Abstract
Thy-1 is a cell surface glycoprotein present on normal lung fibroblasts but absent from the fibroblastic foci of idiopathic pulmonary fibrosis. Thy-1 correlates inversely with fibrogenic phenotypic characteristics and functions as a “fibrosis suppressor.” Promoter region hypermethylation can silence Thy-1 expression in fibroblastic foci, suggesting that epigenetic regulation is important in programming the fibrotic phenotype. We examined whether histone modifications are important in regulating Thy-1 expression in lung fibroblasts. Treatment with the histone deacetylase inhibitor trichostatin A (TSA) restored Thy-1 expression in Thy-1(−) cells in a time-dependent and concentration-dependent fashion and was associated with enrichment of histone acetylation. Chromatin immunoprecipitation demonstrated Thy-1 depletion of trimethylated H3K27 after 24 hours of TSA treatment, concurrent with enrichment of trimethylated H3K4 and acetylated H4. Bisulfite sequencing of the Thy-1 promoter region revealed demethylation of the previously hypermethylated CpG sites after treatment with TSA. Although Thy-1 was hypermethylated in Thy-1(−) lung fibroblasts, we observed that Thy-1(−) cells have lower global DNA methylation compared with Thy-1(+) lung fibroblasts, which was partially reversed by TSA treatment. TSA treatment up-regulates total methyltransferase activity in these cells. Our data indicate that Thy-1 silencing is regulated by histone modifications in addition to promoter hypermethylation in lung fibroblasts. Additionally, our findings indicate that alteration of histone modifications alters DNA methylation. Understanding the molecular hierarchy of events with respect to reactivation of transcription and reversal of histone modification will be critical to understand and modify the regulated expression of Thy-1, a tumor-supressor and fibrosis-suppressor gene.
Keywords: lung fibroblasts, Thy-1 expression, HDAC inhibitor, histone modification, DNA methylation
CLINICAL RELEVANCE.
This research demonstrates the reversibility of epigenetic suppression of Thy-1 in lung fibroblasts in vitro using a histone deacetylase inhibitor that is associated with a change in cell phenotype, suggesting that epigenetic regulation can be an important therapeutic target for lung fibrosis.
DNA methylation and histone modifications are critical modifications to chromatin that regulate gene transcription (1). There are at least eight different classes of histone modifications (2). DNA methylation has been shown to be related to histone acetylation and histone methylation (3). In heterochromatin, the DNA is usually methylated, whereas unmethylated DNA is enriched in association with hyperacetylated histones (4, 5). Increasing evidence shows that DNA methylation and histone modifications collaborate to regulate gene expression (1, 6).
Histone acetyltransferases and histone deacetylases (HDACs) loosen and compact the chromatin structures, respectively, to regulate cell functions (7, 8). Mammalian HDACs consist of four classes based on their sequence similarity, catalytic sites, and cofactor dependency. Class I HDACs are located in the nucleus. Class II HDACs can shuttle between the nucleus and cytoplasm and have cell type–specific patterns of expression. In response to activation of signal transduction pathways, the N-terminal extension in the HDACs links them to specific transcription factors, thus providing a connection between the genome and the extracellular environment (9, 10). HDAC inhibitors (HDACi) are important tools to study the effects of chromatin remodeling on cell functions, and some of them are being used in clinical trials as cancer drugs (11). HDACi are selective inhibitors. Trichostatin A (TSA), the most widely used HDACi, inhibits class I and class II HDACs (12).
Thy-1(CD90) is a glycophosphatidylinositol-linked outer membrane leaflet glycoprotein expressed in normal lung fibroblasts that modulates the profibrotic phenotype of fibroblasts by several mechanisms (13). In fibroblastic foci, the sites of active fibrosis in idiopathic pulmonary fibrosis (IPF), we have demonstrated epigenetic silencing of Thy-1 by promoter region methylation in fibroblasts (14, 15). Here we further our investigation of phenotypic regulation in pulmonary fibroblasts by examining the regulation of Thy-1 expression by histone modifications.
In this study, our findings indicate that TSA can restore Thy-1 expression in Thy-1(−) lung fibroblasts in a concentration- and time-dependent fashion. Thy-1 expression is related to specific histone modifications; changes to histone modifications can affect Thy-1 promoter region DNA methylation status as well as global DNA methylation and related DNA methyltransferase activities. Understanding the epigenetic regulation of Thy-1 may be important in developing new antifibrotic therapies. Treatments that modify the transcriptionally silent state of Thy-1 may reverse the profibrotic phenotype of fibroblasts in IPF and other fibrotic disorders.
MATERIALS AND METHODS
Further details are provided in the online supplement.
Cell Culture and Treatments
Primary rat lung fibroblasts were cultured and sorted as described previously (16). Different concentrations of TSA (Sigma, St. Louis, MO) were added in the culture medium as indicated in the figures after the cells had been seeded for 24 hours.
Cell Proliferation Assay
A proliferation assay kit (MBLI, Woburn, MA) was used to assess the effect of TSA treatment on rat lung fibroblast proliferation.
Total RNA, Genomic DNA Extraction and Modification, PCR, and Real-Time RT-PCR
DNA samples were modified by using the EZ DNA Methylation-Gold kit (Zymo Research, Orange, CA). PCR primers were as previously described (15). Real-time RT-PCR was performed as in previous studies and normalized to glyceraldehyde 3-phosphate dehydrogenase (GAPDH) (17). A linear regression approach was used to estimate input mRNA in comparison to GAPDH without the need for assumption of equal PCR efficiencies (18). More information about total RNA, DNA extraction, and cDNA synthesis is provided in the online supplement.
