Abstract
A novel ArsR-SmtB family transcriptional repressor, KmtR, has been characterized from mycobacteria. Mutants of Mycobacterium tuberculosis lacking kmtR show elevated expression of Rv2025c encoding a deduced CDF-family metal exporter. KmtR-dependent repression of the cdf and kmtR operator-promoters was alleviated by nickel and cobalt in minimal medium. Electrophoretic mobility shift assays and fluorescence anisotropy show binding of purified KmtR to nucleotide sequences containing a region of dyad symmetry from the cdf and kmtR operator-promoters. Incubation of KmtR with cobalt inhibits DNA complex assembly and metal-protein binding was confirmed. KmtR is the second, to NmtR, characterized ArsR-SmtB sensor of nickel and cobalt from M. tuberculosis suggesting special significance for these ions in this pathogen. KmtR-dependent expression is elevated in complete medium with no increase in response to metals, whereas NmtR retains a response to nickel and cobalt under these conditions. KmtR has tighter affinities for nickel and cobalt than NmtR consistent with basal levels of these metals being sensed by KmtR but not NmtR in complete medium. More than a thousand genes encoding ArsR-SmtB-related proteins are listed in databases. KmtR has none of the previously defined metal-sensing sites. Substitution of His88, Glu101, His102, His110, or His111 with Gln generated KmtR variants that repress the cdf and kmtR operator-promoters even in elevated nickel and cobalt, revealing a new sensory site. Importantly, ArsR-SmtB sequence groupings do not correspond with the different sensory motifs revealing that only the latter should be used to predict metal sensing.
Tuberculosis is a leading killer worldwide causing 2 million deaths and 9 million new cases each year. It is estimated that one-third of the world’s population is latently infected with Mycobacterium tuberculosis (1). This organism infects macrophages and somehow survives within phagosomes, despite the antimicrobial mechanisms in this compartment (2). The action of natural resistance-associated macrophage protein 1 (Nramp1, alias SLC11A1) is one such phagosome mechanism that is effective against M. tuberculosis and must be evaded by the more virulent pathogens (3-5). Nramp1 is a divalent cation pump that is recruited to late endosomal-phagosomal membranes (6, 7). The metal substrates, direction of flux, and precise basis of pathogen killing by Nramp1 are not fully understood. To survive inside phagosomes M. tuberculosis must adapt to metal fluxes, and pathogen proteins involved in metal detection and homeostasis are known virulence factors (8-10).
Sensors, such as ArsR-SmtB family repressors, detect surplus metal ions and modulate transcription of genes involved in metal uptake, efflux, sequestration, or detoxification (11, 12). DNA binding by these sensors is weakened upon metal binding, alleviating repression in elevated metal (12). Genes encoding ArsR-SmtB sensors occur in many bacteria, but intriguingly the M. tuberculosis genome encodes an atypically large number (twelve identified by the Pfam data base, HTH_5 family). It is tempting to speculate that these proteins enable this pathogen to respond rapidly to metal-fluxes in the phagosome. Effectors for three of the M. tuberculosis sensors are known: Ni(II)-Co(II) for NmtR (13), Cd(II)-Pb(II) for CmtR (14), and Zn(II) for Rv2358 (herein designated MtSmtB) (15). NmtR and CmtR regulate the nmt and cmt operator-promoters triggering expression of metal transporting P1-type ATPases (13, 14), while MtSmtB regulates the Rv2358-furB operon with FurB (Zur) acting as a sensor of Zn(II) deficiency (11). The effectors of the remaining nine M. tuberculosis sensors are unknown, concealing vital clues about the nature of metal adaptation by this pathogen. Indeed it is unclear at this time whether or not all of these proteins do detect metals.
Prediction of the metals sensed by ArsR-SmtB homologues has often been crudely made, notably in databases, from overall sequence similarity to sensors for which effectors are known. In other bacteria these include: Zn(II) for SmtB and ZiaR (13, 16); As(III), Sb(III), and Bi(III) for ArsR (17); Cd(II), Pb(II), and Zn(II) for CadC and AztR (18, 19); Zn(II) and Co(II) for CzrA (20, 21); and Cu(I), Ag(I), Zn(II), and Cd(II) for BmxR (22). Cyanobacterial SmtB was the first shown to be a winged helix homo-dimer with helices α3 and α4 predicted to form the DNA-associating helix-turn-helix (23). In vitro and in vivo studies revealed two pairs of metal-binding sites per dimer: one pair associated with the α3 helices, including two ligands contributed by the amino-terminal region of the opposing monomer (α3N sites), and a second pair associated with carboxyl-terminal α5 helices (α5 sites) (24-26). Site-directed mutagenesis established that only the α5 sites are required for inducer recognition (24, 25). In contrast to SmtB, cysteine ligands associated with α3 helices (α3 sites) are required for inducer responsiveness of ArsR (27). Further diversity in the effector-binding sites has been described as a “themes and variations” model (28), with variations known on at least three themes: (i) α3 with (CadC and AztR) or without (ArsR) amino-terminal ligands (19, 27, 29), (ii) α5 with (NmtR) or without (SmtB and CzrA) additional carboxyl-terminal ligands (13, 26), and (iii) direct metal binding at α4 helices with an additional ligand from the carboxyl terminus (CmtR) (14). Both α5 and α3N are obligatory for inducer recognition by the Zn(II) sensor ZiaR (16). Permutations in the metal ligand sets (cysteinylthiol, histidine-imidazole, and glutamate/aspartate-carboxyl) and metal coordination geometries (trigonal, tetrahedral, and octahedral) influence which metals are sensed. At least two correct predictions of metal specificity of previously uncharacterized ArsR-SmtB sensors have been made based on deduced metal-sensing sites (30). Do overall sequence similarity and deduced sensory sites always coincide and predict the same metal specificities? Are all of the possible sensory sites now known?
Here we have reduced the catalogue of ArsR-SmtB homologues (in the Pfam HTH_5 family) from 1024 (30) to an ensemble of 554 sequences by systematically excluding the most distant relatives. These form eight major groups based on sequence similarity. Importantly, several groups contain sequences with different metal-binding motifs. Furthermore, sequences that share the same metal-binding motif are scattered among different groups.
