Abstract
All cancers carry somatic mutations. A subset of these somatic alterations, termed driver mutations, confer selective growth advantage and are implicated in cancer development, whereas the remainder are passengers. Here we have sequenced the genomes of a malignant melanoma and a lymphoblastoid cell line from the same person, providing the first comprehensive catalogue of somatic mutations from an individual cancer. The catalogue provides remarkable insights into the forces that have shaped this cancer genome. The dominant mutational signature reflects DNA damage due to ultraviolet light exposure, a known risk factor for malignant melanoma, whereas the uneven distribution of mutations across the genome, with a lower prevalence in gene footprints, indicates that DNA repair has been preferentially deployed towards transcribed regions. The results illustrate the power of a cancer genome sequence to reveal traces of the DNA damage, repair, mutation and selection processes that were operative years before the cancer became symptomatic.
The genomes of all cancer cells carry somatic mutations1. These may include base substitutions, small insertions and deletions (indels), rearrangements and copy number alterations together with epigenetic changes. Some of these somatic alterations, known as driver mutations, confer selective clonal growth advantage and are causally implicated in oncogenesis. By definition, these are found in cancer genes. The remainder are passengers which do not contribute to cancer development. However, passenger mutations bear the imprints of the mutational mechanisms that have generated them, unsullied by processes of selection, and thus provide insights into the aetiology and pathogenesis of cancer.
Over the last quarter of a century several strategies have been used to detect the various classes of somatic mutation in cancer genomes1. As a result, approximately 400 cancer genes have been identified1,2 (http://www.sanger.ac.uk/genetics/CGP/Census/) and somatic mutations from thousands of tumours have provided insights into the mutational processes operative in human cancer1,3.
With the advent of the human genome sequence a new strategy was proposed1,4. Systematic sequencing would identify all somatic mutations of all classes in individual cancer genomes, yielding complete catalogues of somatic mutation. Technological limitations initially constrained this to polymerase chain reaction (PCR)-based resequencing of the coding exons of protein-coding genes in order to find base substitutions and small indels5-10. Recently, however, several novel technologies have been developed11,12. These allow sequencing of randomly generated DNA fragments from cancer genomes and thus detect rearrangements and copy number changes as well as base substitutions and small indels, providing sufficient coverage to identify most somatic mutations in an individual cancer genome. These technologies have previously been used to reveal missense mutations in the coding sequences of two acute myeloid leukaemia genomes13,14. Here, we report the first comprehensive catalogue of somatic mutations from a cancer genome.
The catalogue of somatic mutations
COLO-829 is an immortal, publicly available cancer cell line derived, before treatment, from a metastasis of a malignant melanoma in a 43-year-old male15. No skin primary was identified. Using Illumina GAII genome analysers, we obtained more than 40-fold average haploid genome coverage by aligned sequence from COLO-829 and 32-fold from COLO-829BL, a lymphoblastoid line derived from the same patient (Supplementary Fig. 1). Differences from the reference sequence were called in both cell lines. The variant set obtained from COLO-829BL was then subtracted from that of COLO-829 to establish the catalogue of somatic mutations in COLO-829.
We identified 33,345 somatic base substitutions. A total of 32,325 were single-base and 510 were double-base substitutions (in which two adjacent bases show somatic mutations) (Table 1, Fig. 1 and Supplementary Table 1). Of 48 already known somatic substitutions, 42 were present in the whole-genome catalogue of mutations, a sensitivity of 88%. Of 470 newly found somatic substitutions that were assessed, 454 (97%) were confirmed by conventional sequencing, indicating a 3% false-positive rate.
Table 1. Somatic mutations identified in COLO-829.
| Type of change | Count | Percentage |
|---|---|---|
| Substitutions | 33,345 | 100.0 |
| Coding | 292 | 0.9 |
| Silent | 105 | 0.3 |
| Missense | 172 | 0.5 |
| Truncating | 15 | <0.1 |
| Non-coding | 319 | 1.0 |
| UTR | 205 | 0.6 |
| ncRNA | 113 | 0.3 |
| miRNA | 1 | <0.1 |
| Intronic | 9,543 | 28.6 |
| Splice | 7 | <0.1 |
| Other intronic | 9,536 | 28.6 |
| Intergenic | 23,191 | 69.6 |
| Small insertions and deletions | 66 | 100.0 |
| Coding | 0 | 0.0 |
| UTR | 2 | 3.0 |
| Intronic | 27 | 40.9 |
| Intergenic | 37 | 56.1 |
| Rearrangements | 37 | 100.0 |
| Breakpoints | 74 | |
| Coding | 1 | 1.4 |
| UTR | 0 | 0.0 |
| Intronic | 36 | 48.6 |
| Intergenic | 37 | 50.0 |
| Classes | 37 | 100.0 |
| Intrachromosomal | 34 | 91.9 |
| Deletions | 25 | 67.6 |
| Inversions | 6 | 16.2 |
| Duplications | 2 | 5.4 |
| Other | 1 | 2.7 |
| Interchromosomal | 3 | 8.1 |
miRNA, microRNA; ncRNA, non-coding RNA; UTR, untranslated region.
