasts), with undetectable contamination from higher eukaryotic cell DNA. The relative proportions among yeast, fungal and bacterial DNAs exclusively depend on the cheese source and can be evaluated on the basis of colony forming units. To restrict the analysis to the bacterial metagenome, libraries can then be screened with specific bacterial probes under stringent conditions. A metagenomic library containing the microbiome from water buffalo mozzarella is presently beeing screened in our laboratory with the aim of identifying AbR genes that escape detection with a culture-dependent approach (Devirgiliis et al. 2008). This traditional Italian PDO cheese is consumed fresh within 2 weeks from production and contains high titer of live microflora. In light of the health-promoting functions of commensal bacteria and of the interplay between environmental, food, and gut microbiota (Fig. 1), we are also investigating the presence within the cheese metagenome of metabolic functions responsible for production of bioactive molecules from prebiotic substrates. We believe that food-based metagenomic approaches could be extended to a plethora of fermented dairy products typical of traditional dietary profiles such as the Mediterranean Diet, greatly contributing to deeper understanding of the probiotic, health-promoting functions naturally present within food microbiota.
Fig. 2.
Culture dependent versus culture-independent approaches to study the microbiome of waterbuffalo mozzarella, a fresh Italian PDO cheese containing high titers of live bacteria and yeasts
Acknowledgments
The Authors acknowledge financial support by grant “NUME” (DM 3688/7303/08) from the Italian Ministry of Agriculture, Food and Forestry (MiPAAF).
Open Access
This article is distributed under the terms of the Creative Commons Attribution Noncommercial License which permits any noncommercial use, distribution, and reproduction in any medium, provided the original author(s) and source are credited.
Abbreviations
- AbR
Antibiotic resistance (resistant)
- GI
Gastro-intestinal
- ICE
Integrated conjugative elements
- LAB
Lactic acid bacteria
- NSLAB
Non starter lactic acid bacteria
- PDO
Protected designation of origin
- ROS
Reactive oxygen species
- RPP
Ribosomal protection protein
- SLAB
Starter lactic acid bacteria
References
- Abriouel H, Martin-Platero A, Maqueda M, Valdivia E, Martinez-Bueno M. Biodiversity of the microbial community in a Spanish farmhouse cheese as revealed by culture-dependent and culture-independent methods. Int J Food Microbiol. 2008;127:200–208. doi: 10.1016/j.ijfoodmicro.2008.07.004. [DOI] [PubMed] [Google Scholar]
- Alekshun MN, Levy SB. Molecular mechanisms of antibacterial multidrug resistance. Cell. 2007;128:1037–1050. doi: 10.1016/j.cell.2007.03.004. [DOI] [PubMed] [Google Scholar]
- Allen HK, Moe LA, Rodbumrer J, Gaarder A, Handelsman J. Functional metagenomics reveals diverse beta-lactamases in a remote Alaskan soil. Isme J. 2009;3:243–251. doi: 10.1038/ismej.2008.86. [DOI] [PubMed] [Google Scholar]
- Ammor MS, Florez AB, Mayo B. Antibiotic resistance in non-enterococcal lactic acid bacteria and bifidobacteria. Food Microbiol. 2007;24:559–570. doi: 10.1016/j.fm.2006.11.001. [DOI] [PubMed] [Google Scholar]