The deduced product of M. tuberculosis Rv0827c (hereafter called kmtR) is grouped with NmtR, CmtR, and MtSmtB. KmtR lacks any of the previously described sensory sites. We generated a kmtR mutant of M. tuberculosis and used whole genome microarrays to identify KmtR-regulated genes. KmtR specifically binds to the promoter of Rv2025c (cdf), and its own promoter, but not of Rv0826, which also showed altered expression in the gene profiles. In complex medium KmtR- and MtSmtB-dependent gene expression is elevated and is not responsive to metals. However, in cells grown in metal-limited medium KmtR mediates repression of the cdf and kmtR promoters, and repression is alleviated only by elevated Ni(II) or Co(II), whereas MtSmtB-mediated repression is alleviated by Zn(II). For both proteins, metals corresponding to their in vivo effectors, Co(II) for KmtR and Zn(II) for MtSmtB, impaired DNA binding in vitro. Unexpectedly, KmtR represents the second Ni(II)/Co(II)-sensing ArsR-SmtB repressor, along with NmtR, in M. tuberculosis. However, in contrast to KmtR, NmtR retains Ni(II) and Co(II) responsiveness in the complex medium, which correlates with a lower affinity for these metals.
Site-directed mutagenesis has defined the KmtR residues essential for detecting metals, and these compose an original sensory motif (designated α5-3). This motif is shared among a sub-group of sequences with no previously defined sensory site. Another sequence signature (α2α5) that is common to a large ArsR-SmtB sub-group, not known to detect metals, is also reported. Implications of these findings for identifying ArsR-SmtB metal sensors and correctly predicting the metals they detect are discussed, as are the properties of this new sensor especially in relation to metal adaptation by a devastating pathogen.
EXPERIMENTAL PROCEDURES
Bacterial Strains, Culture Conditions, and DNA Manipulations
M. tuberculosis H37Rv was used as the parental strain to construct the kmtR mutant and for microarray experiments. Mycobacterium smegmatis mc2155 and Mycobacterium bovis BCG4 (Pasteur) were used as mycobacterial hosts for reporter gene assays. Cells were grown at 37 °C with shaking (M. smegmatis and M. bovis) or rolling (M. tuberculosis) in Middlebrook 7H9 medium (Difco) containing 0.05% Tween 80 and 10% oleic acid/albumin/dextrose/catalase enrichment (Difco) or on 7H10 agar medium supplemented with 0.5% glycerol and 10% oleic acid/albumin/dextrose/catalase enrichment. For some reporter gene assays (refer to individual experiments) cells were grown in Sauton medium, supplemented with 0.025% tyloxapol (Sigma), that had been treated overnight at 4 °C with Chelex 100 resin (10 g liter−1) prior to filtration, the addition of MgSO4 (2 mm) and sterilization. Elemental analysis of the media by inductively coupled plasma mass spectrometry revealed: 2.760 and 0.296 μm zinc, 3.078 and 0.027 μm copper, 0.031 and 0.053 μm nickel, 0.012 and 0.005 μm cobalt, 134.981 and 113.099 μm iron, and 0.057 and 0.061 μm manganese in Middlebrook 7H9 media and Chelex-treated Sauton media, respectively. Hygromycin (50 μg ml−1) and kanamycin (25 μg ml−1) were added where appropriate. Escherichia coli strains JM109 (Stratagene) and BL21(DE3) were used and grown at 37 °C in Luria-Bertani broth and agar containing hygromycin (150 μg ml−1), kanamycin (50 μg ml−1), or carbenicillin (200 μg ml−1) where appropriate. Cells were transformed to antibiotic resistance as described previously (31, 32). All generated plasmid constructs were checked by sequence analysis.
Generation of a kmtR Mutant
M. tuberculosis genomic DNA was used as template for PCR with primers I (5′-GAAAAGCTTACCAACGGCACGCACC-3′) and II (5′-GAATCTAGAGGTCCACTATCTGCGTAC-3′) or III (5′-GAATCTAGACCTTAGGGCAGTAGTGCG-3′) and IV (5′-GAAGCGGCCGCTGGGTTACGAATCGCC-3′) to amplify 1024 bp of kmtR upstream sequences (including the first seven codons of kmtR) and 944 bp of kmtR downstream sequences, respectively, and the products were ligated into pGEM-T (Promega, Madison, WI). The kmtR upstream sequences were excised from pGEM-T and ligated into the HindIII/SalI site of p2NIL (33) to generate p2NIL0827A. The kmtR downstream sequences were then excised and ligated into the XbaI/NotI site of p2NIL0827A, generating p2NIL0827B. A hyg-lacZ-sacB marker cassette from pGOAL19 (33) was ligated into the PacI site of p2NIL0827B to form the final deletion construct p2NIL0827C. M. tuberculosis mutant selection was performed as described (33), and deletion of 425 bp, which includes kmtR (but retaining the first seven codons), was confirmed by PCR and Southern blot analyses (31).
RNA Extraction, cDNA Labeling, and Microarray Experiments
RNA was extracted from exponential phase cultures, labeled cDNA and DNA produced by incorporation of either Cy3 or Cy5 dCTP (Amersham Biosciences), and hybridizations on M. tuberculosis H37Rv whole genome PCR-product microarrays (Bacterial Microarray Group at St George’s: TBv2.1.1; ArrayExpress accession no.: A-BUGS-23) performed as previously described (34). Three independent RNA samples were used, and hybridizations were performed in duplicate in competition with labeled genomic DNA. Feature extraction was performed with ImaGene v5.5 (BioDiscovery), and data from multiple photomultiplier amplification settings were processed using the MAVI Pro 2.6.0 software (MWG Biotech). Statistical analyses were performed using Genespring GX v7 (Agilent Technologies) by analysis of variance one-way analysis with a Benjamini and Hochberg false discovery rate of 0.05.