Methylation-Specific PCR and Bisulfite Genomic Sequencing
The methylation status of Thy-1 in the rat Thy-1(+) and Thy-1(–) lung fibroblasts and in TSA-treated Thy-1(−) rat lung fibroblasts were investigated by methylation-specific PCR and bisulfite genomic sequencing as previously described (15).
Flow Cytometry
Cells treated with TSA or control for 24 or 48 hours were analyzed by fluorescence-activated cell sorting using FITC-conjugated anti-CD90 or FITC-conjugated isotype control (16).
Chromatin Immunoprecipitation Assay
The chromatin immunoprecipitation (ChIP) assays were performed as per manufacturer protocol (Epigentek, Brooklyn, NY) with some modifications. ChIP-grade antibodies used were: H3 trimethyl K4 (ab1012; Abcam, Cambridge, MA), H3 trimethyl K27 (ab602), and acetylated H4 (cat. # 06-866; Upstate-Millipore, Billerica, MA). ChIP-DNA was amplified by PCR and real-time PCR with primers indicated in Figure 5. All results for immunoprecipitated DNA are normalized to input DNA.
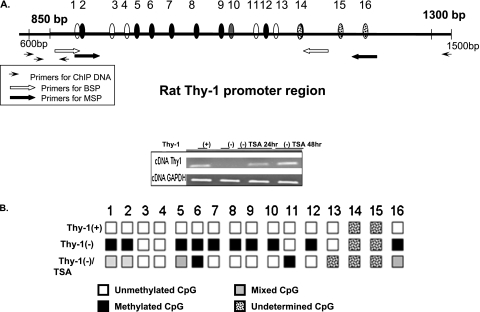
Figure 5.
Methylation status of Thy-1 promoter region in Thy-1(+) and Thy-1(−) rat lung fibroblasts with and without TSA treatment. (A) Examined CpG islands in the Thy-1 promoter region. Sequence numbers are as previously stated (from Ensembl Gene ID: ENSRNOG00000006604) (15). (B) Gel showing representative RT-PCR results for Thy-1 mRNA in cells treated with 500 nm TSA for 24 and 48 hours. Cells treated with glyceraldehyde 3-phosphate dehydrogenase were used as a control. Bisulfite genomic sequencing shows methylation status of cytosine residues in the Thy-1 promoter region of Thy-1(+) and Thy-1(−) cells and Thy-1(−) treated with 500 nm TSA for 48 hours. White boxes: unmethylated CpG; black boxes: methylated CpG; spotted boxes: undetermined sequence; gray box: mixed CpG.
Nuclear Protein Extraction and Immunoblotting
To identify histone modifications, nuclear proteins were extracted by using the EpiQuik Nuclear extraction kit (Epigentek). The following antibodies were used for the immunoblots: anti–acetyl histoneH3 antibody (cat. #06-599; Upstate, Lake Placid, NY) and anti-acetyl histone H4 (cat. #06-866; Upstate); anti-total Histone H3 (cat. #9715; Cell Signaling Technology, Danvers, MA); and anti-total histone H4 (ab10158; Abcam, Cambridge, MA).
Global DNA Methylation Quantification Assay
A Methylamp Global DNA Methylation Quantification Ultra Kit (Epigentek) was used to measure the global DNA methylation. A total of 100 ng of extracted DNA were used for the assay.
Total DNA Methyltransferase Activity Assay
Twenty micrograms of nuclear extracts were used to assay the de novo and maintenance DNA methyltransferase activity with an EpiQuik DNA Methyltransferase Activity Assay Kit (Epigentek).
Immunofluorescence
Immunofluorescence microscopy was performed as previously described (17). Cells on coverslips treated with or without TSA were incubated with FITC-conjugated rat anti-mouse Thy-1 (CD90.1) antibody (cat. # 55897; BD Pharmingen, San Diego, CA) or isotype control at 1:20. α-Smooth muscle actin was labeled with Cy-3–conjugated anti–α-smooth muscle actin (1:100) (C6198; Sigma).
Statistical Analysis
Comparisons involving three or more groups were analyzed using one-way ANOVA or Kruskal-Wallis ANOVA on ranks (Student-Newman-Keuls or Dunnett's methods for multiple comparisons). A P value of 0.05 was used to determine statistical significance.
RESULTS
TSA, a Histone Deacetylase Inhibitor, Causes Reexpression of Thy-1 in Rat Lung Fibroblasts in a Time-Dependent and Concentration-Dependent Fashion
Previously, we demonstrated that rat Thy-1(−) lung fibroblasts are more profibrotic than their Thy-1–expressing counterparts and that the absence of Thy-1 correlates with myofibroblastic differentiation (17). Normal lung fibroblasts express Thy-1, whereas the myofibroblasts in the fibroblastic foci in IPF lack Thy-1 expression (14). The promoter region of the Thy-1 gene is hypermethylated in fibroblasts lacking Thy-1 expression (15). In the current study, we further investigated whether histone modifications regulate the expression of Thy-1 in lung fibroblasts.
We treated Thy-1(−) lung fibroblasts with TSA at different concentrations and time periods. First, we examined the possible cytotoxicity of TSA by performing a cell proliferation assay at various concentrations of TSA over a 48-hour period. There was no obvious toxicity shown with the concentration we used as compared with the control samples (P > 0.05) (Figure 1A). Then, by using real-time RT-PCR, we observed significantly increased (P < 0.05) Thy-1 expression in Thy-1(−) cells after 24 hours; this expression further increased at 48 hours (Figure 1B; see also Figure 5B). As early as 12 hours, we could detect Thy-1 expression in Thy-1(−) cells after treatment with TSA at 100 nm (data not shown). The reexpression of Thy-1 in Thy-1(−) cells by TSA was also concentration dependent. The expression level increased as the TSA concentration increased from 100 nM to 1 μM (Figure 1C). Because the optimal TSA concentration and time of the treatment for reexpression of Thy-1 in Thy-1(−) cells as appear to be 500 nM and 48 hours, respectively, we have chosen these for subsequent experiments.
