Abstract
In captive penguins, avian malaria due to Plasmodium parasites is a well-recognized disease problem as these protozoa may cause severe losses among valuable collections of zoo birds. In blood films from naturally infected birds, identification and differentiation of malaria parasites based on morphological criteria are difficult because parasitaemia is frequently light and blood stages, which are necessary for identification of parasites, are often absent. Post-mortem diagnosis by histological examination of tissue samples is sometimes inconclusive due to the difficulties in differentiating protozoal tissue stages from fragmented nuclei in necrotic tissue. The diagnosis of avian malaria would be facilitated by a technique with the ability to specifically identify developmental stages of Plasmodium in tissue samples. Thus, a chromogenic in-situ hybridization (ISH) procedure with a digoxigenin-labelled probe, targeting a fragment of the 18S rRNA, was developed for the detection of Plasmodium parasites in paraffin wax-embedded tissues. This method was validated in comparison with traditional techniques (histology, polymerase chain reaction), on various tissues from 48 captive penguins that died at the zoological garden Schönbrunn, Vienna, Austria. Meronts of Plasmodium gave clear signals and were easily identified using ISH. Potential cross-reactivity of the probe was ruled out by the negative outcome of the ISH against a number of protozoa and fungi. Thus, ISH proved to be a powerful, specific and sensitive tool for unambiguous detection of Plasmodium parasites in paraffin wax-embedded tissue samples.
Introduction
Avian haemosporidian blood parasites (Sporozoa, Haemosporida) that comprise the genera of Haemoproteus, Plasmodium and Leucocytozoon are responsible for severe disease in domestic, zoo and wild birds and are transmitted by blood-sucking dipteran insect vectors (Valkiūnas, 2005).
These protozoal parasites are cosmopolitan in their distribution over all of the warm continents, show a close genetic relationship (Hellgren et al., 2004) and have been the subject of extensive research for over 100 years.
Parameters defining these genera and also species include the morphology of developmental stages as determined by light microscopy of blood films, characteristic features of life-cycles and host range (Valkiūnas, 2005). The diagnosis of avian malaria has been based traditionally on cytological examination of blood smears, occasionally supported by serology (Atkinson et al. 2001) and histopathology. In blood films from naturally infected birds, identification of malaria parasites based on morphological criteria is difficult because the parasitaemia is frequently light and blood stages, which are necessary for the identification of the parasites, are often absent (Valkiūnas, 2005). Additionally, a blood smear from one bird may contain several species of parasites belonging to one or more genera of haemosporidians, which complicates identification, even for experts (Valkiūnas et al., 2008).
However, none of these methods allows reliable species, and in some cases even genus, discrimination of haemosporidian parasites. Several problems associated with the traditional diagnostic approaches can be solved with molecular methods, which are much more sensitive (Perkins et al., 1998; Richard et al., 2002). Polymerase chain reaction (PCR) techniques—frequently complemented by sequencing—have greatly increased the detection and identification of low-intensity infections, although in cases of simultaneous infections with different haemosporidian parasites, which are common in wildlife, this approach often does not identify all of the involved species (Valkiūnas et al., 2006; Palinauskas et al., 2007).
Infection of wild passerines with Plasmodium and Haemoproteus is prevalent worldwide (Valkiūnas & Iezhova, 2001) with the exception of some arctic and forest tundra regions due to low vector density, and it has been estimated that 68% of all bird species are susceptible to haemosporidians (Atkinson et al., 2000). Large numbers of birds originating from circumpolar regions (families Spheniscidae and Alcidae) kept in zoological collections of temperate parts of the world succumb each year to avian malaria (Fleischmann et al. 1968; Fix et al. 1988; Loupal & Kutzer, 1995; Sturrock et al. 2007). Penguins and other susceptible birds are not adapted evolutionarily and physiologically to the local species of haemosporidians and become infected when they are placed in the nidi of local malaria. The sources of infection are wild passerine birds living free in the zoo, and the parasites are transmitted by insect vectors. Reports of malaria in penguins have implicated three parasite species: Plasmodium relictum (Fix et al. 1988; Cranfield et al. 1990), Plasmodium elongatum (Fleischmann et al. 1968; Cranfield et al. 1990; Graczyk et al. 1994) and Plasmodium juxtanucleare (Grim et al. 2003).