Cloning, Expression, and Purification of M. tuberculosis KmtR, MtSmtB, and NmtR
The kmtR and MtsmtB coding regions were amplified from M. tuberculosis genomic DNA, using primers V (5′-CATATGTACGCAGATAGTGGACCTGACCCGTTGCC-3′) and VI (5′-CCGAATTCTTATTACCCGACATCCTTGGTAGCCG-3′) for kmtR or VII (5′-CATATGGTGACGTCCCCCTCAACG-3′) and VIII (5′-GAATTCTCATATTGCGTCCTCACCGGCGTGCGC-3′) for MtsmtB, ligated to pGEM-T prior to sub-cloning into the NdeI/EcoRI sites of pET29a (Novagen). The proteins were expressed in E. coli BL21(DE3) for 4 h at 37 °C using 1.0 mm or 0.5 mm isopropyl 1-thio-β-d-galactopyranoside for KmtR and MtSmtB, respectively. Crude cell lysates were prepared in buffer A (10 mm HEPES, pH 7.8, 1 mm EDTA, 1 mm DTT,4 and 50 mm NaCl) and applied to a Heparin-affinity column (CL-4B Amersham Biosciences) pre-equilibrated with buffer A, and bound proteins were eluted using a linear gradient to 1 m NaCl. KmtR-containing fractions were then diluted to 50 mm NaCl in 20 mm Tris, pH 8.8, 1 mm EDTA, 1 mm DTT and concentrated using a HiTrap Q HP anion-exchange column (Amersham Biosciences), washed with 20 mm Tris, pH 8.8, 50 mm NaCl, and eluted with 400 mm NaCl (this anion-exchange step was omitted during MtSmtB purification). Fractions containing KmtR or MtSmtB were then applied to a Superdex 75 size-exclusion column (Amersham Biosciences) pre-equilibrated in buffer A containing 200 mm NaCl, prior to concentration using a HiTrap Q HP column. The column was washed with 20 mm Tris, pH 8.8, 50 mm NaCl (for KmtR) or 20 mm HEPES, pH 7.8 (for MtSmtB), and protein was eluted with 400 mm NaCl. NmtR was expressed and purified essentially as described previously (13).
Gel-retardation Assays
Operator-promoter regions were amplified from M. tuberculosis genomic DNA, using primers IX (5′-TACCGGTTCCGGCAGGAACCC-3′) and X (5′-CACCAAGCAGACCTGATC-3′) for PkmtR, XI (5′-GAAGGATCCGGTGATCGTCGTCCGTCC-3′) and XII (5′-GGTACCATCGGGCGCAGGCCCTTTG-3′) for Pcdf, or XIII (5′-GAAGGATCCAAGGGGACACCGGACCAG-3′) and XIV (5′-GAAGGTACCGTATCTTGGGTCACTGGTGG-3′) for PRv0286, and ligated to pGEM-T. Truncated versions of the cdf operator-promoter region were also amplified using primers XI and XV (5′-GACCACCAAGCAAGCTC-3′) for T1, primers XV and XVI (5′-CCGGCGAGAGCATCCGC-3′) for T2, and primers XI and XVII (5′-GATGCTCTCGCCGGTTC-3′) for T3, and ligated to pGEM-T. Competitor DNA and the various operator-promoter regions were amplified from re-circularized pGEM-T or the operator-promoter constructs with primers designed to anneal to the plasmid backbone either site of the cloning site (14). Equal quantities of target and competitor DNA were then used in binding reactions (30). Products were separated on native 9% polyacrylamide gels in TBE buffer (0.089 m Tris, 0.089 m boric acid, 0.002 m EDTA) at 4 °C, stained with ethidium bromide, and visualized under UV light.
Analyses of Nickel, Zinc, and Cobalt Binding
Microtiter plate competition analyses were performed using 2 μm protein incubated with up to 10 μm metal in 10 mm HEPES, pH 7.8, 250 mm NaCl, 1 mm DTT, for 30 min at room temperature, followed by addition of the metallochromic indicator 4-(2-pyridylazo)resorcinol (PAR). PAR-bound metal was detected at 492 nm in a microtiter plate reader. Control reactions lacking protein were performed in parallel. Analysis of metal binding via tryptophan (KmtR) or tyrosine (NmtR) fluorescence was performed using a 1-cm light path, 1-ml cell with a Carey Eclipse fluorescence spectrometer.
Fluorescence Anisotropy Analysis of Protein-DNA Interaction
Complementary oligonucleotides were produced corresponding to regions of dyad symmetry within PkmtR (5′-TCTATTGTTTGCGTATGTACGCAGATAGTGGA-3′), Pcdf (5′-CGCTATTATCTGCGTATGAATGCAGATAAAAGAG-3′), PRv0286 (5′-TCCATAGTGACAACGTGCGTAGTCAGAATTCG-3′), and PMtsmtB (5′-CTTTGACATGCATCATCATGCATGTGACAG-3′). One oligonucleotide of each pair was 5′-labeled with 6-hexachlorofluorescein, and complementary pairs were annealed in 10 mm HEPES, pH 7.8, 150 mm NaCl, by heating to 95 °C followed by cooling slowly to 10 °C. For standard reactions, protein was desalted into anisotropy buffer (10 mm HEPES, pH 7.8, 250 mm NaCl) containing 1 mm DTT using a Sephadex G25 column (Amersham Biosciences), and binding reactions were performed by adding increasing concentrations of protein to 5 nM double-stranded DNA, in anisotropy buffer with 1 mm DTT, in a 1-ml quartz cuvette (10-mm path length). To examine the effects of metal ions on DNA binding, the protein was desalted into anisotropy buffer using a Sephadex G25 column in an anaerobic chamber and incubated overnight at 4 °C under anaerobic conditions with Co(II), Zn(II), or 1 mm EDTA. A gas-tight Hamilton syringe was then used to add increasing concentrations of protein to anaerobic cuvettes containing DNA in anisotropy buffer with 1 mm EDTA or a molar excess of Co(II) or Zn(II). The anisotropy of the solution was measured using an 8100 fluorometer (SLM-Aminco, Urbana, IL) (30).