Figure 1.
The effect of trichostatin A (TSA) on Thy-1 expression by quantitative RT-PCR. (A) The effect of TSA on cell proliferation. The cells were incubated for 48 hours in the presence of TSA at the indicated concentrations. Proliferation assay was done in triplicate. No significant difference was observed (P > 0.05). (B and C) Histograms of real-time RT-PCR results for Thy-1 expression. (B) TSA-treated (500 nM) Thy-1(−) rat lung fibroblasts at 24 and 48 hours compared with untreated Thy-1(−) and Thy-1(+) rat lung fibroblasts. (C) TSA-treated (100 nM, 500 nM, and 1 μm treated Thy-1(−) rat lung fibroblasts at 48 hours, compared with untreated Thy-1(−) and hy-1(+) cells. Control wells showed the same results when harvested at 24 or 48 hours. In this study, unless otherwise stated, all controls were collected at 48 hours. All experiments were done in sorted (based on Thy-1 expression) rat lung fibroblast cell lines. Results are averages of at least three independent experiments. Data are expressed as the ratio of Thy-1 to glyceraldehyde 3-phosphate dehydrogenase by the standard curve method. *P < 0.001 versus Thy-1(−) cells untreated by ANOVA. Bars represent mean ± SD.
To confirm the reexpression of Thy-1 protein in TSA-treated Thy-1(−) cells, we performed flow cytometry to examine the cell surface expression of Thy-1 (Figure 2). The percentage of cells expressing Thy-1 increased significantly after treatment with TSA at 500 nM for 48 hours.
Figure 2.
Thy-1 expression changes by FACS. Thy-1(+) and Thy-1(–) rat lung fibroblasts cultured for 48 hours. Thy-1(−) cells were treated with 500 nm TSA for 24 hours and 48 hours. Thy-1 expression was analyzed by flow cytometry by staining the cells with anti–Thy-1 FITC. (A) Graph showing the representative changes of cell surface Thy-1 expression in Thy-1(−) cells after treatment with 500 nm TSA for 24 and 48 hours and in comparison to Thy-1(+) cells. (B) Histogram showing the mean FITC fluorescence of the positive population (average of n = 3 per condition and expressed relative to value for Thy-1(+) cells). Bars represent mean ± SD from the average of three independent experiments. *P < 0.01 versus Thy-1(−).
Thy-1 Reexpression in Thy-1(−) Cells after TSA Treatment Is Correlated with Changes in Histone Modifications
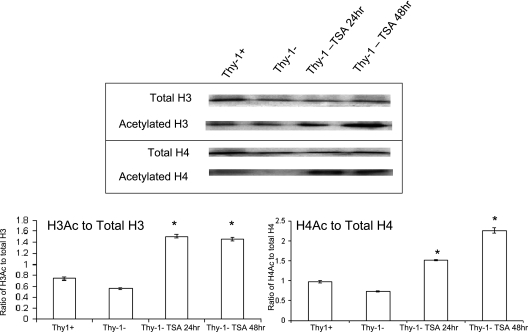
Cellular levels of acetyl histones.
Because treatment with TSA partially restored expression of Thy-1 in Thy-1(−) cells, we examined if the inhibition of histone deacetylase by TSA resulted in a global increase in histone acetylation. We determined total histone H3 and H4, as well as acetylated histone H3 and H4 levels, before and after treatment with TSA for 24 and 48 hours at 500 nM. Increases in acetylated H3 and H4 were observed in TSA-treated Thy-1(−) cells compared with the untreated cells. The untreated Thy-1(+) cells had a slightly higher level of acetylated H3 and H4 than the nontreated Thy-1(−) cells, but the difference was not as significant as with the TSA-treated Thy-1(−) cells compared with untreated Thy-1(−) cells, which indicates that other mechanisms may account for the majority of the baseline difference (Figure 3).
Figure 3.
Western blots of H3/Ac H3 and H4/Ac H4. Acetylated histone H3 and H4 are increased by treatment with TSA in Thy-1(−) cells. Nuclear extracts from cells that are untreated or treated with 500 nM TSA for 24 or 48 hours were immunoblotted with specific antibodies against total H3, H4, acetylated histone H3 (AcH3), and acetylated histone H4 (Ac H4). Representative blots are shown. Blots were probed first with antiacetylated forms and then stripped and reprobed with antibody against total H3 and H4. Histograms indicate ratio of AcH3 and AcH4 to total H3 or H4, respectively, based on scanning densitometry. Bars represent mean ± SD from the average of three independent experiments. *P < 0.001 versus untreated Thy-1(−) cells.
Changes in other histone modifications related with Thy-1 expression.
We examined additional specific histone modifications in relation to the expression of Thy-1. Because acetylation of H4 (H4Ac) and trimethylation of H3K4 (H3K4Me3) are known markers of active chromatin structure, whereas trimethylation of H3K27 (H3K27Me3) is one of the primary marks of inactive chromatin structure (1), we examined the association of the Thy-1 promoter region with these chromatin markers using ChIP assays. Figure 4 shows that acetylated H4 is associated with the Thy-1 promoter region in Thy-1(−) cells; Thy-1(−) cells have a lower amount of precipitated DNA compared with the Thy-1(+) cells and with Thy-1(−) cells treated with TSA. Similarly, the amount of H3K4Me3 associated with Thy-1 promoter region was also reduced in Thy-1(−) cells compared with the Thy-1+ and TSA-treated Thy-1(−) cells (Figure 4B). These results suggest that enrichment of markers associated with open chromatin structure is associated with transcriptionally active Thy-1. On the other hand, the closed chromatin structure marked by H3K27Me3 is associated with transcriptionally repressed Thy-1. The Thy-1 promoter in Thy-1(−) cells demonstrates enrichment by immunoprecipitation of this histone modification, whereas with the TSA treatment, the Thy-1 promoter DNA associated with this protein is reduced (Figure 4C).