Any naïve penguin can contract the disease, but chicks and juveniles are most susceptible. Owing to low parasitaemia, the destruction of red blood cells usually does not cause clinical anaemia in penguins, and they often die without detectable parasitaemia (Stoskopf & Beier, 1979). However, the most common clinical signs are anorexia, depression, dyspnoea and regurgitation (Stoskopf & Beier, 1979).
The typical findings at necropsy and histological examination include splenomegaly, pulmonary oedema, hydropericardium and the presence of parasites in the reticuloendothelial system (Grim et al. 2003).
In tissue samples of dead birds the diagnosis of infection may prove complex because histological identification of meronts in tissue samples, especially in cases of low parasite numbers, may be difficult and long searches may be required. On the other hand, fragmented nuclei within necrotic tissue may be erroneously considered as meronts.
To overcome the problems with unequivocal identification of tissue stages, the aim of the present study was to establish a specific detection method for avian Plasmodium species in birds and to relate tissue lesions with the presence of these protozoal parasites.
Materials and Methods
Probe design.
An oligonucleotide probe labelled with digoxigenin at the 3′end (Eurofins MWG Operon, Ebersberg, Germany) was designed to detect a part of the small subunit (18S) ribosomal RNA sequence of all avian Plasmodium species that were available from the GenBank database.
These sequences were aligned using the Sci Ed Central software package (Scientific & Educational software, Cary, North Carolina, USA) and a region of homology was selected as the probe sequence. The probe sequence was 5′-TTTAATAACTCGTTATATATATCAGTG-TAGCAC-3′. To ensure probe specificity and to exclude unintended cross-hybridization with other organisms, the sequence was submitted to the Basic Local Alignment Search Tool (BLAST; http://www.ncbi.nlm.nih.gov/blast.cgi). The sequence was 100% complementary to a segment of 18S ribosomal RNA of a number of avian Plasmodium species (including P. relictum, P. juxtanucleare, Plasmodium reichenowi, Plasmodium gallinaceum and many not further characterized avian Plasmodium species), and there was only one nucleotide difference to avian P. elongatum and Plasmodium lophurae as well as to many simian and human Plasmodium species, including Plasmodium vivax and Plasmodium falciparum. The probe showed no complementarity with other related protozoa and is thus considered highly specific for Plasmodium, and cross-hybridization is unlikely to occur.
Tissue samples and histopathology.
Formalin-fixed, paraffin wax-embedded tissues including the spleen, liver, kidneys, lung, skeletal muscle, small intestine and brain from 48 captive penguins (33 Humboldt penguins [Spheniscus humboldti], 10 rockhopper penguins [Eudyptes chrysocome] and five king penguins [Aptenodytes patagonicus]) that had died between 2000 and 2008 at the zoological garden Schönbrunn, Vienna, Austria were examined in this retrospective study. Five of these penguins (four Humboldt penguins, one rockhopper penguin) had a tentative diagnosis of infection with avian malaria parasites by standard histological investigation.
Supplementary tissue sections from a bobwhite quail (Colinus virginianus) infected with Haemoproteus lophortyx (courtesy of H. L. Shivaprasad, Davis, California, USA) and cultured P. falciparum parasites (courtesy of F. Petry, Mainz, Germany) were examined using the Plasmodium probe. The protozoal culture containing an unknown number of P. falciparum parasites was fixed in 10% buffered formalin and embedded in paraffin wax. Prior to embedding, the culture was soaked with rice starch (3.3 mg/ml) for 5 h and centrifuged at 6000 × g for 10 min to produce a pellet. The pellet was overlaid with agar, hardened at 4°C, carefully removed from the tube and finally embedded in paraffin wax.