Construction of Promoter-lacZ Fusions, Site-directed Mutagenesis, and β-Galactosidase Assays
kmtR upstream sequences and coding region (1378 bp) were amplified from M. tuberculosis genomic DNA, using primers XVIII (5′-GAAGTCGACGACACTCGTCGCGAGATCC-3′) and XIX (5′-GAAGGATCCAAGCTTCACTACTGCCCTAAGGTCTGACC-3′), and ligated to pGEM-T generating pGEM-TkmtR.Pcdf (159 bp) was amplified using primers XX (5′-GAAAAGCTTGGTGATCGTCGTCCGTCC-3′) and XXI (5′-GAAGGATCCATCGGGCGCAGGCCCTTTG-3′) and ligated into the HindIII/BamHI site of pGEM-TkmtR, immediately downstream of kmtR, generating pGEM-TkmtR-Pcdf. The kmtR and Pcdf sequences were then released and ligated into the ScaI/BamHI site of pJEM15 (32) creating pJEM15kmtR-Pcdf. QuikChange mutagenesis (Stratagene) was subsequently used to generate derivatives of pJEM15kmtR-Pcdf with codon substitutions in kmtR: Met24 to a UAG stop codon; Glu41, Glu101, His88, His102, His110, and His111 to Gln, Asp95 to Ala, and Cys16 to Ser. To generate a construct with kmtR fused to lacZ (pJEM15kmtR), QuikChange was used to introduce a BamHI site immediately after the kmtR stop codon in pJEM15kmtR-Pcdf, Pcdf was released, and the construct was re-ligated. Constructs pJEM15kmtR-tT4-Pcdf and pJEM15kmtR-tT4-PRv0286 were also generated containing the transcriptional terminator of coliphage T4 (tT4) immediately downstream of kmtR. For the former, tT4 was amplified using primers XXII (5′-GCCAAGCTTATGACCTTTAATAGATTATATTACTAATTAATTGGGGACCCTAGAGGTC-3′) and XXIII (5′-GCCAAGCTTTATGCTTGTAAACCG-3′) with pJEM15 as template and ligated into the HindIII site of pGEM-TkmtR-Pcdf, between kmtR and Pcdf. The kmtR, tT4, and Pcdf sequences were then released and ligated into the ScaI/BamHI site of pJEM15. For pJEM15kmtR-tT4-PRv0286, an SnaBI site was introduced at the 5′-end of the kmtR sequences in pGEM-TkmtR, creating pGEM-TkmtR2, and a fragment containing tT4 and PRv0286 was amplified from M. tuberculosis DNA with a primer incorporating the tT4 sequence (5′-GCCTACGTAAAGCTTATGACCTTTAATAGATTATATTACTAATTAATTGGGGACCCTAGGGTCCCCTTTTTTATTTTAAAAATTTTTTCACAAAACGGTTTACAAGCATAAAGGGGACACCGGACCAGCGG-3′) and primer XIV, and ligated into the HindIII/BamHI site of pGEM-TkmtR2, immediately downstream of kmtR. The DNA fragment containing kmtR, tT4, and PRv0286 was released and ligated into the ScaI/BamHI site of pJEM15. Derivatives of pJEM15kmtR, pJEM15kmtR-tT4-Pcdf, and pJEM15kmtR-tT4-PRv0286 were also generated in which Met24 within kmtR was substituted with a UAG stop codon by QuikChange. To construct an MtsmtB-lacZ fusion, M. tuberculosis DNA was used as template with primers 5′-GAAGATATCACTCCCTTCGAGGGATCG-3′ and 5′-GAAGGATCCGGACACCGGCTGCACTC-3′, and the amplification product was ligated to pGEM-T prior to subcloning into the ScaI/BamHI site of pJEM15. The lacZ fusion constructs were introduced into M. smegmatis mc2155 or M. bovis BCG. M. smegmatis mc2155 containing nmtR and the nmtA operator-promoter in pJEM15 (13) was also used to examine expression from PnmtA. β-Galactosidase assays were performed as described (13) in triplicate on at least three separate occasions. The medium was supplemented with various concentrations of metals (described in individual experiments) for ~20 h immediately prior to assays. The metal salts used were ZnSO4, CoSO4, NiSO4, CdCl2, CuSO4, Pb(NO3)2, AgNO3, Bi(NO3)2, MnCl2, and FeSO4.
Sequence Analysis of ArsR-SmtB Family Sensors
The presence or absence of metal-binding ligands at known sensory site locations was previously determined (30) for the 1024 protein sequences in the HTH_5 family of the Pfam data base. The CLANS program (35), which performs sequence clustering based on pairwise sequence similarities established by BLAST matches, was used to exclude proteins from this group that possessed least similarity and were unlikely to sense metal ions (at least by ligands at known sensory site locations). This was achieved by using gradually lower threshold BLAST E-values (from 10−1 to 10−15) as long as excluded sequences did not possess any known (30) metal binding pattern. A dendrogram of the resulting sequences was constructed from a ClustalW alignment and displayed in a tree-like representation using DRAWTREE from the PHYLIP package (36).
RESULTS
Sensory Sites Are Dispersed among Eight ArsR-SmtB Groups
Candidate metal sensory motifs associated with predicted elements of secondary structure have previously (30) been documented for 1024 deduced ArsR-SmtB sequences. Do the sensory motifs match sequence similarities? This family of proteins is heterogeneous and includes some single domain and multidomain proteins sharing relatively low sequence similarity. The initial ensemble was therefore reduced to 554 sequences, using the CLANS program, to exclude proteins with weakest similarity. The resulting consensus sequence derived from a ClustalW alignment has 57% identity and 73% similarity with an alignment of reference (30) built with known ArsR-SmtB family metal sensors, confirming the consistency of the ensemble. A tree diagram constructed from the alignment (Fig. 1) reveals eight major sequence groups. It is noted that, although the tree diagram provides a representation of sequence similarity among aligned sequences, it is not a phylogenetic tree and should not be interpreted as such. Importantly, several metal-binding motifs map to multiple sequence groups, and several groups contain proteins with more than one metal-binding motif. This strongly argues against a dogma in which proteins with overall sequence similarity to ArsR are presumed to sense arsenite, those similar to CadC cadmium, because the ArsR and CadC sensory motifs are dispersed across different sequence groups, interspersed with other sensory motifs.
FIGURE 1. Tree diagram constructed from the alignment of 554 sequences derived from the HTH_5 family of the Pfam data base.