Figure 4.
Histone modifications in the Thy-1 promoter region by chromatin immunoprecipitation (ChIP) assays. Histone modifications associated with the promoter region of the rat Thy-1 gene. The quantitative ChIP assay was performed to analyze the histone modifications H4Ac (A), H3K4Me3 (B), and H3K27Me3 (C) associated with the promoter region of Thy-1 gene. Bars represent the relative levels of PCR product of the region of Thy-1 (as indicated in Figure 5A) associated with a specific histone modification state under the indicated conditions, after chromatin immunoprecipitation with specific antibodies, normalized to total input DNA. IgG served as a negative control for immunoprecipitation. Shown at the bottom is a representative PCR gel picture of the ChIP-DNA and input DNA. Bars represent mean ± SD from the average of three independent experiments. *P < 0.001 versus untreated Thy-1(−) cells.
Changes in Histone Modifications by Histone Deacetylase Inhibition Affect Thy-1 DNA Methylation Status
TSA treatment demethylates some hypermethylated CpG sites in the Thy-1 promoter region.
The combined repressive effects of histone modifiers and DNA methyltransferases on gene silencing are largely unknown. However, it has been reported that the HDAC inhibitor TSA exhibits DNA-demethylating activities in mammalian cells (19, 20). In this study, we examined TSA for its role in DNA methylation in Thy-1(−) fibroblasts. Thy-1(−) “profibrotic” lung myofibroblasts are hypermethylated at the Thy-1 promoter region (15). After treatment with TSA for 48 hours at 500 nM, we examined the DNA methylation status in the Thy-1 gene promoter region. Bisulfite sequencing was performed to examine the changes of methylation status in specific CpG sites. The diagram in Figure 5 shows methylation changes in CpG islands in the Thy-1 promoter region. The bisulfite sequencing revealed that some of the previously methylated CpG sites are demethylated. Some CpG sites are completely demethylated, but some are a mixture of demethylated and methylated copies. Some of the CpG sites in the examined region remained hypermethylated when compared with the untreated Thy-1(−) cells, which may explain why the TSA-treated Thy-1(−) cells did not reach the full expression levels of Thy-1 mRNA as in the Thy-1(+) cells (see Figure 1).
TSA treatment partially reversed the global DNA methylation status as well as DNA methyltransferase activity in Thy-1(−) lung fibroblasts.
Hypermethylation of CpG islands in specific genes and global DNA hypomethylation are critical epigenetic alterations in many cancers (21, 22). Reduced levels of DNA methylation are thought to be associated with genomic instability (23). These alterations have not been studied extensively in cells associated with fibrotic responses. Global DNA methylation quantification (Figure 6A) showed that Thy-1(+) lung fibroblasts have a higher percentage of methylated DNA compared with Thy-1(−) cells (P < 0.05). After treating Thy-1(−) cells with TSA, the percentage of total DNA methylation was higher compared with the nontreated group. These results suggest that histone acetylation differentially affects global DNA methylation compared with specific gene hypermethylation in fibroblasts. Next, we examined the total DNA methyltransferase (DNMT) activity (including de novo and maintenance methyltransferases). Total DNMT activity was lower in Thy-1(−) cells than in Thy-1(+) cells. After TSA treatment, DNMT activity increased but not to the same levels seen in Thy-1(+) cells (Figure 6B) (P < 0.05). This is similar to what is observed with the total DNA methylation change (Figure 6A).
Figure 6.
The effects of TSA on global DNA methylation and total DNA methyltransferease (DNMT) activity. (A) Global DNA methylation in untreated Thy-1(+) and Thy-1(−) cells and TSA-treated Thy-1(−) cells. Methylcytosine levels were measured as percentage of total cytosine, which was determined using the Methylamp Global DNA Methylation Quantification Kit (Epigentek). Relative to Thy-1(+) cells, mean global DNA methylcytosine levels in Thy-1(−) cells were significantly reduced (*P < 0.05 versus Thy-1(+)). A significant increase in methylation was observed in the 48-hour TSA-treated Thy-1(−) cells versus untreated Thy-1(−) cells (**P < 0.05). Values are expressed as mean ± SD of at least three independent experiments. (B) DNMT activity (total) was measured in untreated Thy-1(+) and Thy-1(−) cells and in TSA-treated Thy-1(−) cells as described in Materials and Methods. Relative to Thy-1(+) cells, mean DNMT activity in Thy-1(−) cells was significantly reduced (*P < 0.001 versus Thy-1(+)). A significant increase in activity was observed in 48-hour TSA-treated Thy-1(−) cells versus untreated Thy-1(−) cells, **P < 0.05. Values are expressed as mean ± SD of at least three independent experiments.
Cell Phenotype Alteration after Treatment with TSA
With the reexpression of Thy-1 in Thy-1(−) lung fibroblasts by TSA, we anticipated that the Thy-1(−) cell phenotype would be changed to one similar to that of Thy-1(+) cells. Thus, after we treated the Thy-1(−) cells with TSA for 24 and 48 hours, we performed immunohistochemistry to observe whether reexpression of Thy-1 is associated with decreased expression of α-smooth muscle actin (SMA), a key marker of the myofibroblast phenotype. After 24 hours of treatment, we began to detect Thy-1 in the previously negative Thy-1 cells, as we had observed in measuring cell surface expression of Thy-1 by FACS (see Figure 2). The staining of α-SMA stress fibers was strong in Thy-1(−) cells; however, after treatment with TSA for 48 hours, α-SMA was reduced in cells expressing Thy-1 (Figure 7). We confirmed this finding by Western blots (Figures 7D and 7E).