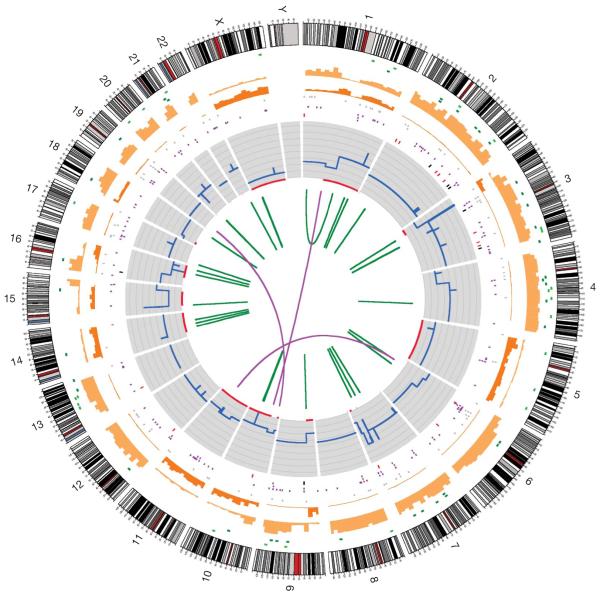
Figure 1. The catalogue of somatic mutations in COLO-829.
Chromosome ideograms are shown around the outer ring and are oriented pter–qter in a clockwise direction with centromeres indicated in red. Other tracks contain somatic alterations (from outside to inside): validated insertions (light-green rectangles); validated deletions (dark-green rectangles); heterozygous (light-orange bars) and homozygous (dark-orange bars) substitutions shown by density per 10 megabases; coding substitutions (coloured squares: silent in grey, missense in purple, nonsense in red and splice site in black); copy number (blue lines); regions of LOH (red lines); validated intrachromosomal rearrangements (green lines); validated interchromosomal rearrangements (purple lines).
A total of 680 small deletions and 303 small insertions were predicted, of which 182 were evaluated and 66 (36%) confirmed. Thus the false-positive rate for insertions and deletions was higher than for substitutions. We were unable to estimate sensitivity as only a single small deletion had previously been reported in COLO-829. This was not called in the whole-genome sequence, although it was present in a subset of reads. Most confirmed somatic insertions and deletions were of single bases (Supplementary Table 2).
To detect rearrangements we searched for fragments in which the sequence reads from the two ends mapped discordantly to the genome16. A total of 51 somatic rearrangements were predicted. We assessed the sensitivity of detection by evaluating the proportion of somatic changes in genomic copy number for which a rearrangement was found. Thirty out of forty-one (73%) copy number changes examined were detected as rearrangements. However, an additional seven (17%) were at centromeres or other regions where alignment is expected to be difficult. Of the 51 rearrangements, 75% were confirmed as somatic by PCR across the breakpoint junction or the presence of a copy number change in the appropriate position. Of the 37 somatic rearrangements confirmed by PCR and mapped to the base-pair level (Table 1, Fig. 1 and Supplementary Table 3), 3 were interchromosomal and 34 were intrachromosomal, including 25 deletions, 6 inversions, 2 duplications and 1 large intrachromosomal event.
All well-supported somatic copy number changes and regions of loss of heterozygosity (LOH) detected using genotyping arrays were found by whole-genome sequencing (Fig. 1). Two regions of high copy number genomic amplification, on chromosomes 3p and 15p, and two homozygous deletions, on chromosomes 10q and 16q, were observed. Somatic mutations of mitochondria in cancer have previously been reported, although their role in oncogenesis is unclear17. No somatic mutations present in more than 20% of mitochondrial sequences were observed in COLO-829.
Mutations of coding genes and microRNAs
A total of 292 somatic base substitutions were in protein-coding sequences (Table 1 and Supplementary Table 4). Of these, 187 caused amino acid changes (non-synonymous), including 172 that were missense and 15 nonsense, and 7 affected highly conserved bases at splice sites. There were 105 silent (synonymous) substitutions. One somatic substitution was found in the microRNA hsa-mir-518d in the central region of the stem structure. None of the 66 confirmed small insertions and deletions was in coding sequences.
On the assumption that most silent mutations are biologically inert, the ratio of non-synonymous to synonymous substitutions in protein-coding sequences can be used to estimate the extent of selection overall on non-synonymous changes6,18. The ratio in COLO-829 was 1.78, not significantly different from that expected by chance (P=0.5). Thus, the substantial majority of mutations do not seem to be subject to positive or negative selection. However, this test will be insensitive to small numbers of selected mutations.