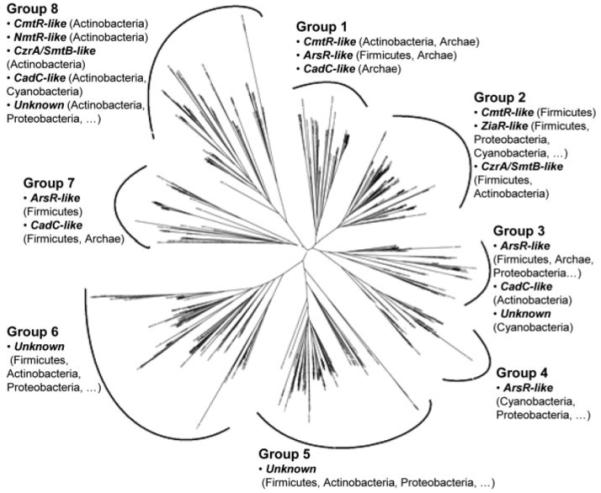
The tree diagram shows eight major groups. The main metal binding motifs occurring in proteins within each group are indicated: CmtR-like sequences possess the CXXXC metal-binding pattern in the α4 helix and potential ligands in the carboxyl-terminal region; CzrA/SmtB-like and NmtR-like possess the pattern DXHX(10)HXX(E/H) in the α5 helix, with the latter also possessing carboxyl-terminal ligands; ArsR-like possess either one or more of (i) CXCXXC, (ii) CXC, or (iii) CXXD in the α3 helix; CadC-like and ZiaR-like possess either one or more of i, ii, and iii in addition to potential ligands in the amino-terminal region, with ZiaR-like also possessing the DXHX(10)HXX(E/H) pattern in α5; the unknown lack these patterns. For each motif, the main bacterial phyla are shown in parentheses (a series of periods indicates sequences are also present from other phyla), and sequences with the motif may be from more than one branch (sub-group).
Sequences with no defined metal-binding motif (unknowns) are present in four groups (3, 5, 6, and 8). KmtR from M. tuberculosis is an unknown of group 8. There are no clues as to the metals sensed by KmtR or the identities of its promoter targets.
KmtR Binds a Region within Its Own and the Rv2025c, cdf, Operator-Promoter Regions
Which genes are regulated by KmtR? We constructed a mutant of M. tuberculosis lacking the kmtR coding region and profiled transcript abundance. Expression of only two open reading frames, Rv2025c (genome location 2270.750 to 2271.748 kb) and Rv0826 (adjacent to kmtR, Fig. 2A), is significantly altered in ΔkmtR relative to wild type with a 53- and 15-fold increase in transcript abundance, respectively. Fully annotated microarray data has been deposited in the Bacterial Microarray Group at St George’s data base (accession Number: E-BUGS-49) and ArrayExpress (E-BUGS-49) data base. The deduced product of Rv2025c shares sequence similarity with transmembrane cation diffusion facilitator (CDF) family metal ion transporters, whereas the deduced product of Rv0826 is a conserved hypothetical cytoplasmic protein lacking similarity to previously characterized proteins. The operator-promoter regions of Rv2025c (hereafter referred to as cdf) and Rv0826 therefore represent candidates for KmtR binding. In addition, by analogy to other ArsR-SmtB regulators, KmtR may also repress expression from its own operator-promoter region, which would not be detectable in the array experiments performed with ΔkmtR.
FIGURE 2. KmtR binds to the kmtR and cdf operator promoter regions.
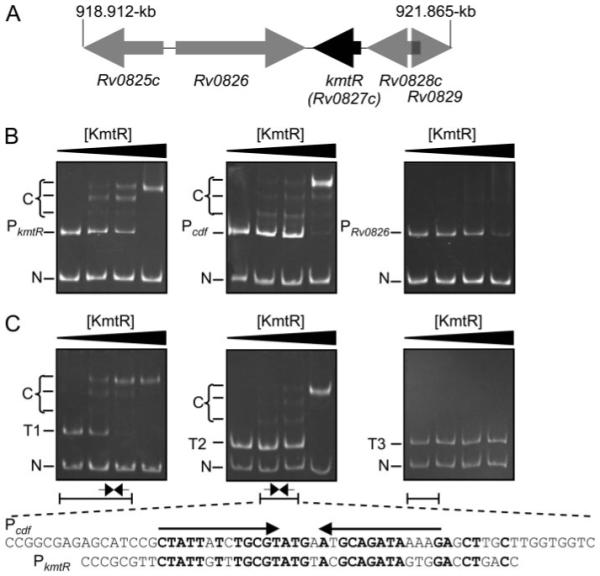
A, physical map of the kmtR gene region and position in the M. tuberculosis genome (vertical lines). B, gel-retardation assays used (from left to right) 0, 0.25, 0.5, and 1 μm KmtR with a 310-bp DNA fragment containing PkmtR (including the first 18 codons of kmtR), a 292-bp DNA fragment containing Pcdf (including the cdf start codon), or a 292-bp DNA fragment containing PRv0286 (including the first 5 codons of Rv0826) as probe and a 136-bp fragment of nonspecific competitor DNA (N). The latter contained identical sequences to the probes but lacks the operator-promoter regions. C, gel-retardation assays used (from left to right) 0, 0.5, 1, and 2 μm KmtR with probes T1 (241 bp), T2 (198 bp), or T3 (190 bp) and nonspecific competitor DNA. Predominant complexes (C) are indicated. Diagrammatic representations of the Pcdf sequences within T1, T2, and T3 are shown with the position of a degenerate 13-4-13 hyphenated inverted repeat. The sequence of the Pcdf fragment within T2 is shown in full, with arrows indicating the inverted repeat, with a similar repeat identified within PkmtR (conserved nucleotides are shown in bold).
KmtR was expressed as a non-fusion protein, purified, and used in gel-retardation assays with DNA fragments containing the operator-promoter regions of kmtR, cdf, and Rv0826. Retarded complexes were formed with probes containing PkmtR and Pcdf and increasing concentrations of KmtR (Fig. 2B), with a concomitant decrease in the amount of free probe but no shift of nonspecific competitor DNA, demonstrating specific binding of KmtR to sequences within PkmtR and Pcdf. More than one retarded band was detected with each promoter indicative of high order KmtR complexes forming with DNA. No specific complexes were detected with PRv0286, although a slight decrease in the amount of free probe, but not control DNA, was noted at the highest concentration of KmtR (Fig. 2B). To map the KmtR·DNA binding site, assays were repeated using probes containing truncations (T1–T3) of Pcdf. Retarded complexes were detected with T1 and T2 but not with T3 (Fig. 2C), confirming KmtR binding to sequences contained within a 62-bp DNA fragment that includes a degenerate 13-4-13 hyphenated-inverted repeat. A similar repeat was detected within PkmtR, overlapping the kmtR start codon (Fig. 2C), but not within PRv0286. Furthermore, a search of the M. tuberculosis genome with the consensus inverted repeat (5′-CTATTNTNTGCGTNNNNANGCAGATANNNG-3′) exclusively identified the cdf and kmtR operator-promoter regions.