- Ammor MS, Gueimonde M, Danielsen M, Zagorec M, van Hoek AH, de Los Reyes-Gavilan CG, Mayo B, Margolles A. Two different tetracycline resistance mechanisms, plasmid-carried tet(L) and chromosomally located transposon-associated tet(M), coexist in Lactobacillus sakei Rits 9. Appl Environ Microbiol. 2008;74:1394–1401. doi: 10.1128/AEM.01463-07. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Andriulli A, Neri M, Loguercio C, Terreni N, Merla A, Cardarella MP, Federico A, Chilovi F, Milandri GL, De Bona M, Cavenati S, Gullini S, Abbiati R, Garbagna N, Cerutti R, Grossi E. Clinical trial on the efficacy of a new symbiotic formulation, Flortec, in patients with irritable bowel syndrome: a multicenter, randomized study. J Clin Gastroenterol. 2008;42:S218–S223. doi: 10.1097/MCG.0b013e31817fadd6. [DOI] [PubMed] [Google Scholar]
- Backhed F, Manchester JK, Semenkovich CF, Gordon JI. Mechanisms underlying the resistance to diet-induced obesity in germ-free mice. Proc Natl Acad Sci USA. 2007;104:979–984. doi: 10.1073/pnas.0605374104. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bajzer M, Seeley RJ. Physiology: obesity and gut flora. Nature. 2006;444:1009–1010. doi: 10.1038/4441009a. [DOI] [PubMed] [Google Scholar]
- Beaber JW, Burrus V, Hochhut B, Waldor MK. Comparison of SXT and R391, two conjugative integrating elements: definition of a genetic backbone for the mobilization of resistance determinants. Cell Mol Life Sci. 2002;59:2065–2070. doi: 10.1007/s000180200006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Beres SB, Musser JM. Contribution of exogenous genetic elements to the group A Streptococcus metagenome. PLoS One. 2007;2:e800. doi: 10.1371/journal.pone.0000800. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bernardeau M, Vernoux JP, Henri-Dubernet S, Gueguen M. Safety assessment of dairy microorganisms: the Lactobacillus genus. Int J Food Microbiol. 2008;126:278–285. doi: 10.1016/j.ijfoodmicro.2007.08.015. [DOI] [PubMed] [Google Scholar]
- Bik EM. Composition and function of the human-associated microbiota. Nutr Rev. 2009;67(Suppl 2):S164–S171. doi: 10.1111/j.1753-4887.2009.00237.x. [DOI] [PubMed] [Google Scholar]
- Boguslawska J, Zycka-Krzesinska J, Wilcks A, Bardowski J. Intra- and interspecies conjugal transfer of Tn916-like elements from Lactococcus lactis in vitro and in vivo. Appl Environ Microbiol. 2009;75:6352–6360. doi: 10.1128/AEM.00470-09. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Borgo F, Ricci G, Arends K, Schiwon K, Grohmann E, Fortina MG. Evaluation of plasmid content and tetracycline resistance conjugative transfer in Enterococcus italicus strains of dairy origin. Curr Microbiol. 2009;59:261–266. doi: 10.1007/s00284-009-9428-5. [DOI] [PubMed] [Google Scholar]
- Casewell M, Friis C, Marco E, McMullin P, Phillips I. The European ban on growth-promoting antibiotics and emerging consequences for human and animal health. J Antimicrob Chemother. 2003;52:159–161. doi: 10.1093/jac/dkg313. [DOI] [PubMed] [Google Scholar]
- Chouraqui JP, Grathwohl D, Labaune JM, Hascoet JM, de Montgolfier I, Leclaire M, Giarre M, Steenhout P. Assessment of the safety, tolerance, and protective effect against diarrhea of infant formulas containing mixtures of probiotics or probiotics and prebiotics in a randomized controlled trial. Am J Clin Nutr. 2008;87:1365–1373. doi: 10.1093/ajcn/87.5.1365. [DOI] [PubMed] [Google Scholar]
- Clewell DB, Flannagan SE, Jaworski DD. Unconstrained bacterial promiscuity: the Tn916-Tn1545 family of conjugative transposons. Trends Microbiol. 1995;3:229–236. doi: 10.1016/S0966-842X(00)88930-1. [DOI] [PubMed] [Google Scholar]
- Comunian R, Daga E, Dupre I, Paba A, Devirgiliis C, Piccioni V, Perozzi G, Zonenschain D, Rebecchi A, Morelli L, De Lorentiis A, Giraffa G. Susceptibility to tetracycline and erythromycin of Lactobacillus paracasei strains isolated from traditional Italian fermented foods. Int J Food Microbiol. 2009;138:151–156. doi: 10.1016/j.ijfoodmicro.2009.11.018. [DOI] [PubMed] [Google Scholar]