Several individual substitutions highlighted candidate novel cancer genes. For example, two heterozygous missense mutations (het, p.S229L; het, p.D283H) were in SPDEF, which encodes a member of the ETS transcription factor family19. SPDEF expression is associated with disease progression in prostate, breast and ovarian cancer20,21. Sequencing SPDEF through 48 additional, untreated metastatic melanomas revealed a further somatic mutation (p.W158*). A missense mutation (het, p.G297E) was identified in the matrix metalloproteinase gene MMP28. Missense mutations in the family of matrix metalloproteinases were recently reported in melanoma, including several in MMP28 (ref. 22). A missense mutation (het, p.N561K) was also identified in UVRAG, which was originally identified in a complementation assay for ultraviolet light sensitivity in xeroderma pigmentosum group C cells, has recently been shown to have an important role in autophagy and has been proposed as a tumour suppressor gene23,24. To determine the significance of these and other mutations, analysis of additional melanomas will be required.
Of the 37 somatic rearrangements mapped to the base pair, 19 interrupted protein-coding genes. No in-frame fusion genes were predicted, but one gene (MAGI2) had an exonic deletion that creates a predicted in-frame rearranged transcript. PCR with reverse transcription (RT–PCR) across the exon–exon rearrangement boundary showed that the chimaeric transcript is expressed, but the biological significance of this rearrangement remains to be clarified.
Rearrangements occur at a high frequency in cancer over fragile sites. In COLO-829, rearrangements were found in FHIT and WWOX, which overlie the fragile sites FRA3B and FRA16D, respectively. Both rearrangements are contained within individual introns, however, and are not predicted to alter the sequences of the encoded proteins.
There are several modest copy-number reductions and increases that affect large genomic regions and which alter the copy number of many protein-coding genes (Fig. 1). Of particular interest, however, is the relatively restricted region of high (8- to 12-fold) copy number increase on chromosome 3p which contains four complete genes: RARB, TOP2B, NGLY1 and KS (OXSM). A ~0.5-megabase (Mb) region on chromosome 15 is also amplified to 4–6 copies and contains MKRN3 and NDN. None of these genes has previously been implicated in cancer development through amplification, although another member of the retinoic receptor family, RARA, is consistently rearranged in acute promyelocytic leukaemia25. A 12-kilobase (kb) internal homozygous deletion of the recessive cancer gene PTEN is predicted to cause premature termination and is presumably implicated in the development of COLO-829. However, the significance of a 40-kb homozygous deletion that removes the start ATG codon of CNOT1, a repressor of transcriptional activation by oestrogen receptor26, is unknown.
Of the three previously identified driver mutations in COLO-829, genome-wide sequencing revealed BRAF V600E in addition to the PTEN deletion described above. A 2-bp deletion in CDKN2A was not detected, but was found in sequence reads when a targeted search was made.
A mutational signature of ultraviolet exposure
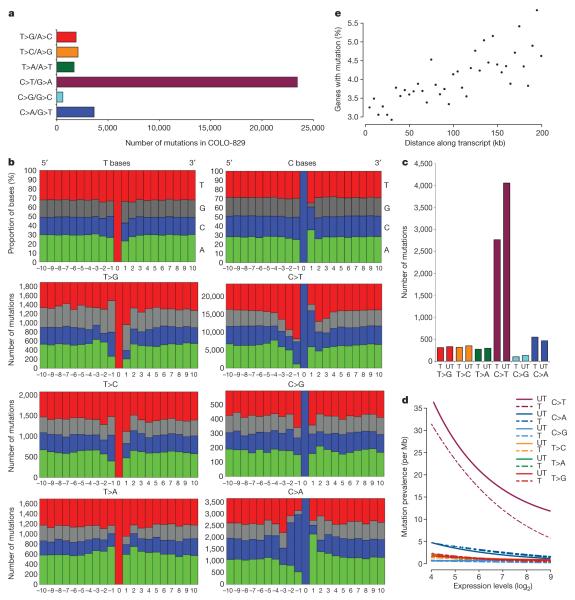
Most somatic base substitutions in COLO-829 were C>T/G>A transitions (Fig. 2a). Of the 510 dinucleotide substitutions, 360 were CC>TT/GG>AA changes. This mutational spectrum is reminiscent of that previously associated with ultraviolet light exposure, a known environmental risk factor in the development of malignant melanoma27-30.