In addition, archived paraffin wax-embedded tissues from different species naturally infected with protozoal parasites of the genera Cryptosporidium, Sarcocystis, Eimeria, Toxoplasma, Giardia, Entamoeba, or fungi (Aspergillus, Candida, Encephalitozoon) and viruses (canine adenovirus 2, canine parvovirus 2, West Nile virus, Usutu virus) were investigated with the Plasmodium probe in order to exclude cross-hybridization. Application of an irrelevant oligonucleotide probe (Leishmania spp. probe) to positive control slides was carried out as additional negative control.
All sections used for in-situ hybridization (ISH) were also stained by Giemsa and with haematoxylin and eosin to demonstrate parasites.
In-situ hybridization.
The paraffin wax-embedded tissue samples were sectioned (∼ 3 μm) and placed on Superfrost Plus slides. They were dewaxed in Neoclear and rehydrated in a series of graded alcohols (100%, 96%, 70%) and distilled water. Proteolytic treatment was done with proteinase K 6 μg/ml (Roche, Basel, Switzerland) in Tris-buffered saline at 37°C for 50 min and the slides were rinsed with distilled water and dehydrated in 96% ethanol and 100% isopropanol followed by air-drying.
Afterwards the slides were covered with hybridization mixture, 100 μl of which were composed of 12 μl distilled water, 20 μl of 20x standard sodium citrate (SSC), 50 μl formamide (50%), 2 μl Denhardt's solution, 10 μl dextran sulphate (50%, w/v), 5 μl herring sperm DNA (50 mg/ml) and 1 μl Plasmodium probe at a concentration of 100 ng/ml.
The slides were then incubated at 95°C for 6 min and immediately afterwards placed on crushed ice. After cooling, the slides were hybridized overnight in a humid chamber at 40°C On the second day the slides were washed in 2x SSC, 1x SSC and 0.1x SSC at room temperature.
The slides were then incubated with anti-digoxigenin-AP Fab fragments (Roche) (dilution 1:200) for 1 h at room temperature. After washing, the signal was visualized with 5-bromo-4-chloro-3-indolyl phosphate and 4-nitro-blue tetrazolium chloride (Roche) for 1 h at room temperature in the dark. The staining reaction was terminated by placing the slides in tris-ethylendiaminetetraacetic acid (TE) buffer (pH 8.0) for 10 min.
Finally, the slides were counterstained with haematoxylin and mounted under coverslips with Aquatex (VWR International, Vienna, Austria).
Polymerase chain reaction.
For confirmation of all ISH results, PCR assays (Richard et al. 2002; Martínez et al. 2009) targeting part of the parasite cytochrome b gene were applied.
Amplification of parasite DNA was attempted using the primer pair Palu-F/Palu-R (Martínez et al. 2009), which amplified a 390-base-pair sequence from a conserved region of the cytochrome b gene of Plasmodium and Haemoproteus. In case this PCR amplification was successful, a second PCR assay was carried out using the primers 621/983 (Richard et al. 2002) that amplified a 341-base-pair sequence of the cytochrome b gene of Plasmodium.
For PCR amplification, paraffin wax sections (10 μm) were dewaxed in xylene and afterwards washed in ethanol and dried. DNA was extracted using Nexttec Clean Columns (Nexttec, Leverkusen, Germany) according to the manufacturer's instructions.
The 25 μl reaction mixture was composed of 10 μl HotMasterMix (5Prime; Eppendorf, Hamburg, Germany), 1 μl each forward and reverse primer (10 pM), 11 μl distilled water and 2 μl template DNA. The PCR amplification started with denaturation for 2 min at 94°C, was followed by 40 cycles of heat denaturation at 94°C for 30 sec, primer annealing at 50°C for primers 621/983 or 56°C for primers Palu-F/Palu-R for 30 sec and DNA extension at 72°C for 1 min, and ended with a final extension at 72°C for 10 min. A 10 μl aliquot of each PCR product was analysed by gel electrophoresis on a 2% Tris-acetate–ethylenediamine tetraacetic acid–agarose gel. The agarose gel was stained with ethidium bromide and visualized with a BioSens SC-Series 710 gel documentation system using the BioSens gel imaging system software (GenXpress, Wiener Neudorf, Austria). Positive PCR controls were not used. The negative control was a PCR mixture containing laboratory grade water instead of template DNA.