- De Filippo C, Cavalieri D, Di Paola M, Ramazzotti M, Poullet JB, Massart S, Collini S, Pieraccini G, Lionetti P. Impact of diet in shaping gut microbiota revealed by a comparative study in children from Europe and rural Africa. Proc Natl Acad Sci USA. 2010;107:14691–14696. doi: 10.1073/pnas.1005963107. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Devirgiliis C, Caravelli A, Coppola D, Barile S, Perozzi G. Antibiotic resistance and microbial composition along the manufacturing process of Mozzarella di Bufala Campana. Int J Food Microbiol. 2008;128:378–384. doi: 10.1016/j.ijfoodmicro.2008.09.021. [DOI] [PubMed] [Google Scholar]
- Devirgiliis C, Coppola D, Barile S, Colonna B, Perozzi G. Characterization of the Tn916 conjugative transposon in a food-borne strain of Lactobacillus paracasei. Appl Environ Microbiol. 2009;75:3866–3871. doi: 10.1128/AEM.00589-09. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Devirgiliis C, Barile S, Caravelli A, Coppola D, Perozzi G. Identification of tetracycline- and erythromycin-resistant Gram-positive cocci within the fermenting microflora of an Italian dairy food product. J Appl Microbiol. 2010;109:313–323. doi: 10.1111/j.1365-2672.2010.04661.x. [DOI] [PubMed] [Google Scholar]
- Diaz-Torres ML, Villedieu A, Hunt N, McNab R, Spratt DA, Allan E, Mullany P, Wilson M. Determining the antibiotic resistance potential of the indigenous oral microbiota of humans using a metagenomic approach. FEMS Microbiol Lett. 2006;258:257–262. doi: 10.1111/j.1574-6968.2006.00221.x. [DOI] [PubMed] [Google Scholar]
- Donato JJ, Moe LA, Converse BJ, Smart KD, Berklein FC, McManus PS, Handelsman J. Metagenomic analysis of apple orchard soil reveals antibiotic resistance genes encoding predicted bifunctional proteins. Appl Environ Microbiol. 2010;76:4396–4401. doi: 10.1128/AEM.01763-09. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ercolini D, Mauriello G, Blaiotta G, Moschetti G, Coppola S. PCR-DGGE fingerprints of microbial succession during a manufacture of traditional water buffalo mozzarella cheese. J Appl Microbiol. 2004;96:263–270. doi: 10.1046/j.1365-2672.2003.02146.x. [DOI] [PubMed] [Google Scholar]
- Feld L, Schjorring S, Hammer K, Licht TR, Danielsen M, Krogfelt K, Wilcks A. Selective pressure affects transfer and establishment of a Lactobacillus plantarum resistance plasmid in the gastrointestinal environment. J Antimicrob Chemother. 2008;61:845–852. doi: 10.1093/jac/dkn033. [DOI] [PubMed] [Google Scholar]
- Feld L, Bielak E, Hammer K, Wilcks A. Characterization of a small erythromycin resistance plasmid pLFE1 from the food-isolate Lactobacillus plantarum M345. Plasmid. 2009;61:159–170. doi: 10.1016/j.plasmid.2009.01.002. [DOI] [PubMed] [Google Scholar]
- Florez AB, Ammor MS, Mayo B. Identification of tet(M) in two Lactococcus lactis strains isolated from a Spanish traditional starter-free cheese made of raw milk and conjugative transfer of tetracycline resistance to lactococci and enterococci. Int J Food Microbiol. 2008;121:189–194. doi: 10.1016/j.ijfoodmicro.2007.11.029. [DOI] [PubMed] [Google Scholar]
- Foulquie Moreno MR, Sarantinopoulos P, Tsakalidou E, De Vuyst L. The role and application of enterococci in food and health. Int J Food Microbiol. 2006;106:1–24. doi: 10.1016/j.ijfoodmicro.2005.06.026. [DOI] [PubMed] [Google Scholar]
- Frank DN, St Amand AL, Feldman RA, Boedeker EC, Harpaz N, Pace NR. Molecular-phylogenetic characterization of microbial community imbalances in human inflammatory bowel diseases. Proc Natl Acad Sci USA. 2007;104:13780–13785. doi: 10.1073/pnas.0706625104. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Franz CM, Stiles ME, Schleifer KH, Holzapfel WH. Enterococci in foods–a conundrum for food safety. Int J Food Microbiol. 2003;88:105–122. doi: 10.1016/S0168-1605(03)00174-0. [DOI] [PubMed] [Google Scholar]