Figure 2. Patterns of somatic substitution.
a, Mutation spectrum. b, Mutation sequence context compared to random T bases and C bases (top two panels). c, Mutation counts by transcribed (T) and untranscribed (UT) strands. d, Effect of gene expression on mutation prevalence. Lines are parametrically fitted curves to the data. Mutation prevalence is expressed as the number of mutations per Mb of each class. T, transcribed strands; UT, untranscribed strands. e, Effect of distance along the transcript on mutation prevalence. Each dot represents a 5-kb bin along gene footprints, from transcription start sites to 200 kb. The y axis shows the fraction of genes in each bin carrying a somatic mutation.
DNA damage due to ultraviolet light leads to the formation of covalent links between two adjacent pyrimidines31. Consequently, C>T mutations due to ultraviolet light usually occur at dipyrimidine sequences. Therefore, to evaluate further the role of ultraviolet light in the pathogenesis of somatic mutations in COLO-829, we examined the sequence context of C>T substitutions (Fig. 2b). A total of 92% of C>T mutations occurred at the 3′ base of a pyrimidine dinucleotide compared with 53% expected by chance (P<0.0001). CC>TT mutations exhibited a similar pattern (82% were 3′ to a pyrimidine, P<0.0001). The frequency of C>Tand CC>TT mutations due to ultraviolet light exposure is also known to be higher at CpG dinucleotides27. In COLO-829, both C>T substitutions (7.7%, P<0.0001) and CC>TT double substitutions (10.0%, P=0.014) showed elevated frequencies at CpG dinucleotides compared to that expected by chance (4.4%). Therefore, the mutation spectrum and sequence context indicate that most C>T/G>A somatic substitutions in COLO-829 are attributable to ultraviolet-light-induced DNA damage.
Expression-directed DNA repair
Ultraviolet-light-induced DNA damage is predominantly repaired by nucleotide excision repair (NER)32,33. NER is a nonspecific repair process activated by sensing of DNA distortion caused by the DNA modification induced by a mutagen. Over the whole genome, DNA damage is sensed by a specific protein complex and NER is then implemented through excision of an oligonucleotide carrying the damage and filling of the gap by replicative polymerases. The main purpose of this form of NER, known as global genome repair, may be prevention of somatic mutations that would otherwise be transmitted to daughter cells through mitosis. There is, however, an additional form of NER that is selectively directed at the transcribed strand of genes34,35. This transcription-coupled repair is sensed by the stalling of RNA polymerase II when it encounters a bulky DNA adduct. The purpose of transcription-coupled repair may primarily be to reduce interference with transcription caused by DNA damage.
A consequence of transcription-coupled repair is that DNA damage on the transcribed strand is repaired more efficiently than damage on the non-transcribed strand. Thus, fewer mutations accumulate on the transcribed strand. We investigated whether transcription-coupled repair had been operative in COLO-829 by comparing the number of C>T mutations found on the transcribed strands of all 21,417 protein-coding genes to the number of C>T mutations on the non-transcribed strands. There were 2,773 C>T changes on the transcribed strands and 4,058 on the non-transcribed strands (P<0.0001) (Fig. 2c), with a similar pattern for CC>TT double substitutions (48 on transcribed strands and 75 on non-transcribed strands). The results are therefore consistent with the past operation of transcription-coupled repair on ultraviolet-light-induced DNA damage in COLO-829.
The involvement of transcription-coupled repair predicts an overall lower prevalence of C>T/G>A substitutions over genic than intergenic DNA. Indeed, only 10,004 (30%) somatic C>T changes were found over the combined genomic footprints of transcribed genes compared with 13,164 (40%) expected if the mutations had been randomly distributed (Table 1, P<0.0001). However, the effect of transcription-coupled repair accounts only for one-third of the deficit of mutations over protein-coding gene footprints. This suggests the existence of an additional class of NER which is preferentially deployed to both transcribed and non-transcribed strands of genes compared to intergenic DNA. A form of NER with these features has been previously proposed but is not extensively characterized32. We also observed a reduction in mutation prevalence in exons (8.33 per Mb) compared to introns (9.93 per Mb, P=0.0001). Preferential targeting of NER to exons has not been reported and it is conceivable that this effect may, in part, be attributable to negative selection on coding sequence mutations.
To investigate further the relationship between transcription and NER we examined the correlation between mutation prevalence and gene expression (Fig. 2d). Genes expressed at a high level in COLO-829 showed a lower prevalence of somatic mutations compared to genes expressed at a low level, on both the transcribed and non-transcribed strands. The effect of transcript levels on mutation prevalence was most pronounced for C>T/G>A mutations (P<0.0001) but was also observed in other mutational classes. A trend towards a higher prevalence of somatic substitutions at the 3′ compared to the 5′ ends of genes was also observed (Fig. 2e). This may be due to aborted transcription, such that 3′ ends are overall less transcribed than 5′ ends, with the consequence that expression-related repair processes are deployed less at 3′ ends and hence the mutation prevalence is higher.