Formation of KmtR·DNA complexes were monitored by fluorescence anisotropy with fluorescein-labeled oligonucleotides corresponding to 32 and 34 bp of PkmtR and Pcdf, respectively, centered on the identified inverted repeat sequences in Fig. 2C and to a 32-bp region of PRv0286 containing a degenerate 12-2-12 hyphenated inverted repeat. A decrease in rotation and increase in polarization and robs, with Pcdf and PkmtR sequences, confirmed the formation of KmtR·DNA complexes, with KKmtR calculated to be 1.2 × 10−7 m and 2.4 × 10−7 m, respectively, based on a 1:1 model (Figs. 3, A and B). It is noted that the DNA binding isotherm must be a complex function of more than one protein oligomerization event, with more than one DNA affinity, as also evidenced by the multiple bands in gel retardation assays (Fig. 2), and Δrobs ~ 0.1 is consistent with five dimers binding (25). No increase in robs with PRv0286 sequences (Fig. 3C) is consistent with no specific binding of KmtR to PRv0286. Rv0826 is located adjacent, but convergent, to kmtR within the M. tuberculosis genome (Fig. 2A). It is therefore possible that kmtR transcription interferes with Rv0826 transcription, and hence the increased Rv0826 expression in ΔkmtR relative to wild-type cells, detected in microarray experiments, is a result of the local changes in DNA structure and loss of kmtR transcription in ΔkmtR. These data (Fig. 3) are thus wholly consistent with the outcome of gel-retardation assays (Fig. 2).
FIGURE 3. KmtR binding to PkmtR and Pcdf as measured by fluorescence anisotropy.
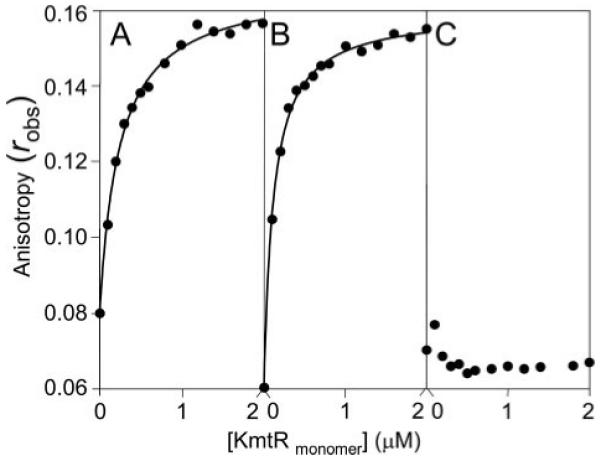
6-Hexachloroflourescein-labeled PkmtR 32-mer (A), Pcdf 34-mer (B), or PRv0286 32-mer (C) at 5 nm were titrated with KmtR and anisotropy, robs, measured. The curves were calculated based on a 1:1 model using SigmaPlot, KKmtR = 0.24 μm and 0.12 μm for PkmtR and Pcdf, respectively.
No Metal Enhances Expression from a KmtR-regulated Promoter in Complete Medium
To determine which, if any, metals are sensed by KmtR and induce transcription from a KmtR-regulated promoter, β-galactosidase activity was measured in M. smegmatis cells containing kmtR and the cdf operator-promoter region fused to lacZ in plasmid pJEM15 and grown (20 h) in standard Middlebrook 7H9 medium supplemented with maximum permissive concentrations of a broad range of different metal ions. No substantial increase in β-galactosidase activity was detected in response to any of the metal ions tested (Fig. 4A), although an unexpected reduction in activity was detected in cells exposed to Zn(II) or Cu(II). Expression from Pcdf was therefore examined in response to a range of concentrations of Zn(II), up to inhibitory levels. Reduced β-galactosidase activity was detected in response to increasing Zn(II) concentrations (Fig. 4B) below inhibitory levels (inset in Fig. 4B). A similar trend is observed with copper (not shown). PAR, 4-(2-pyridylazo)-resorcinol, has been used previously to detect nanomolar to picomolar Zn(II)-protein dissociation constants, because, under conditions of surplus PAR (37), a PAR2·Zn(II) complex is formed, which absorbs at 500 nm (Δε = 66,000 m−1 cm−1) with an overall conditional stability constant documented to be 3.85 × 1012 m−2. Purified KmtR was capable of withholding Zn(II) from PAR (Fig. 4C).
FIGURE 4. No metal enhances expression from a KmtR-regulated promoter in a mycobacterium grown in Middlebrook 7H9 medium.
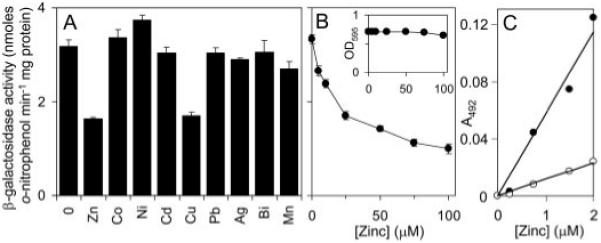
A and B, β-galactosidase activity was measured in M. smegmatis mc2155 containing kmtR and Pcdf fused to lacZ (in pJEM15kmtR-Pcdf) grown with no metal supplement and maximum permissive concentrations of Zn(II) (70 μm), Co(II) (15 μm), Ni(II) (35 μm), Cd(II) (75 nm), Cu(II) (50 μm), Pb(II) (10 μm), Ag(I) (0.75 μm), Bi(III) (0.75 μm), or Mn(II) (0.75 μm) (A), or up to inhibitory [Zn(II)] (B). Inset, growth (A595) of cultures against added [Zn(II)]. C, microtiter plate assays of Zn(II)-PAR formation (400 μm PAR), measured at 492 nm and zeroed against apo-PAR, as a function of [Zn(II)] in the absence (closed symbols) or presence (open symbols) of KmtR (2 μm).