- Gilmore MS, Ferretti JJ. Microbiology. The thin line between gut commensal and pathogen. Science. 2003;299:1999–2002. doi: 10.1126/science.1083534. [DOI] [PubMed] [Google Scholar]
- Green BD, Keller M. Capturing the uncultivated majority. Curr Opin Biotechnol. 2006;17:236–240. doi: 10.1016/j.copbio.2006.05.004. [DOI] [PubMed] [Google Scholar]
- Hammerum AM, Heuer OE, Emborg HD, Bagger-Skjot L, Jensen VF, Rogues AM, Skov RL, Agerso Y, Brandt CT, Seyfarth AM, Muller A, Hovgaard K, Ajufo J, Bager F, Aarestrup FM, Frimodt-Moller N, Wegener HC, Monnet DL. Danish integrated antimicrobial resistance monitoring and research program. Emerg Infect Dis. 2007;13:1632–1639. doi: 10.3201/eid1311.070421. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hansen EB. Commercial bacterial starter cultures for fermented foods of the future. Int J Food Microbiol. 2002;78:119–131. doi: 10.1016/S0168-1605(02)00238-6. [DOI] [PubMed] [Google Scholar]
- Hehemann JH, Correc G, Barbeyron T, Helbert W, Czjzek M, Michel G. Transfer of carbohydrate-active enzymes from marine bacteria to Japanese gut microbiota. Nature. 2010;464:908–912. doi: 10.1038/nature08937. [DOI] [PubMed] [Google Scholar]
- Hummel A, Holzapfel WH, Franz CM. Characterisation and transfer of antibiotic resistance genes from enterococci isolated from food. Syst Appl Microbiol. 2007;30:1–7. doi: 10.1016/j.syapm.2006.02.004. [DOI] [PubMed] [Google Scholar]
- Jernberg C, Lofmark S, Edlund C, Jansson JK. Long-term impacts of antibiotic exposure on the human intestinal microbiota. Microbiology. 2010;156:3216–3223. doi: 10.1099/mic.0.040618-0. [DOI] [PubMed] [Google Scholar]
- Jokovic N, Nikolic M, Begovic J, Jovcic B, Savic D, Topisirovic L. A survey of the lactic acid bacteria isolated from Serbian artisanal dairy product kajmak. Int J Food Microbiol. 2008;125:305–311. doi: 10.1016/j.ijfoodmicro.2008.07.026. [DOI] [PubMed] [Google Scholar]
- Kazimierczak KA, Scott KP. Antibiotics and resistance genes: influencing the microbial ecosystem in the gut. Adv Appl Microbiol. 2007;62:269–292. doi: 10.1016/S0065-2164(07)62009-7. [DOI] [PubMed] [Google Scholar]
- Kazimierczak KA, Scott KP, Kelly D, Aminov RI. Tetracycline resistome of the organic pig gut. Appl Environ Microbiol. 2009;75:1717–1722. doi: 10.1128/AEM.02206-08. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Klare I, Konstabel C, Werner G, Huys G, Vankerckhoven V, Kahlmeter G, Hildebrandt B, Muller-Bertling S, Witte W, Goossens H. Antimicrobial susceptibilities of Lactobacillus, Pediococcus and Lactococcus human isolates and cultures intended for probiotic or nutritional use. J Antimicrob Chemother. 2007;59:900–912. doi: 10.1093/jac/dkm035. [DOI] [PubMed] [Google Scholar]
- Kohanski MA, DePristo MA, Collins JJ. Sublethal antibiotic treatment leads to multidrug resistance via radical-induced mutagenesis. Mol Cell. 2010;37:311–320. doi: 10.1016/j.molcel.2010.01.003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Liu SQ. Practical implications of lactate and pyruvate metabolism by lactic acid bacteria in food and beverage fermentations. Int J Food Microbiol. 2003;83:115–131. doi: 10.1016/S0168-1605(02)00366-5. [DOI] [PubMed] [Google Scholar]
- Mahillon J, Chandler M. Insertion sequences. Microbiol Mol Biol Rev. 1998;62:725–774. doi: 10.1128/mmbr.62.3.725-774.1998. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Maietti L, Bonvini B, Huys G, Giraffa G. Incidence of antibiotic resistance and virulence determinants among Enterococcus italicus isolates from dairy products. Syst Appl Microbiol. 2007;30:509–517. doi: 10.1016/j.syapm.2007.05.002. [DOI] [PubMed] [Google Scholar]