Thus, the catalogue of somatic mutations in COLO-829 has exposed multiple levels of selective application of DNA repair: preferential targeting to transcribed compared to untranscribed genomic regions; to exons compared to introns; to transcribed DNA strands compared to non-transcribed strands; and to the 5′ compared to the 3′ ends of genes.
Evidence for other mutational processes
C>A/G>T transversions constitute the second commonest class of substitution in COLO-829 and exhibit a distinctive sequence context (Fig. 2b). C>A/G>T mutations also show evidence of transcription-coupled repair (P=0.002) (Fig. 2c, d). However, the bias is against G>T mutations on the transcribed strand, suggesting that the original DNA damage was predominantly on guanine (Fig. 2c). There is substantial evidence that mutagens such as reactive oxygen species predominantly cause damage to guanine which ultimately results in G>T changes. Indeed, results from experimental systems suggest that the sequence context observed in COLO-829 for C>A/G>T transversions is optimal for this class of mutagen36. Thus, in addition to the direct effects of ultraviolet light, the somatic mutational catalogue of COLO-829 may harbour traces of subsidiary mechanisms of DNA damage.
Timing of somatic mutations
The catalogue of somatic mutations is a cumulative record of the mutational events that have occurred during the lineage of cell divisions starting from the fertilized egg and ending in the cancer cell. By combining information on chromosome copy number change with base substitutions the relative order of some mutations on this mitotic lineage can be established.
Several genomic regions in COLO-829 show evidence of loss of one parental chromosome, leading to LOH, followed by re-duplication of the remaining copy. In these regions, mutations which occurred before the re-duplication event will be homozygous, whereas those arising after re-duplication will be heterozygous. In most such regions, a small fraction of mutations are heterozygous, indicating relatively late re-duplication (Fig. 1). However, in a region of LOH on chromosome 1q, there are more heterozygote substitutions than homozygote, suggesting earlier re-duplication (Fig. 1). In this region we were therefore able to compare somatic substitutions that occurred relatively early in the evolution of the cancer (homozygous mutations) with those that occurred later (heterozygous mutations), and found differences in their mutational spectra. C>T mutations account for a higher proportion of early (82%) compared to late mutations (53%, P<0.0001). By contrast, C>A/G>T changes account for a higher proportion of late (19%) compared to early (2%) mutations. The results suggest that exposure to ultraviolet light was extinguished after the 1q re-duplication event, perhaps when the melanoma metastasized, resulting in a reduced rate of C>T/G>A mutation. Moreover, the increased proportion of C>A/G>T in the later phase suggests that the process underlying these mutations is, at least in part, unrelated to ultraviolet light exposure.
Discussion
We have generated the first comprehensive catalogue of somatic mutations from a human cancer genome. The catalogue includes the overwhelming majority of mutations present in COLO-829, although there remains a small fraction of base substitutions and rearrangements that have eluded detection and a small number of false positives. For insertions and deletions the sensitivity is lower and the false-positive rate is higher, indicating the need for additional computational approaches to their discovery. Epigenetic changes have not been included as the technologies for their exhaustive detection are still under development.
The catalogue of mutations in COLO-829 carries the imprint of past ultraviolet-light-induced DNA damage together with evidence for auxiliary, independent mechanisms of damage. The catalogue also bears traces of the DNA repair processes that have been operative, including transcription-coupled NER and other, less well characterized patterns of NER deployment. Buried within it are most of the driver mutations that have conferred selective growth advantage on this melanoma. The total number of drivers for this, or any other, cancer is currently unknown. Some may be among the 187 non-synonymous substitutions in protein-coding sequences. However, the possibility of drivers in non-coding RNAs, regulatory or currently cryptic functional regions of the genome can now be explored.
In future, the generation of thousands of comprehensive, high-quality catalogues of somatic mutation will provide powerful insights into the processes of DNA damage, mutation, repair and selection that underlie the evolution of all human cancers. These will form the basis of our understanding of cancer causation and development, providing the foundation for prevention and treatment.
METHODS SUMMARY.
We sequenced 75 bases from both ends of 200-bp and 400-bp libraries constructed from the genomes of COLO-829 and COLO-829BL using Illumina GAII Genome Analysers. To study structural variation, these data were supplemented with paired 50 base reads from 2-kb, 3-kb and 4-kb mate-pair libraries11. Sequences were aligned to the reference human genome (NCBI36) using ELAND11, MAQ37 and BWA38. Differences from the reference sequence were called in both cell lines. The variant set obtained from COLO-829BL was then subtracted from that of COLO-829 to establish the catalogue of somatic mutations in COLO-829.
Acknowledgements
The authors acknowledge the support of C. Henry, S. Kahn, D. Evers, G. Smith, K. Hall and the technical and administrative support staff at Illumina. I.V. is supported by the Human Frontiers Science Program and P.J.C. by the Kay Kendall Leukaemia Fund. We would like to acknowledge the Wellcome Trust for support under grant reference 077012/Z/05/Z.