Gene sequence analysis.
PCR products using the primers Palu-F and Palu-R were sequenced according to Bakonyi et al. (2004), except that DNA purification after amplification was carried out using the QIAquick gel extraction kit (QIAGEN, Hilden, Germany) in accordance with the manufacturer's instructions instead of ethanol precipitation. The obtained nucleotide sequences were identified by a BLAST search against the GenBank database.
GenBank accession numbers.
The nucleotide sequences determined in the present study were deposited in the GenBank database under the accession numbers HQ404522 to HQ404534.
Results
Tissue samples and histopathology.
Lesions typical of avian malaria, including splenomegaly, hepatomegaly and severe pulmonary oedema were present in nine out of 48 penguins.
Histologically, in five of these nine penguins (four Humboldt penguins, one rockhopper penguin) numerous protozoal meronts were evident in the capillary endothelium of various tissues, especially the lung, liver, spleen and brain by standard haematoxylin and eosin staining. Severe hyperaemia and fibrinoid necrosis was observed in the lung; in endothelial cells of the capillaries, numerous roundish or oval exoerythrocytic meronts, containing a variable number of merozoites, were present (Figure 1a). In the brain, characteristic histological lesions were absent, although many elongated exoerythrocytic meronts (phanerozoites) were observed in the capillary endothelium (Figure 1b). The spleen showed focal necrosis and karyorrhexis as well as the formation of microthrombi and several exoerythrocytic meronts in the endothelial cells of the capillaries. In some cases, it was difficult to distinguish between nuclear fragments and meronts, which made their unequivocal identification challenging. In the liver, perivascular non-suppurative inflammatory infiltrates containing mainly plasma cells were found. Additionally, meronts were observed in the sinusoidal endothelium and the cytoplasm of hepatocytes (Figure 1c). In the remaining four of these nine penguins, which had a tentative diagnosis of avian malaria, neither histological lesions nor the presence of avian malaria parasites were noticed in any of the tissues investigated.
Figure 1.
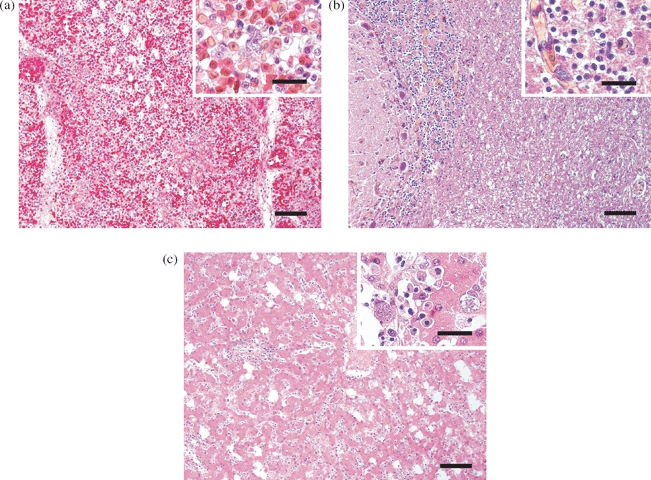
Histological sections of (1a) lung, (1b) brain and (1c) liver of penguins naturally infected with P. elongatum parasites. Several exoerythrocytic meronts in the capillary endothelium are present; in the brain, elongated exoerythrocytic meronts are present. Haematoxylin and eosin staining, bar = 150 μm. Inset: bar = 40 μm.
In none of the 39 penguins without macroscopic changes suggestive of avian malaria, characteristic histological lesions or presence of meronts were noticed.
In-situ hybridization.