Figure 7.
Restored expression of Thy-1 in rat lung fibroblasts correlates with decreased expression of α-smooth muscle actin (SMA). Rat Thy-1(−) lung fibroblasts were treated with TSA for 48 hours at 500 nM. Cells were grown on coverslips, fixed, permeabilized, and stained with FITC-conjugated Thy-1 antibody and Cy3-conjugated α-SMA antibody. Nuclei were counterstained with Hoechst reagent. (A) Thy-1(+) cells have thinner α-SMA fibers (stained in red; yellow arrow). Thy-1 is stained in green (white arrow). (B) Thy-1(−) cells have thicker α-SMA fibers (stained in red; yellow arrow). (C) Thy-1(−) cells treated with TSA at 500 nM for 48 hours. Some Thy-1(−) cells showed restored Thy-1 expression (stained in green; white arrow) and less striking α-SMA fibers (stained in red; yellow arrow). (D) Representative Western blot (anti–α-SMA or anti–β-actin) of Thy-1(−) cells without and with TSA for 250 or 500 nM for 48 hours. (E) Bars represent mean ± SD from average of three independent experiments of Western blots from (D) (α-SMA expression ration to β-actin). *P < 0.001 versus untreated Thy-1(−) cells.
DISCUSSION
Previous studies from our laboratory (15) and others (24) have shown that Thy-1 silencing is correlated with CpG hypermethylation in its promoter region. In this study, we examined the hypothesis that Thy-1 gene silencing is mediated by histone modifications as well. Chromatin histone modifications and DNA methylation are important components in the epigenome that regulate gene expression and biological processes in somatic cells (22, 25). Increasing data suggest that DNA methylation and histone deacetylation are coordinately regulated to modify gene expression (26). The hyperacetylation effect of HDACi would be predicted to result in a general increase in gene expression as the treatment opens up the chromatin structure. However, studies have shown that treatment with HDAC inhibitors, like TSA, only significantly altered 2 to 5% of the genes (27). TSA can affect the methylation status of some genes, but not the others (20).
In this report, we showed that the histone deacetylation inhibitor TSA can induce histone acetylation, restores the expression of the fibrosis suppressor gene Thy-1 associated with both changes in chromatin marks and demethylation of the Thy-1 promoter region, and may increase genomic stability by increasing global DNA methylation. These data suggest that histone modifications and DNA methylation are coordinately regulated to change the biological behavior of fibrotic lung fibroblasts. In epigenetic regulation, histone modifications and DNA methylation can work together to silence or activate a gene. Histone deacetylase inhibitors, as well as DNA methyltransferase inhibitors, can be used together to achieve maximal effects in restoring expression of some genes. In our model, Thy-1 reexpression in Thy-1(−) cells after treatment with TSA never reached the same levels as in Thy-1(+) cells even with the addition of the demethylation agent 5-aza-2′deoxycytidine (AZA) (see Figure E1 in the online supplement). We previously showed (15) that AZA can partially restore Thy-1 expression in fibrotic lung fibroblasts. The addition of TSA in the culture together with AZA appears to be additive but does not restore Thy-1 expression to levels seen in Thy-1(+) cells (Figure E1). There are likely to be other mechanisms, transcriptional or posttranscriptional, that suppress Thy-1 expression in Thy-1(−) fibroblasts. For example, our earlier studies (14) indicated that shedding can be an important mechanism for posttranscriptional regulation of Thy-1 expression.
There is abundant evidence showing that histone acetylation and DNA methylation interact (28, 29). Some studies have shown that TSA had no effect on DNA methylation (30, 31). Other studies have shown that TSA induces global and gene-specific DNA demethylation in human cancer cells (32). In the latter study, the authors noted that the demethylation effect of TSA is gene selective. In another report, TSA and the DNMT inhibitor 5-azaCdR restored the expression of lysozyme, a secretory protein mainly expressed in glandular epithelial cells, but TSA did not change its DNA methylation status (33). The authors conclude that DNA methylation and histone acetylation are simultaneously and independently operative in their model. TSA has been reported to induce promoter region demethylation in Neurspora crassa (20) and in mammalian cells (19). Although the exact mechanisms are unclear, in addition to TSA, other HDAC inhibitors have been reported to have demethylation activity (34, 35). In human endometrial cells, TSA and butyrate can suppress DNMT3B expression by decreasing the stability of DNMT3B mRNA (35). Another HDACi that has much stronger activity than that of TSA, depsipeptide, has been found to decrease the binding of DNMT1 to the promoter of genes that were silent but not to affect the expression of DNMT1 (34). Although we did not measure the activity and expression of each of the specific DNA methyltransferases, our data indicated that the overall activity of total DNMTs in treated Thy-1(−) cells was changed.
In human cancer genomes, a hallmark alteration is global hypomethylation, resulting in genomic instability, and tumor suppressor gene hypermethylation, resulting in gene silencing (36). IPF has been proposed as a neoproliferative disorder of the lung, based on the fundamental pathological characteristics of IPF and cancer (37). Thy-1(−) fibroblasts are more profibrotic (17) and thus may have similar alterations to some cancer cells. We suspect that the higher overall DNA methylation levels observed in Thy-1(+) cells correspond with a more physiologically stable phenotype and are consistent with the observed higher overall DNMT activity (Figure 6B). TSA appears to partially reverse the Thy-1(−) cells' total DNA methylation levels as well as the total DNMT activity (Figure 6). The exact mechanism remains unclear; DNA methylation is a complex process and involves multiple molecular interactions. Taken together, this study and our previous study (15) showed that the expression of Thy-1 can be restored through DNA demethylation of the promoter region by the DNMT inhibitor 5-aza-2′deoxycytidine or by the histone deacetylation inhibitor TSA.