- Manichanh C, Rigottier-Gois L, Bonnaud E, Gloux K, Pelletier E, Frangeul L, Nalin R, Jarrin C, Chardon P, Marteau P, Roca J, Dore J. Reduced diversity of faecal microbiota in Crohn’s disease revealed by a metagenomic approach. Gut. 2006;55:205–211. doi: 10.1136/gut.2005.073817. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Martins dos Santos V, Muller M, de Vos WM. Systems biology of the gut: the interplay of food, microbiota and host at the mucosal interface. Curr Opin Biotechnol. 2010;21:539–550. doi: 10.1016/j.copbio.2010.08.003. [DOI] [PubMed] [Google Scholar]
- Mathur S, Singh R. Antibiotic resistance in food lactic acid bacteria-a review. Int J Food Microbiol. 2005;105:281–295. doi: 10.1016/j.ijfoodmicro.2005.03.008. [DOI] [PubMed] [Google Scholar]
- Mayrhofer S, van Hoek AH, Mair C, Huys G, Aarts HJ, Kneifel W, Domig KJ. Antibiotic susceptibility of members of the Lactobacillus acidophilus group using broth microdilution and molecular identification of their resistance determinants. Int J Food Microbiol. 2010;144:81–87. doi: 10.1016/j.ijfoodmicro.2010.08.024. [DOI] [PubMed] [Google Scholar]
- Morea M, Baruzzi F, Cocconcelli PS. Molecular and physiological characterization of dominant bacterial populations in traditional mozzarella cheese processing. J Appl Microbiol. 1999;87:574–582. doi: 10.1046/j.1365-2672.1999.00855.x. [DOI] [PubMed] [Google Scholar]
- Mori T, Mizuta S, Suenaga H, Miyazaki K. Metagenomic screening for bleomycin resistance genes. Appl Environ Microbiol. 2008;74:6803–6805. doi: 10.1128/AEM.00873-08. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nawaz M, Wang J, Zhou A, Ma C, Wu X, Moore JE, Cherie Millar B, Xu J. Characterization and transfer of antibiotic resistance in lactic acid bacteria from fermented food products. Curr Microbiol. 2010;62:1081–1089. doi: 10.1007/s00284-010-9856-2. [DOI] [PubMed] [Google Scholar]
- Neyrinck AM, Delzenne NM. Potential interest of gut microbial changes induced by non-digestible carbohydrates of wheat in the management of obesity and related disorders. Curr Opin Clin Nutr Metab Care. 2010;13:722–728. doi: 10.1097/MCO.0b013e32833ec3fb. [DOI] [PubMed] [Google Scholar]
- Ogier JC, Serror P. Safety assessment of dairy microorganisms: the Enterococcus genus. Int J Food Microbiol. 2008;126:291–301. doi: 10.1016/j.ijfoodmicro.2007.08.017. [DOI] [PubMed] [Google Scholar]
- Ouoba LI, Lei V, Jensen LB. Resistance of potential probiotic lactic acid bacteria and bifidobacteria of African and European origin to antimicrobials: determination and transferability of the resistance genes to other bacteria. Int J Food Microbiol. 2008;121:217–224. doi: 10.1016/j.ijfoodmicro.2007.11.018. [DOI] [PubMed] [Google Scholar]
- Paulsen IT, Banerjei L, Myers GS, Nelson KE, Seshadri R, Read TD, Fouts DE, Eisen JA, Gill SR, Heidelberg JF, Tettelin H, Dodson RJ, Umayam L, Brinkac L, Beanan M, Daugherty S, DeBoy RT, Durkin S, Kolonay J, Madupu R, Nelson W, Vamathevan J, Tran B, Upton J, Hansen T, Shetty J, Khouri H, Utterback T, Radune D, Ketchum KA, Dougherty BA, Fraser CM. Role of mobile DNA in the evolution of vancomycin-resistant Enterococcus faecalis. Science. 2003;299:2071–2074. doi: 10.1126/science.1080613. [DOI] [PubMed] [Google Scholar]
- Pembroke JT, MacMahon C, McGrath B. The role of conjugative transposons in the Enterobacteriaceae. Cell Mol Life Sci. 2002;59:2055–2064. doi: 10.1007/s000180200005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Phillips I. Withdrawal of growth-promoting antibiotics in Europe and its effects in relation to human health. Int J Antimicrob Agents. 2007;30:101–107. doi: 10.1016/j.ijantimicag.2007.02.018. [DOI] [PubMed] [Google Scholar]