Plasmodium tissue stages (meronts) were readily identified by a distinct purple to black signal within the capillary endothelium. Clearly stained parasites were found in various tissues, especially the lung (Figure 2a), brain (Figure 2b), liver (Figure 2c) and spleen of 13 penguins (11 Humboldt penguins, two rockhopper penguins). These included all five histologically diagnosed cases, the four birds with macroscopic changes suggestive of avian malaria but without histological lesions and four additional penguins without macroscopic and histological alterations. In the latter four cases the quantity of protozoa was markedly lower. Cultured P. falciparum parasites were clearly labelled with the Plasmodium probe.
Figure 2.
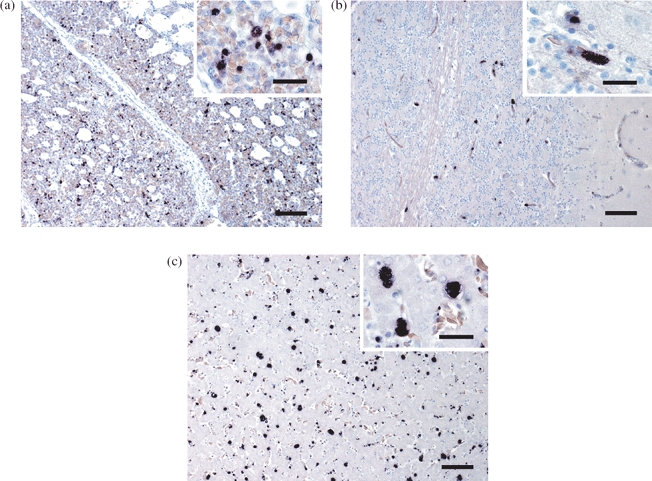
ISH of (1a) lung, (1b) brain and (1c) liver shows numerous Plasmodium meronts that are readily discernible by their distinct purple to black signal. Bar = 150 μm. Inset: bar = 40 μm.
There was no cross-reactivity with H. lophortyx (Figure 3), other protozoal parasites, fungi and viruses.
Figure 3.
By ISH, H. lophortyx megaloschizonts in skeletal muscle of a bobwhite quail (C. virginianus) show no specific signal. Bar = 40 μm.
Polymerase chain reaction.
The PCRs using the primers Palu-F/Palu-R and 621/983 yielded distinct amplification products of the expected sizes from various DNA extracts of the lung, brain, liver and spleen of the 13 ISH-positive penguins and confirmed the tentative diagnosis of an infection with avian malaria parasites. In the 35 ISH-negative samples, parasite DNA was not detectable using the primer pair Palu-F/Palu-R.
Gene sequence analysis.
DNA sequences from the 13 penguins were edited and aligned using the Sci Ed Central software package. In all these cases no double peaks could be observed in the electropherograms; the presence of double infections therefore seemed unlikely.
DNA sequences from nine penguins proved to be 99 to 100% identical with published sequences from unambiguously determined P. elongatum (DQ368381, DQ659588, AY733089; Valkiūnas et al. 2008). Sequences from the other four birds were 99 to 100% identical with published sequences from avian Plasmodium spp. (DQ847263, EU810610, DQ991068, DQ368384).