METHODS
Sequencing
Construction of short (200/400 bp) and mate-pair (2/3/4 kb) paired end libraries, flowcell preparation and cluster generation was as previously described11. Paired end sequencing was performed on Illumina GAIIx genome analysers as described in the Illumina Genome Analyser operating manual. Primary data analysis including image analysis, base-calling and alignment was carried out with the Illumina pipeline.
Substitution detection
The tumour and normal genomes were built and SNP-called separately. Short insert 2*75 bp paired end reads were aligned to NCBI36 using ELAND11 (v.1.1.1.3;). SNPs were called by CASAVA11 (v1.2) but with an additional parameter: the 75 base reads were divided into equal thirds (bins); we then only called substitutions where the non-reference base was observed at least once either in all three bins, or at least twice in the first bin and once in the second bin. This parameter removes most false-positive SNPs that are due to insertions and deletions (indels).
Somatic substitutions were identified as alleles that were called in the tumour genome but not in the germ line. A depth of at least 10× was required in the germ line. In order to allow for any under-called positions in the germ line, no observations of that allele were permitted in the germ line, although one call was permitted if the depth was ≥30×. Substitutions corresponding to known SNP positions (dbSNP 129) were excluded. Substitutions were annotated using Ensembl version 52.
Insertion and deletion detection
Indels in the tumour and normal genomes were called using Pindel39, BWA38 and GROUPER (A. J. Cox et al., manuscript in preparation) on the NCBI36 genome build. Pindel identifies singleton reads (aligned reads whose mate does not align), splits the unaligned read and attempts to align it in two portions. BWA aligns reads allowing gaps, and then samtools (http://samtools.sourceforge.net/) can be used to call indels from the alignments. GROUPER identifies clusters of singleton reads (minimum two), performs a local assembly of unmapped reads and then maps the contig back to the same region to identify the indel. Somatic indels were identified by subtracting all indels called using any of the three methods in the normal genome from indels called in the tumour (minimum of three reads) with Pindel. A minimum of 10× normal coverage was also required. Indels were annotated based on Ensembl version 52.
Structural variant detection
Abnormal read pairs that mapped to the genome at an unexpected distance or orientation were identified, grouped and filtered as previously described16. Structural variants were called from the long insert data using MAQ alignments, requiring at least ten independent read pairs in the tumour and no read pairs representing the rearrangement in the normal. Structural variants were called from the short insert data using both MAQ and ELAND alignments. When a structural variant was identified, all reads that were predicted to overlap the breakpoint region based on the location of their paired read and insert size were used to attempt an assembly across the breakpoint with Velvet40. Assembled contigs were aligned back to the reference genome to annotate the precise breakpoint.
Confirmation by capillary sequencing
Substitutions and insertions and deletions were confirmed by capillary sequencing across the region of the mutation. Structural variants were confirmed by PCR across the breakpoint and capillary sequence. All confirmations were done in both the tumour and normal to determine if the variants were somatic or germ line.
Copy number determination
Reads were counted in windows across the genome, corrected for genome uniqueness as previously described16. A GC correction was applied to each bin count based on a linear correction for its GC content. An HMM was used to segment the data, taking into account known breakpoint locations, and to estimate the integer copy number for each segment (Supplementary Table 5).
Loss of heterozygosity detection
The zygosity of each of ~924,000 known SNP positions (corresponding to SNPs used on the Affymetrix SNP 6.0 array) was determined in both the normal and the tumour. Regions of LOH (where SNPs were heterozygous in the normal but homozygous in the tumour) were identified using an HMM (Supplementary Table 6).
Substitution analysis
For mutation context, the bases±10 bp from the mutation were extracted and the number of each base counted. Context of equivalent changes (for example, C>T and G>A) were combined. The background context was determined from 100,000 random positions of the equivalent base type from chromosome 2. The calculation of mutation frequency in transcribed and untranscribed regions was corrected for sequence gaps and regions of low coverage. Strand bias was calculated based on annotating each mutation as to whether it fell on the transcribed or untranscribed strand in Ensembl 52. Gene expression data were derived from the Affymetrix U133 Plus 2.0 array, run in triplicate, normalized and averaged. Poisson regression was used for the analysis of the effects of gene expression on mutation prevalence, incorporating the number of at-risk bases in each gene footprint as the offset, allowing quadratic terms for the relationship between expression and mutation prevalence, and using a dummy variable for transcribed versus non-transcribed strand mutations. Duplication timing was determined by calculating the frequency of heterozygous and homozygous mutations per Mb throughout LOH regions of at least 5 Mb in size, excluding gaps in the reference sequence. The image of the entire COLO-829 genome was produced using Circos41.