- Rizzotti L, La Gioia F, Dellaglio F, Torriani S. Molecular diversity and transferability of the tetracycline resistance gene tet(M), carried on Tn916–1545 family transposons, in enterococci from a total food chain. Antonie Van Leeuwenhoek. 2009;96:43–52. doi: 10.1007/s10482-009-9334-7. [DOI] [PubMed] [Google Scholar]
- Roberts MC. Update on acquired tetracycline resistance genes. FEMS Microbiol Lett. 2005;245:195–203. doi: 10.1016/j.femsle.2005.02.034. [DOI] [PubMed] [Google Scholar]
- Roberts MC. Update on macrolide-lincosamide-streptogramin, ketolide, and oxazolidinone resistance genes. FEMS Microbiol Lett. 2008;282:147–159. doi: 10.1111/j.1574-6968.2008.01145.x. [DOI] [PubMed] [Google Scholar]
- Sekirov I, Russell SL, Antunes LC, Finlay BB. Gut microbiota in health and disease. Physiol Rev. 2010;90:859–904. doi: 10.1152/physrev.00045.2009. [DOI] [PubMed] [Google Scholar]
- Settanni L, Moschetti G. Non-starter lactic acid bacteria used to improve cheese quality and provide health benefits. Food Microbiol. 2010;27:691–697. doi: 10.1016/j.fm.2010.05.023. [DOI] [PubMed] [Google Scholar]
- Seville LA, Patterson AJ, Scott KP, Mullany P, Quail MA, Parkhill J, Ready D, Wilson M, Spratt D, Roberts AP. Distribution of tetracycline and erythromycin resistance genes among human oral and fecal metagenomic DNA. Microb Drug Resist. 2009;15:159–166. doi: 10.1089/mdr.2009.0916. [DOI] [PubMed] [Google Scholar]
- Simon C, Daniel R. Achievements and new knowledge unraveled by metagenomic approaches. Appl Microbiol Biotechnol. 2009;85:265–276. doi: 10.1007/s00253-009-2233-z. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Slattery L, O’Callaghan J, Fitzgerald GF, Beresford T, Ross RP. Invited review: Lactobacillus helveticus-a thermophilic dairy starter related to gut bacteria. J Dairy Sci. 2010;93:4435–4454. doi: 10.3168/jds.2010-3327. [DOI] [PubMed] [Google Scholar]
- Sonnenburg JL. Microbiology: genetic pot luck. Nature. 2010;464:837–838. doi: 10.1038/464837a. [DOI] [PubMed] [Google Scholar]
- Streit WR, Schmitz RA. Metagenomics- the key to the uncultured microbes. Curr Opin Microbiol. 2004;7:492–498. doi: 10.1016/j.mib.2004.08.002. [DOI] [PubMed] [Google Scholar]
- Szczepanowski R, Linke B, Krahn I, Gartemann KH, Gutzkow T, Eichler W, Puhler A, Schluter A. Detection of 140 clinically relevant antibiotic-resistance genes in the plasmid metagenome of wastewater treatment plant bacteria showing reduced susceptibility to selected antibiotics. Microbiology. 2009;155:2306–2319. doi: 10.1099/mic.0.028233-0. [DOI] [PubMed] [Google Scholar]
- Teuber M. Veterinary use and antibiotic resistance. Curr Opin Microbiol. 2001;4:493–499. doi: 10.1016/S1369-5274(00)00241-1. [DOI] [PubMed] [Google Scholar]
- Teuber M, Perreten V. Role of milk and meat products as vehicles for antibiotic-resistant bacteria. Acta Vet Scand Suppl. 2000;93:75–87. [PubMed] [Google Scholar]
- Teuber M, Schwarz F, Perreten V. Molecular structure and evolution of the conjugative multiresistance plasmid pRE25 of Enterococcus faecalis isolated from a raw-fermented sausage. Int J Food Microbiol. 2003;88:325–329. doi: 10.1016/S0168-1605(03)00195-8. [DOI] [PubMed] [Google Scholar]
- Thaker M, Spanogiannopoulos P, Wright GD. The tetracycline resistome. Cell Mol Life Sci. 2010;67:419–431. doi: 10.1007/s00018-009-0172-6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Toomey N, Bolton D, Fanning S. Characterisation and transferability of antibiotic resistance genes from lactic acid bacteria isolated from Irish pork and beef abattoirs. Res Microbiol. 2010;161:127–135. doi: 10.1016/j.resmic.2009.12.010. [DOI] [PubMed] [Google Scholar]