Discussion
The present report describes an ISH procedure with a digoxigenin-labelled oligonucleotide probe for the detection of avian malaria parasites in tissue sections of penguins. Identification of these protozoal parasites is usually based on microscopic examination of stained blood smears, or on the use of PCR assays. However, none of these techniques is able to specifically identify haemosporidians in histological slides. We thus attempted to achieve this goal using a technique that had previously successfully identified protozoa of different phyla (Chvala et al. 2006; Liebhart et al. 2006: Richter et al. 2008; Mostegl et al. 2010). In the present project, the approach to design species-specific or even genus-specific oligonucleotide probes was expected to be complex due to the fact that morphologically based taxonomy distinguishes a few hundred haemosporidian species. This enormous number, however, largely overestimates the number of genetically defined species, because morphologically identical parasites were given different names when observed in different bird species (Garnham, 1966). Hence, identification to species level is a challenge and requires screening of many parasite cells in a blood smear, because morphological characteristics may overlap for some species (Valkiūnas, 2005). These difficulties obviously led to some incorrect linkages of morphology and DNA sequences that unfortunately are also deposited in GenBank (Valkiūnas et al. 2008). Thus, a strategy to minimize incorrect species identification has been recommended by Valkiūnas et al. (2008). They suggest vigorous comparison of morphological criteria of parasites and their DNA lineages with the expertise of taxonomists, and suggest establishing a database for reference DNA sequences from unambiguously identified parasites. A further challenge was that the required Plasmodium probe had to be designed against 18S rRNA sequences, because only these target molecules seem to be present in sufficient abundance to allow robust labelling of the respective parasite cells. This is clearly documented by the successful application of this approach for a number of other protozoa (Chvala et al. 2006; Liebhart et al. 2006; Richter et al. 2008; Mostegl et al. 2010). Unfortunately only very few reliable (i.e. convincingly identified to species level) rRNA gene sequences from avian Plasmodium species are found in GenBank. So, probe design had to be based on these fragmentary genetic data. From parasites belonging to Haemoproteus and Leucocytozoon genera, there are no 18S rRNA gene sequences available in GenBank. For this reason, hypothetical cross-reactivity of the Plasmodium probe could not be evaluated in silico. The negative outcome of the ISH of a bobwhite quail (C. virginianus) infected with H. lophortyx, however, suggests that the probe does not cross-react with members of the genus Haemoproteus and may thus be considered specific for Plasmodium.
For species identification of haemosporidians, geneticists predominantly rely on cytochrome b gene sequences; thus there is an abundance of such data. Unfortunately, ISH using these sequences as the target produced no staining (Dinhopl, N., personal observation, unpublished).
For genetic identification of the involved Plasmodium species, we relied on comparing our cytochrome b sequences with GenBank entries. To avoid incorrect assignments we only accepted sequences as valid when they were derived from organisms that had been identified by both morphological and molecular methods or when they showed a high degree of homology to such sequences. Sequences with a higher amount of nucleotide differences and sequences with very close genetic relationships to other than the indicated species were not considered valid and were not used for the alignments on which species identification of parasites were based. In our cases, nine strongly similar sequences were closely related to P. elongatum and another four sequences of high similarity were closely related to P. elongatum-like, unclassified Plasmodium species. As only these two Plasmodium variants were present in several independent malaria outbreaks over a time span of 9 years, it seems that the natural avian hosts in and around Vienna are predominantly infected with these P. elongatum-like parasites. This aspect will be further investigated in the future.
PCR detects amplified parasite DNA and not necessarily the intact parasite, and does not reflect the stage of disease or the severity of infection. Additionally, the localization of the parasites within tissue lesions and simultaneous evaluation of their morphology and distribution is not feasible with this method.
Histological identification of meronts in tissue samples, especially in cases with low parasite numbers, may be cumbersome and long searches may be required.
Other objects viewed by light microscopy may also be erroneously considered as meronts. In contrast, meronts were easily identified by ISH, even at low magnification and in different penguin tissues. This method has the advantage of high specificity due to nucleic acid detection and offers the possibility of correlating the presence of parasites with associated lesions. Unfortunately, the ISH technique described here does not allow discrimination of different subgenera or species of haemosporidians, especially due to the previously mentioned lack of 18S rRNA gene sequence information of a large number of defined Plasmodium species. Future directions of the ongoing research are generating rRNA gene sequences of defined species of the genera Haemoproteus and Leucocytozoon in order to design specific probes that may allow distinguishing between genera and subgenera, and perhaps discrimination to species level.
In any case, the ISH procedure described here proved to be a powerful tool for unambiguous detection of Plasmodium parasites in paraffin wax-embedded tissues, which will also be widely applicable for post-mortem examination of suspicious cases.
Acknowledgements
The authors thank the staff of the Zoo Schönbrunn for submission of the dead birds and for their good cooperation. Thanks are due to Prof. H.L. Shivaprasad (Davis, California, USA) for the generous gift of tissue sections from a bobwhite quail (C. virginianus) infected with H. lophortyx, and to Dr F. Petry (Mainz, Germany) for the generous gift of cultured P. falciparum parasites. The authors are grateful to Klaus Bittermann for the professional digital artwork and wish to thank Nora Nedorost and Anton Maderner for their excellent technical support. The present study was funded by the Austrian Science Fund (FWF) grant P20926.