Because TSA is a histone deacetylase inhibitor and not a demethylation agent, the demethylation effect shown in our model may be the indirect effect of chromatin remodeling, which may alter the susceptibility of DNA to methylation. We further investigated which chromatin modifications are associated with Thy-1 before and after treatment with TSA. Figure 3 clearly shows that acetylated H3 and H4 are significantly higher in the TSA-treated cells than in the untreated control cells. By ChIP assay, we observed that acetylated histone H4 (H4Ac) and trimethylated histone H3 Lys-4 (H3K4Me3) directly correlated and that trimethylated histone H3 Lys-27 (H3K27Me3) inversely correlated with Thy-1 expression and changed after treatment with TSA. We do not know the exact mechanisms of the changes associated with histone modifications after treatment with TSA. The mechanism of decreased Thy-1 association with H3K27Me3 and enriched association with H3K4 me3 in TSA-treated cells is not clear because the TSA can induce histone acetylation but not methylation. Reports have shown that modification at one site of the histone tail can influence modification of a second site (38, 39); thus, it is possible that acetylated H3 and H4 may affect the other forms of modification of histone H3 and H4. It is also possible that TSA can directly affect histone-related enzymes. A study in human lung cancer cells using depsipeptide indicates that depsipeptide can suppress the expression of methyltransferases of H3K9 (34). We do not know if the dissociation H3K27 from Thy-1 in TSA-treated Thy-1(−) cells is a direct or indirect effect of TSA on H3K27 methyltransferases; this deserves further exploration. In another report (3) focused on the DNA repair protein MGMT, its methylation-mediated silencing seems to be associated with decreased acetylation of H3 and H4. Their study indicates a multiprotein complex containing methylation binding proteins and histone methyltransferases interacting with the gene promoter region resulting in a closed chromatin structure, thus inactivating the gene. This may be similar to what we have observed regarding Thy-1 gene expression in lung fibroblasts. As with increased acetylation of H3 and H4 by TSA treatment (Figure 3), Thy-1 expression was restored and was associated with a relaxed chromatin structure (Figure 4).
Although it is known that histone deacetylation and cytosine methylation communicate with each other and cause transcriptional silencing, it is not certain which is the initiating event. DNA methylation may be the main marker for gene silencing that triggers the events resulting in a repressive chromatin state, or the loss of histone acetylation may be the initial event followed by changes in DNA methyltransferase/demethylase activities to induce local gene hypermethylation and cause the silencing of the gene (25, 40–43). A study using the histone deacetylase inhibitor valproic acid (VPA) (41) examined DNA methylation changes in some specific genes and noted that, after treatment with VPA, certain CpG sites of some genes showed increased methylation, whereas other CpG sites were demethylated. Together with our own data demonstrating methylation changes in a specific gene and global DNA methylation, this may suggest that HDACi are able to reprogram the gene epigenetic makeup through histone acetylation. It would be very useful to identify the specific HDAC involved in Thy-1 expression. However, TSA is a broad HDACi that inhibits class I and II HDACs. The possible specific HDAC(s) that are involved in the regulation of Thy-1 expression can be explored in the future by methods such as ChIP when the specific antibodies become available.
Finally, this study demonstrates that epigenetic modification using TSA results in reprogramming of at least one element of the profibrotic phenotype, the expression of α-SMA (Figure 7). This result may be mediated via direct effects on the α-SMA promoter, as previous studies showed that TSA can affect α-SMA expression (44, 45), or this could be the result of effects on transcription of multiple profibrotic genes, though we have previously demonstrated that restoration of Thy-1 expression alone is sufficient to reverse the myofibroblastic phenotype of lung fibroblasts (17).
Thy-1 has been proposed as a “fibrosis suppressor” gene (14). This report showing that a HDACi can restore Thy-1 expression in fibrotic fibroblasts and change the phenotype of the cells suggests the possibility that drugs in this category could be used in fibrotic lung diseases such as IPF. Because other drugs in this family are in use for treatment of other diseases, this approach could offer a very exciting alternative for the treatment of this deadly disease.
Supplementary Material
Acknowledgments
The authors thank Drs. Namasivayam Ambalavanan and J.P. Clancy for helpful discussion and data interpretation and Wei Zhang for technical assistance.
This work was supported in part by National Heart, Lung and Blood Institute grant R01 HL082818 (J.S.H.), by American Heart Association grant 09SDG2260095 (Y.Y.S), and by National Center for Research Resources Research Facilities Improvement Program Grant C06RR 15,490.
This article has an online supplement, which is accessible from this issue's table of contents at www.atsjournals.org
Originally Published in Press as DOI: 10.1165/rcmb.2010-0154OC on August 19, 2010
Author Disclosure: J.S.H. received expert witness fees from Robinson, Smith and Wells for up to $1,000 and consultancy fees from University of Alabama-Birmingham for $10,001 to $50,000. J.S.H. received sponsored grants from DHHS and NIH for more than $100,001 each from and Kaul Pediatric Research Institute for $10,001 to $50,000. Y.S. received sponsored grants from American Heart Association and NIH for more than $100,001 each. T.T. received a sponsored grant from NIH for more than $100,001. B.V. received a sponsored grant from the American Thoracic Society for $10,001 to $50,000 for the ATS Fellows Career Development Award.