Zinc Only Alleviates MtSmtB-mediated Repression in Minimal Medium
The product of M. tuberculosis Rv2358 (herein designated MtSmtB) acts as a Zn(II)-responsive repressor of its own expression in mycobacterial cells (15). The unexpected Zn(II)-mediated repression of expression from Pcdf encouraged us to further characterize MtSmtB for subsequent use as a control in studies examining metal-responsiveness of KmtR. Consistent with previous findings (15), purified MtSmtB retarded a DNA fragment that included 30 bp from the MtsmtB operator-promoter region with the formation of a single complex (supplemental Fig. S1). The effects of Zn(II) on DNA binding were examined by fluorescence anisotropy, in which fluorescently labeled oligonucleotides corresponding to the MtSmtB·DNA binding site (30 bp, centered on a hyphenated inverted repeat) were titrated with MtSmtB in the presence of a molar excess of EDTA or Zn(II), the latter to ensure saturation of the Zn(II) sites, under anaerobic conditions. An increase in mass due to MtSmtB·DNA complex formation was detected with apo-MtSmtB in the presence of EDTA (Fig. 5A), and Δrobs ~ 0.02 is consistent with binding of a single dimer. No increase was detected with Zn(II)·MtSmtB (Fig. 5A), confirming that Zn(II) inhibits MtSmtB·DNA binding. Zn(II) association with MtSmtB was confirmed using the metallochromic indicator PAR. The association of Zn(II) (up to 2 μm) with PAR (400 μm) was monitored in the absence or presence of MtSmtB (2 μm) (Fig. 5B). Zn(II)-dependent PAR absorbance is reduced by the addition of MtSmtB.
FIGURE 5. MtSmtB binds its own operator-promoter and responds to zinc in cells grown in Chelex-treated Sauton medium.
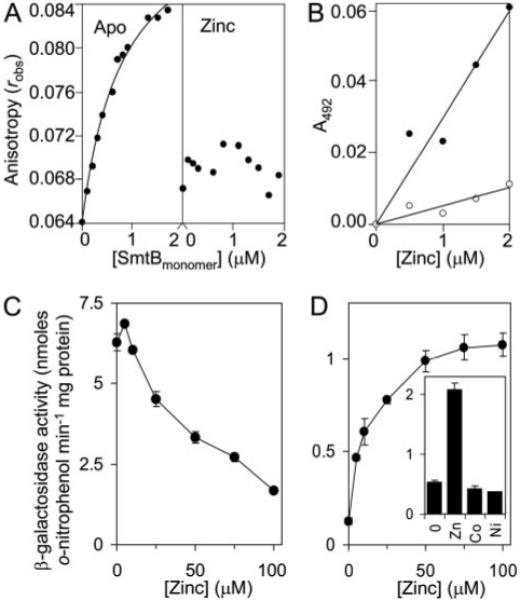
A, MtSmtB (50 μm) was incubated overnight with either 1 mm EDTA (left) or 20 μm Zn(II) (right) and increasing concentrations added to 6-hexachloroflourescein-labeled PMtSmtB 30-mer (5 nm) in the presence of 1 mm EDTA or 35 μm ZnCl2 (final [Zn(II)] = 35–35.84 μm), respectively, under anaerobic conditions and anisotropy, robs, measured. The curve represents a 1:1 binding model, KMtSmtB = 0.81 μm. B, microtiter plate assays of Zn(II)-PAR formation (400 μm PAR), measured at 492 nm and zeroed against apo-PAR, as a function of [Zn(II)] in the absence (closed symbols) or presence (open symbols) of MtSmtB (2 μm). C and D, β-galactosidase activity measured in M. smegmatis mc2155 containing MtsmtB-regulated lacZ grown up to inhibitory [Zn(II)] in Middlebrook 7H9 medium (C) or metal-depleted Sauton medium (D). Inset, β-galactosidase activity in cells grown with no metal supplement or with maximum permissive concentrations of Zn(II) (70 μm), Co(II) (15 μm), or Ni(II) (30 μm).
To examine Zn(II) responsiveness of MtSmtB in mycobacterial cells, a 624-bp DNA fragment, including the MtsmtB operator-promoter region and the entire MtsmtB coding region, were fused to a promoterless lacZ in plasmid pJEM15 and introduced into M. smegmatis. β-Galactosidase activity was measured following growth (~20 h) of these cells in Middlebrook 7H9 medium supplemented with Zn(II) up to inhibitory levels. Counter to expectations, no substantial increase in expression was detected at any viable concentration of Zn(II), whereas a reduction in expression from PMtsmtB was detected in medium supplemented with >10 μm Zn(II) (Fig. 5C). Maximal expression of MtsmtB (Rv2358) was previously (15, 38) observed at ~100 μm Zn(II), but notably, these studies had used M. smegmatis cells grown under metal-limiting conditions prior to assay. We therefore re-examined the expression from PMtsmtB in cells grown for several passages in Chelex-treated Sauton minimal medium prior to exposure to a range of concentrations of Zn(II), up to inhibitory levels (100 μm). Under these conditions, expression from PMtsmtB was substantially reduced in the absence of added Zn(II), but increases of ~8-fold in response to Zn(II) with maximal expression were detected at 100 μm (Fig. 5D). No increase in expression from PMtsmtB was detected with Co(II) or Ni(II), at maximum permissive concentrations (Fig. 5D, inset), indicating specificity of MtSmtB for Zn(II).
Nickel and Cobalt Alleviate KmtR-mediated Repression in Minimal Medium
Having established that Zn(II)-induced transcription from PMtsmtB is only detectable in cells that have been “starved” of divalent metal ions prior to assay, metal-responsive expression from a kmtR-regulated promoter (Pcdf) was re-examined in M. smegmatis following growth in Chelex-treated Sauton minimal medium. Then, elevated β-galactosidase activity was detected in response to exposure (~20 h) to maximum permissive concentrations of Co(II) or Ni(II) but no other metals (Fig. 6A). Furthermore, in the absence of added metal ions, elevated β-galactosidase activity was detected from an analogous construct in which codon Met24 within the kmtR coding region was converted to a stop codon (Fig. 6B), confirming that KmtR acts negatively toward expression. However, a reduction in β-galactosidase activity was detected from this construct in cells grown in 7H9 medium supplemented with increasing levels of Zn(II) or Cu(II) (data not shown), demonstrating that loss of expression from Pcdf in the presence of elevated levels of these metal ions is independent of KmtR. The inhibitory effects of Zn(II) and Cu(II) on Pcdf (and PMtsmtB) expression therefore most likely relate to metal toxicity causing some reduction in transcriptional and/or translational activity in these cells or directly inhibiting β-galactosidase. Such an effect is most apparent in conditions where promoter expression is de-repressed.