References
- Atkinson C.T., Dusek R.J., Lease J.K. Serological response and immunity to superinfection with avian malaria in experimentallyinfected Hawaii amakihi. Journal of Wildlife Diseases. 2001;37:20–27. doi: 10.7589/0090-3558-37.1.20. [DOI] [PubMed] [Google Scholar]
- Atkinson C.T., Dusek R.J., Woods K.L., Iko W.M. Pathogenicity of avian malaria in experimentally infected Hawaii amakihi. Journal of Wildlife Diseases. 2000;36:197–204. doi: 10.7589/0090-3558-36.2.197. [DOI] [PubMed] [Google Scholar]
- Bakonyi T., Gould E.A., Kolodziejek J., Weissenböck H., Nowotny N. Complete genome analysis and molecular characterization of Usutu virus that emerged in Austria in 2001: comparison with the South African strain SAAR-1776 and other flaviviruses. Virology. 2004;328:301–310. doi: 10.1016/j.virol.2004.08.005. [DOI] [PubMed] [Google Scholar]
- Chvala S., Fragner K., Hackl R., Hess M., Weissenböck H. Cryptosporidium infection in domestic geese (Anser anser f. domestica) detected by in-situ hybridization. Journal of Comparative Pathology. 2006;134:211–218. doi: 10.1016/j.jcpa.2005.11.002. [DOI] [PubMed] [Google Scholar]
- Cranfield M.R., Shaw M., Beall F., Skjoldager M., laleggio M. A review and update of avian malaria in the African penguin (Spheniscus demersus) Proceedings of the American Association of Zoo Veterinarians. 1990;21:243–248. [Google Scholar]
- Fix A.S., Waterhouse C., Greiner E.C., Stoskopf M.K. Plasmodium relictum as a cause of avian malaria in wild-caught Magellanic penguins (Spheniscus magellanicus) Journal of Wildlife Diseases. 1988;24:610–619. doi: 10.7589/0090-3558-24.4.610. [DOI] [PubMed] [Google Scholar]
- Fleischmann R.W., Sladen W.J.L., Melby E.C. Malaria (Plasmodium elongatum) in captive African penguins (Spheniscus demersus) Journal of the American Veterinary Medical Association. 1968;153:928–935. [PubMed] [Google Scholar]
- Garnham P.C.C. Malaria Parasites and Other Haemosporidia. Oxford: Blackwell Scientific Publications; 1966. [Google Scholar]
- Graczyk T.K., Cranfield M.R., McCutchan T.F., Bicknese E.J. Characteristics of naturally acquired avian malaria infections in naïve juvenile African black-footed penguins (Spheniscus demersus) Parasitology Research. 1994;80:634–637. doi: 10.1007/BF00932944. [DOI] [PubMed] [Google Scholar]
- Grim C.K., van der Merwe E., Sullivan M., Parson N., McCutchan T.F., Cranfield M. Plasmodium juxtanucleare associated with mortality in black footed penguins (Spheniscus demersus) admitted to a rehabilitation center. Journal of Zoo and Wildlife Medicine. 2003;34:250–255. doi: 10.1638/02-070. [DOI] [PubMed] [Google Scholar]
- Hellgren O., Waldenström I., Bensch S. A new PCR assay for simultaneous studies of Leucocytozoon, Plasmodium, and Haemoproteus from avian blood. Journal of Parasitology. 2004;90:797–802. doi: 10.1645/GE-184R1. [DOI] [PubMed] [Google Scholar]
- Liebhart D., Weissenböck H., Hess M. In-situ hybridization for the detection and identification of Histomonas meleagridis in tissues. Journal of Comparative Pathology. 2006;135:237–242. doi: 10.1016/j.jcpa.2006.08.002. [DOI] [PubMed] [Google Scholar]
- Loupal G., Kutzer E. Infektionen mit Plasmodium spec. bei Papageitauchern (Fratercula arctica) Kleintierpraxis. 1995;41:901–906. (in German). [Google Scholar]