References
- 1.Berger SL. The complex language of chromatin regulation during transcription. Nature 2007;447:407–412. [DOI] [PubMed] [Google Scholar]
- 2.Kouzarides T. Chromatin modifications and their function. Cell 2007;128:693–705. [DOI] [PubMed] [Google Scholar]
- 3.Danam RP, Howell SR, Brent TP, Harris LC. Epigenetic regulation of O6-methylguanine-DNA methyltransferase gene expression by histone acetylation and methyl-CpG binding proteins. Mol Cancer Ther 2005;4:61–69. [PubMed] [Google Scholar]
- 4.Shi X, Hong T, Walter KL, Ewalt M, Michishita E, Hung T, Carney D, Pena P, Lan F, Kaadige MR, et al. ING2 PHD domain links histone H3 lysine 4 methylation to active gene repression. Nature 2006;442:96–99. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Shogren-Knaak M, Ishii H, Sun JM, Pazin MJ, Davie JR, Peterson CL, et al. Histone H4–K16 acetylation controls chromatin structure and protein interactions. Science 2006;311:844–847. [DOI] [PubMed] [Google Scholar]
- 6.Fuks F. DNA methylation and histone modifications: teaming up to silence genes. Curr Opin Genet Dev 2005;15:490–495. [DOI] [PubMed] [Google Scholar]
- 7.Chang S, Young BD, Li S, Qi X, Richardson JA, Olson EN. Histone deacetylase 7 maintains vascular integrity by repressing matrix metalloproteinase 10. Cell 2006;126:321–334. [DOI] [PubMed] [Google Scholar]
- 8.McKinsey TA, Zhang CL, Olson EN. Control of muscle development by dueling HATs and HDACs. Curr Opin Genet Dev 2001;11:497–504. [DOI] [PubMed] [Google Scholar]
- 9.de Ruijter AJ, van Gennip AH, Caron HN, Kemp S, van Kuilenburg AB. Histone deacetylases (HDACs): characterization of the classical HDAC family. Biochem J 2003;370:737–749. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Martin M, Kettmann R, Dequiedt F. Class IIa histone deacetylases: regulating the regulators. Oncogene 2007;26:5450–5467. [DOI] [PubMed] [Google Scholar]
- 11.Minucci S, Pelicci PG. Histone deacetylase inhibitors and the promise of epigenetic (and more) treatments for cancer. Nat Rev Cancer 2006;6:38–51. [DOI] [PubMed] [Google Scholar]
- 12.Yoshida M, Kijima M, Akita M, Beppu T. Potent and specific inhibition of mammalian histone deacetylase both in vivo and in vitro by trichostatin A. J Biol Chem 1990;265:17174–17179. [PubMed] [Google Scholar]
- 13.Rege TA, Hagood JS. Thy-1, a versatile modulator of signaling affecting cellular adhesion, proliferation, survival, and cytokine/growth factor responses. Biochim Biophys Acta 2006;1763:991–999. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Hagood JS, Prabhakaran P, Kumbla P, Salazar L, MacEwen MW, Barker TH, Ortiz LA, Schoeb T, Siegal GP, Alexander CB, et al. Loss of fibroblast Thy-1 expression correlates with lung fibrogenesis. Am J Pathol 2005;167:365–379. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Sanders YY, Pardo A, Selman M, Nuovo GJ, Tollefsbol TO, Siegal GP, Hagood JS. Thy-1 promoter hypermethylation: a novel epigenetic pathogenic mechanism in pulmonary fibrosis. Am J Respir Cell Mol Biol 2008;39:610–618. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Hagood JS, Pardo A, Selman M, Nuovo GJ, Tollefsbol TO, Siegal GP, Hagood JS. Differential expression of platelet-derived growth factor-alpha receptor by Thy-1(-) and Thy-1(+) lung fibroblasts. Am J Physiol 1999;277:L218–L224. [DOI] [PubMed] [Google Scholar]
- 17.Sanders YY, Kumbla P, Hagood JS. Enhanced myofibroblastic differentiation and survival in Thy-1(-) lung fibroblasts. Am J Respir Cell Mol Biol 2007;36:226–235. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Ramakers C, Ruijter JM, Deprez RH, Moorman AF. Assumption-free analysis of quantitative real-time polymerase chain reaction (PCR) data. Neurosci Lett 2003;339:62–66. [DOI] [PubMed] [Google Scholar]
- 19.Hu JF, Pham J, Dey I, Li T, Vu TH, Hoffman AR. Allele-specific histone acetylation accompanies genomic imprinting of the insulin-like growth factor II receptor gene. Endocrinology 2000;141:4428–4435. [DOI] [PubMed] [Google Scholar]
- 20.Selker EU. Trichostatin A causes selective loss of DNA methylation in Neurospora. Proc Natl Acad Sci USA 1998;95:9430–9435. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Guerrero-Preston R, Santella RM, Blanco A, Desai M, Berdasco M, Fraga M. Global DNA hypomethylation in liver cancer cases and controls: a phase I preclinical biomarker development study. Epigenetics 2007;2:223–226. [DOI] [PubMed] [Google Scholar]
- 22.Piyathilake CJ, Frost AR, Bell WC, Oelschlager D, Weiss H, Johanning GL, Niveleau A, Heimburger DC, Grizzle WE. Altered global methylation of DNA: an epigenetic difference in susceptibility for lung cancer is associated with its progression. Hum Pathol 2001;32:856–862. [DOI] [PubMed] [Google Scholar]
- 23.Eden A, Gaudet F, Waghmare A, Jaenisch R. Chromosomal instability and tumors promoted by DNA hypomethylation. Science 2003;300:455. [DOI] [PubMed] [Google Scholar]
- 24.Lung HL, Bangarusamy DK, Xie D, Cheung AK, Cheng Y, Kumaran MK, Miller L, Liu ET, Guan XY, Sham JS, et al. THY1 is a candidate tumour suppressor gene with decreased expression in metastatic nasopharyngeal carcinoma. Oncogene 2005;24:6525–6532. [DOI] [PubMed] [Google Scholar]