- Tsai F, Coyle WJ. The microbiome and obesity: is obesity linked to our gut flora? Curr Gastroenterol Rep. 2009;11:307–313. doi: 10.1007/s11894-009-0045-z. [DOI] [PubMed] [Google Scholar]
- Tsukumo DM, Carvalho BM, Carvalho-Filho MA, Saad MJ. Translational research into gut microbiota: new horizons in obesity treatment. Arq Bras Endocrinol Metabol. 2009;53:139–144. doi: 10.1590/S0004-27302009000200004. [DOI] [PubMed] [Google Scholar]
- Tuohy KM, Gougoulias C, Shen Q, Walton G, Fava F, Ramnani P. Studying the human gut microbiota in the trans-omics era-focus on metagenomics and metabonomics. Curr Pharm Des. 2009;15:1415–1427. doi: 10.2174/138161209788168182. [DOI] [PubMed] [Google Scholar]
- Turnbaugh PJ, Gordon JI. The core gut microbiome, energy balance and obesity. J Physiol. 2009;587:4153–4158. doi: 10.1113/jphysiol.2009.174136. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Turpin W, Humblot C, Thomas M, Guyot JP. Lactobacilli as multifaceted probiotics with poorly disclosed molecular mechanisms. Int J Food Microbiol. 2010;143:87–102. doi: 10.1016/j.ijfoodmicro.2010.07.032. [DOI] [PubMed] [Google Scholar]
- Van Hoorde K, Verstraete T, Vandamme P, Huys G. Diversity of lactic acid bacteria in two Flemish artisan raw milk Gouda-type cheeses. Food Microbiol. 2008;25:929–935. doi: 10.1016/j.fm.2008.06.006. [DOI] [PubMed] [Google Scholar]
- Vaughan EE, de Vries MC, Zoetendal EG, Ben-Amor K, Akkermans AD, de Vos WM. The intestinal LABs. Antonie Van Leeuwenhoek. 2002;82:341–352. doi: 10.1023/A:1020672724450. [DOI] [PubMed] [Google Scholar]
- Ventura M, O’Flaherty S, Claesson MJ, Turroni F, Klaenhammer TR, van Sinderen D, O’Toole PW. Genome-scale analyses of health-promoting bacteria: probiogenomics. Nat Rev Microbiol. 2009;7:61–71. doi: 10.1038/nrmicro2047. [DOI] [PubMed] [Google Scholar]
- Wegener HC. Antibiotics in animal feed and their role in resistance development. Curr Opin Microbiol. 2003;6:439–445. doi: 10.1016/j.mib.2003.09.009. [DOI] [PubMed] [Google Scholar]
- Werner G, Coque TM, Hammerum AM, Hope R, Hryniewicz W, Johnson A, Klare I, Kristinsson KG, Leclercq R, Lester CH, Lillie M, Novais C, Olsson-Liljequist B, Peixe LV, Sadowy E, Simonsen GS, Top J, Vuopio-Varkila J, Willems RJ, Witte W, Woodford N. Emergence and spread of vancomycin resistance among enterococci in Europe. Euro Surveill. 2008;13:1–11. [PubMed] [Google Scholar]
- Witte W. Selective pressure by antibiotic use in livestock. Int J Antimicrob Agents. 2000;16(Suppl 1):S19–S24. doi: 10.1016/s0924-8579(00)00301-0. [DOI] [PubMed] [Google Scholar]
- Wright GD. Mechanisms of resistance to antibiotics. Curr Opin Chem Biol. 2003;7:563–569. doi: 10.1016/j.cbpa.2003.08.004. [DOI] [PubMed] [Google Scholar]
- Zago M, Huys G, Giraffa G. Molecular basis and transferability of tetracycline resistance in Enterococcus italicus LMG 22195 from fermented milk. Int J Food Microbiol. 2010;142:234–236. doi: 10.1016/j.ijfoodmicro.2010.05.022. [DOI] [PubMed] [Google Scholar]
- Zhang H, DiBaise JK, Zuccolo A, Kudrna D, Braidotti M, Yu Y, Parameswaran P, Crowell MD, Wing R, Rittmann BE, Krajmalnik-Brown R. Human gut microbiota in obesity and after gastric bypass. Proc Natl Acad Sci USA. 2009;106:2365–2370. doi: 10.1073/pnas.0812600106. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zonenschain D, Rebecchi A, Morelli L. Erythromycin- and tetracycline-resistant lactobacilli in Italian fermented dry sausages. J Appl Microbiol. 2009;107:1559–1568. doi: 10.1111/j.1365-2672.2009.04338.x. [DOI] [PubMed] [Google Scholar]