Footnotes
Full Methods and any associated references are available in the online version of the paper at www.nature.com/nature.
The authors declare no competing financial interests.
Supplementary Information is linked to the online version of the paper at www.nature.com/nature.
References
- 1.Stratton MR, Campbell PJ, Futreal PA. The cancer genome. Nature. 2009;458:719–724. doi: 10.1038/nature07943. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Futreal PA, et al. A census of human cancer genes. Nature Rev. Cancer. 2004;4:177–183. doi: 10.1038/nrc1299. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Pfeifer GP, Besaratinia A. Mutational spectra of human cancer. Hum. Genet. 2009;125:493–506. doi: 10.1007/s00439-009-0657-2. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Futreal PA, et al. Cancer and genomics. Nature. 2001;409:850–852. doi: 10.1038/35057046. [DOI] [PubMed] [Google Scholar]
- 5.The Cancer Genome Atlas Research Network Comprehensive genomic characterization defines human glioblastoma genes and core pathways. Nature. 2008;455:1061–1068. doi: 10.1038/nature07385. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Greenman C, et al. Patterns of somatic mutation in human cancer genomes. Nature. 2007;446:153–158. doi: 10.1038/nature05610. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Wood LD, et al. The genomic landscapes of human breast and colorectal cancers. Science. 2007;318:1108–1113. doi: 10.1126/science.1145720. [DOI] [PubMed] [Google Scholar]
- 8.Ding L, et al. Somatic mutations affect key pathways in lung adenocarcinoma. Nature. 2008;455:1069–1075. doi: 10.1038/nature07423. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Jones S, et al. Core signaling pathways in human pancreatic cancers revealed by global genomic analyses. Science. 2008;321:1801–1806. doi: 10.1126/science.1164368. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Parsons DW, et al. An integrated genomic analysis of human glioblastoma multiforme. Science. 2008;321:1807–1812. doi: 10.1126/science.1164382. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Bentley DR, et al. Accurate whole human genome sequencing using reversible terminator chemistry. Nature. 2008;456:53–59. doi: 10.1038/nature07517. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Margulies M, et al. Genome sequencing in microfabricated high-density picolitre reactors. Nature. 2005;437:376–380. doi: 10.1038/nature03959. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Ley TJ, et al. DNA sequencing of a cytogenetically normal acute myeloid leukaemia genome. Nature. 2008;456:66–72. doi: 10.1038/nature07485. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Mardis ER, et al. Recurring mutations found by sequencing an acute myeloid leukemia genome. N. Engl. J. Med. 2009;361:1058–1066. doi: 10.1056/NEJMoa0903840. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Morse HG, Moore GE. Cytogenetic homogeneity in eight independent sites in a case of malignant melanoma. Cancer Genet. Cytogenet. 1993;69:108–112. doi: 10.1016/0165-4608(93)90083-x. [DOI] [PubMed] [Google Scholar]
- 16.Campbell PJ, et al. Identification of somatically acquired rearrangements in cancer using genome-wide massively parallel paired-end sequencing. Nature Genet. 2008;40:722–729. doi: 10.1038/ng.128. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Chatterjee A, Mambo E, Sidransky D. Mitochondrial DNA mutations in human cancer. Oncogene. 2006;25:4663–4674. doi: 10.1038/sj.onc.1209604. [DOI] [PubMed] [Google Scholar]
- 18.Greenman C, Wooster R, Futreal PA, Stratton MR, Easton DF. Statistical analysis of pathogenicity of somatic mutations in cancer. Genetics. 2006;173:2187–2198. doi: 10.1534/genetics.105.044677. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Oettgen P, et al. PDEF, a novel prostate epithelium-specific ets transcription factor, interacts with the androgen receptor and activates prostate-specific antigen gene expression. J. Biol. Chem. 2000;275:1216–1225. doi: 10.1074/jbc.275.2.1216. [DOI] [PubMed] [Google Scholar]