- Martínez I, Martínez-de la Puente J., Herrero J., Del Cerro S., Lobato E., Rivero-de Aguilar J., et al. A restriction site to differentiate Plasmodium and Haemoproteus infections in birds: on the inefficiency of general primers for detection of mixed infection. Parasitology. 2009;136:713–722. doi: 10.1017/S0031182009006118. [DOI] [PubMed] [Google Scholar]
- Mostegl M.M., Richter B., Nedorost N., Maderner A., Dinhopl N., Kulda J., et al. Design and validation of an oligonucleotide probe for the detection of protozoa from the order Trichomonadida using chromogenic in situ hybridization. Veterinary Parasitology. 2010;171:1–6. doi: 10.1016/j.vetpar.2010.03.022. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Palinauskas V., Kosarev V., Shapoval A., Bensch S., Valkiūnas G. Comparison of mitochondrial cytochrome b lineages and morphospecies of two avian malaria parasites of the subgenera Haemamoeba and Giovannolaia (Haemosporida: Plasmodiidae) Zootaxa. 2007;1626:39–50. [Google Scholar]
- Perkins S.L., Osgood S.M., Schall J.J. Use of PCR for detection of subpatent infections of lizard malaria: implications for epizootiology. Molecular Ecology. 1998;7:1587–1590. [Google Scholar]
- Richard A.F., Sehgal R.N.M., Jones H.I., Smith T.B. A comparative analysis of PCR-based detection methods for avian Malaria. Journal of Parasitology. 2002;88:819–822. doi: 10.1645/0022-3395(2002)088[0819:ACAOPB]2.0.CO;2. [DOI] [PubMed] [Google Scholar]
- Richter B., Kübber-Heiss A., Weissenböck H., Schmidt P. Detection of Cryptosporidium spp., Entamoeba spp. and Monocercomonas spp. in the gastrointestinal tract of snakes by in-situ hybridization. Journal of Comparative Pathology. 2008;138:63–71. doi: 10.1016/j.jcpa.2007.11.001. [DOI] [PubMed] [Google Scholar]
- Stoskopf M.K., Beier J. Avian malaria in African black-footed penguins. Journal of the American Veterinary Medicine Association. 1979;175:944–947. [PubMed] [Google Scholar]
- Sturrock H.J.W., Tompkins D.M. Avian malaria (Plasmodium spp.) in yellow-eyed penguins: investigating the cause of high seroprevalence but low observed infection. New Zealand Veterinary Journal. 2007;55:158–160. doi: 10.1080/00480169.2007.36761. [DOI] [PubMed] [Google Scholar]
- Valkiūnas G. Avian Malaria Parasites and Other Haemosporidia. Boca Raton, FL: CRC Press; 2005. [Google Scholar]
- Valkiūnas G, lezhova T.A. A comparison of the blood parasites in three subspecies of the yellow wagtail Motacilla flava. Journal of Parasitology. 2001;87:930–934. doi: 10.1645/0022-3395(2001)087[0930:ACOTBP]2.0.CO;2. [DOI] [PubMed] [Google Scholar]
- Valkiūnas G, Bensch S., lezhova T.A., Križanauskiene A., Hellgren O., Bolshakov C.V. Nested cytochrome b polymerase chain reaction diagnostics underestimate mixed infections of avian blood haemosporidian parasites: microscopy is still essential. Journal of Parasitology. 2006;92:418–122. doi: 10.1645/GE-3547RN.1. [DOI] [PubMed] [Google Scholar]
- Valkiūnas G, Zehtindjiev P., Dimitrov D., Križanauskiene A., lezhova T.A., Bensch S. Polymerase chain reaction-based identification of Plasmodium (Huffia) elongatum, with remarks on species identity of haemosporidian lineages deposited in GenBank. Parasitology Research. 2008;102:1185–1193. doi: 10.1007/s00436-008-0892-9. [DOI] [PubMed] [Google Scholar]