- 25.Ben-Porath I, Cedar H. Epigenetic crosstalk. Mol Cell 2001;8:933–935. [DOI] [PubMed] [Google Scholar]
- 26.Gius D, Cui H, Bradbury CM, Cook J, Smart DK, Zhao S, Young L, Brandenburg SA, Hu Y, Bisht KS, et al. Distinct effects on gene expression of chemical and genetic manipulation of the cancer epigenome revealed by a multimodality approach. Cancer Cell 2004;6:361–371. [DOI] [PubMed] [Google Scholar]
- 27.Van Lint C, Emiliani S, Verdin E. The expression of a small fraction of cellular genes is changed in response to histone hyperacetylation. Gene Expr 1996;5:245–253. [PMC free article] [PubMed] [Google Scholar]
- 28.Vaissiere T, Sawan C, Herceg Z. Epigenetic interplay between histone modifications and DNA methylation in gene silencing. Mutat Res 2008;659:40–48. [DOI] [PubMed] [Google Scholar]
- 29.Mutskov VJ, Farrell CM, Wade PA, Wolffe AP, Felsenfeld G. The barrier function of an insulator couples high histone acetylation levels with specific protection of promoter DNA from methylation. Genes Dev 2002;16:1540–1554. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Enright BP, Kubota C, Yang X, Tian XC. Epigenetic characteristics and development of embryos cloned from donor cells treated by trichostatin A or 5-aza-2′-deoxycytidine. Biol Reprod 2003;69:896–901. [DOI] [PubMed] [Google Scholar]
- 31.Liu S, Liu Z, Xie Z, Pang J, Yu J, Lehmann E, Huynh L, Vukosavljevic T, Takeki M, Klisovic RB, et al. Bortezomib induces DNA hypomethylation and silenced gene transcription by interfering with Sp1/NF-kappaB-dependent DNA methyltransferase activity in acute myeloid leukemia. Blood 2008;111:2364–2373. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Ou JN, Torrisani J, Unterberger A, Provencal N, Shikimi K, Karimi M, Ekstrom TJ, Szyf M. Histone deacetylase inhibitor Trichostatin A induces global and gene-specific DNA demethylation in human cancer cell lines. Biochem Pharmacol 2007;73:1297–1307. [DOI] [PubMed] [Google Scholar]
- 33.Claus R, Fliegauf M, Stock M, Duque JA, Kolanczyk M, Lubbert M. Inhibitors of DNA methylation and histone deacetylation independently relieve AML1/ETO-mediated lysozyme repression. J Leukoc Biol 2006;80:1462–1472. [DOI] [PubMed] [Google Scholar]
- 34.Wu LP, Wang X, Li L, Zhao Y, Lu S, Yu Y, Zhou W, Liu X, Yang J, Zheng Z, et al. Histone deacetylase inhibitor depsipeptide activates silenced genes through decreasing both CpG and H3K9 methylation on the promoter. Mol Cell Biol 2008;28:3219–3235. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Xiong Y, Dowdy SC, Podratz KC, Jin F, Attewell JR, Eberhardt NL, Jiang SW. Histone deacetylase inhibitors decrease DNA methyltransferase-3B messenger RNA stability and down-regulate de novo DNA methyltransferase activity in human endometrial cells. Cancer Res 2005;65:2684–2689. [DOI] [PubMed] [Google Scholar]
- 36.Cheung HH, Lee TL, Rennert OM, Chan WY. DNA methylation of cancer genome. Birth Defects Res C Embryo Today 2009;87:335–350. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Vancheri C, Failla M, Crimi N, Raghu G. Idiopathic pulmonary fibrosis: a disease with similarities and links to cancer biology. Eur Respir J 2010;35:496–504. [DOI] [PubMed] [Google Scholar]
- 38.Edmondson DG, Davie JK, Zhou J, Mirnikjoo B, Tatchell K, Dent SY. Site-specific loss of acetylation upon phosphorylation of histone H3. J Biol Chem 2002;277:29496–29502. [DOI] [PubMed] [Google Scholar]
- 39.Cheung P, Tanner KG, Cheung WL, Sassone-Corsi P, Denu JM, Allis CD. Synergistic coupling of histone H3 phosphorylation and acetylation in response to epidermal growth factor stimulation. Mol Cell 2000;5:905–915. [DOI] [PubMed] [Google Scholar]
- 40.Mutskov V, Felsenfeld G. Silencing of transgene transcription precedes methylation of promoter DNA and histone H3 lysine 9. EMBO J 2004;23:138–149. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Milutinovic S, D'Alessio AC, Detich N, Szyf M. Valproate induces widespread epigenetic reprogramming which involves demethylation of specific genes. Carcinogenesis 2007;28:560–571. [DOI] [PubMed] [Google Scholar]
- 42.Keshet I, Lieman-Hurwitz J, Cedar H. DNA methylation affects the formation of active chromatin. Cell 1986;44:535–543. [DOI] [PubMed] [Google Scholar]
- 43.Dong E, Guidotti A, Grayson DR, Costa E. Histone hyperacetylation induces demethylation of reelin and 67-kDa glutamic acid decarboxylase promoters. Proc Natl Acad Sci USA 2007;104:4676–4681. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Glenisson W, Castronovo V, Waltregny D. Histone deacetylase 4 is required for TGFbeta1-induced myofibroblastic differentiation. Biochim Biophys Acta 2007;1773:1572–1582. [DOI] [PubMed] [Google Scholar]
- 45.Guo W, Shan B, Klingsberg RC, Qin X, Lasky JA. Abrogation of TGF-beta1-induced fibroblast-myofibroblast differentiation by histone deacetylase inhibition. Am J Physiol Lung Cell Mol Physiol 2009;297:L864–L870. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.