- 20.Sood AK, et al. Expression characteristics of prostate-derived Ets factor support a role in breast and prostate cancer progression. Hum. Pathol. 2007;38:1628–1638. doi: 10.1016/j.humpath.2007.03.010. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Ghadersohi A, et al. Prostate-derived Ets transcription factor as a favorable prognostic marker in ovarian cancer patients. Int. J. Cancer. 2008;123:1376–1384. doi: 10.1002/ijc.23667. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Palavalli LH, et al. Analysis of the matrix metalloproteinase family reveals that MMP8 is often mutated in melanoma. Nature Genet. 2009;41:518–520. doi: 10.1038/ng.340. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Teitz T, et al. Isolation by polymerase chain reaction of a cDNA whose product partially complements the ultraviolet sensitivity of xeroderma pigmentosum group C cells. Gene. 1990;87:295–298. doi: 10.1016/0378-1119(90)90316-j. [DOI] [PubMed] [Google Scholar]
- 24.Liang C, et al. Autophagic and tumour suppressor activity of a novel Beclin1-binding protein UVRAG. Nature Cell Biol. 2006;8:688–698. doi: 10.1038/ncb1426. [DOI] [PubMed] [Google Scholar]
- 25.Scaglioni PP, Pandolfi PP. The theory of APL revisited. Curr. Top. Microbiol. Immunol. 2007;313:85–100. doi: 10.1007/978-3-540-34594-7_6. [DOI] [PubMed] [Google Scholar]
- 26.Winkler GS, Mulder KW, Bardwell VJ, Kalkhoven E, Timmers HT. Human Ccr4-Not complex is a ligand-dependent repressor of nuclear receptor-mediated transcription. EMBO J. 2006;25:3089–3099. doi: 10.1038/sj.emboj.7601194. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Pfeifer GP, You YH, Besaratinia A. Mutations induced by ultraviolet light. Mutat. Res. 2005;571:19–31. doi: 10.1016/j.mrfmmm.2004.06.057. [DOI] [PubMed] [Google Scholar]
- 28.Armstrong BK, Kricker A. The epidemiology of UV induced skin cancer. J. Photochem. Photobiol. B. 2001;63:8–18. doi: 10.1016/s1011-1344(01)00198-1. [DOI] [PubMed] [Google Scholar]
- 29.Leiter U, Garbe C. Epidemiology of melanoma and nonmelanoma skin cancer—the role of sunlight. Adv. Exp. Med. Biol. 2008;624:89–103. doi: 10.1007/978-0-387-77574-6_8. [DOI] [PubMed] [Google Scholar]
- 30.Giglia-Mari G, Sarasin A. TP53 mutations in human skin cancers. Hum. Mutat. 2003;21:217–228. doi: 10.1002/humu.10179. [DOI] [PubMed] [Google Scholar]
- 31.Daya-Grosjean L, Sarasin A. The role of UV induced lesions in skin carcinogenesis: an overview of oncogene and tumor suppressor gene modifications in xeroderma pigmentosum skin tumors. Mutat. Res. 2005;571:43–56. doi: 10.1016/j.mrfmmm.2004.11.013. [DOI] [PubMed] [Google Scholar]
- 32.Nouspikel T. DNA repair in mammalian cells: Nucleotide excision repair: variations on versatility. Cell. Mol. Life Sci. 2009;66:994–1009. doi: 10.1007/s00018-009-8737-y. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Shuck SC, Short EA, Turchi JJ. Eukaryotic nucleotide excision repair: from understanding mechanisms to influencing biology. Cell Res. 2008;18:64–72. doi: 10.1038/cr.2008.2. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Hanawalt PC, Spivak G. Transcription-coupled DNA repair: two decades of progress and surprises. Nature Rev. Mol. Cell Biol. 2008;9:958–970. doi: 10.1038/nrm2549. [DOI] [PubMed] [Google Scholar]
- 35.Fousteri M, Mullenders LH. Transcription-coupled nucleotide excision repair in mammalian cells: molecular mechanisms and biological effects. Cell Res. 2008;18:73–84. doi: 10.1038/cr.2008.6. [DOI] [PubMed] [Google Scholar]
- 36.Kawanishi S, Hiraku Y, Oikawa S. Mechanism of guanine-specific DNA damage by oxidative stress and its role in carcinogenesis and aging. Mutat. Res. 2001;488:65–76. doi: 10.1016/s1383-5742(00)00059-4. [DOI] [PubMed] [Google Scholar]
- 37.Li H, Ruan J, Durbin R. Mapping short DNA sequencing reads and calling variants using mapping quality scores. Genome Res. 2008;18:1851–1858. doi: 10.1101/gr.078212.108. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Li H, Durbin R. Fast and accurate short read alignment with Burrows-Wheeler transform. Bioinformatics. 2009;25:1754–1760. doi: 10.1093/bioinformatics/btp324. [DOI] [PMC free article] [PubMed] [Google Scholar]
References
- 39.Ye K, et al. Pindel: a pattern growth approach to detect break points of large deletions and medium sized insertions from paired-end short reads. Bioinformatics. 2009;25:2865–2871. doi: 10.1093/bioinformatics/btp394. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Zerbino DR, Birney E. Velvet: algorithms for de novo short read assembly using de Bruijn graphs. Genome Res. 2008;18:821–829. doi: 10.1101/gr.074492.107. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Krzywinski M, et al. Circos: an information aesthetic for comparative genomics. Genome Res. 2009;19:1639–1645. doi: 10.1101/gr.092759.109. [DOI] [PMC free article] [PubMed] [Google Scholar]