FIGURE 6. KmtR responds to cobalt and nickel in a mycobacterium grown in Chelex-treated Sauton medium.
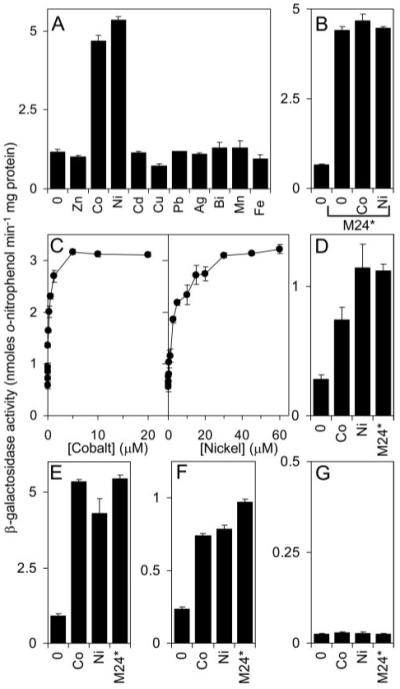
A–C, β-galactosidase activity measured in M. smegmatis mc2155 containing kmtR and Pcdf fused to lacZ (in pJEM15kmtR-Pcdf) or the stop codon derivative (M24*) following growth in medium with no metal supplement or maximum permissive concentrations of Zn(II), Co(II), Ni(II), Cd(II), Cu(II), Pb(II), Ag(I), Bi(III), Mn(II), or Fe(III) (A and B), or up to inhibitory concentrations of Co(II) or Ni(II) (C). D, β-galactosidase activity measured in M. bovis BCG containing pJEM15kmtR-Pcdf grown with no metal supplement or with maximum permissive concentrations of Co(II) or Ni(II), or containing the stop codon derivative (M24*) and grown with no metal supplement. E–G, β-galactosidase activity measured in M. smegmatis mc2155 containing kmtR (in pJEM15kmtR) (E), kmtR and Pcdf separated by tT4 (in pJEM15kmtR-tT4-Pcdf) (F), or kmtR and PRv0286 separated by tT4 (in pJEM15kmtR-tT4-PRv0286) (G), fused to lacZ, grown with no metal supplement or with maximum permissive concentrations of Co(II) or Ni(II), or in cells containing the stop codon derivatives of these constructs (M24*) and grown with no metal supplement.
Exposure of metal-starved M. smegmatis to a range of concentrations of Co(II) and Ni(II) (Fig. 6C) revealed KmtR-mediated repression is alleviated by these metals in a concentration-dependent manner with maximal expression detected at ~5 μm Co(II) and ~30 μm Ni(II). KmtR-mediated repression was similarly alleviated by elevated concentrations of Co(II) and Ni(II) in the non-pathogenic (attenuated) endogenous BCG host after growth in the minimal medium (Fig. 6D).
To determine whether KmtR acts similarly at the PkmtR and Pcdf promoters, β-galactosidase activity was examined in cells containing the kmtR operator-promoter region and coding region fused to lacZ in plasmid pJEM15 (which will reveal activity from PkmtR) and in cells containing these sequences but with a transcriptional terminator, tT4, and Pcdf located between kmtR and lacZ, thus enabling expression from Pcdf to be measured while ensuring no transcriptional read-through from kmtR. In both cases, elevated expression was observed in cells grown in the minimal medium in response to Co(II) and Ni(II) (Fig. 6, E and F) and in the absence of added metal ions elevated expression is observed in cells containing equivalent constructs with a stop codon introduced into the kmtR coding region. KmtR therefore acts as a Co(II)- and Ni(II)-responsive repressor at both PkmtR and Pcdf. In contrast, expression from PRv0286 remained low in cells exposed to maximum permissive concentrations of Co(II) or Ni(II) and in cells lacking functional kmtR (Fig. 6G), consistent with a lack of KmtR binding to this promoter (Figs. 2B and 3C).
Association of apo-KmtR or pre-formed Co(II)-KmtR with DNA (fluorescent PkmtR 32-mer) showed a large increase in robs upon titration with apo-KmtR, whereas Co(II) inhibited the formation of these DNA complexes (Fig. 7A). The large Δrobs is consistent with formation of high order apo-KmtRn·DNA complexes. Metal-KmtR binding was investigated by competition against PAR. The extinction coefficient for Co(II)-PAR was too small to allow reliable analysis of Co(II) binding (data not shown), but formation of a PAR2·Ni(II) complex (39) was readily detected at 492 nm (Fig. 7B). KmtR withholds Ni(II) from PAR preventing the increase in Ni(II)-dependent PAR absorbance, confirming the formation of metal (Ni(II))·KmtR complexes.
FIGURE 7. Cobalt destabilizes KmtR·DNA complexes.
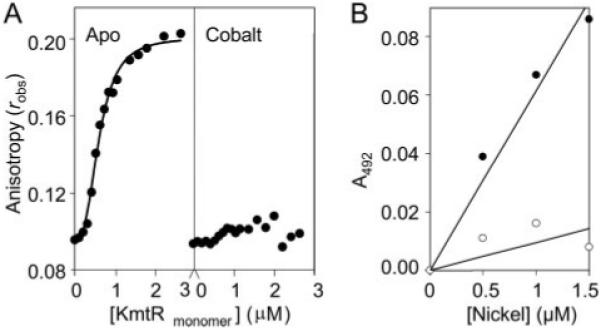
A, KmtR (13.3 μm) was incubated overnight with either 1 mm EDTA (left) or 35 μm Co(II) (right), increasing concentrations were added to 6-hexachloroflourescein-labeled PkmtR 32-mer (5 nm) in the presence of 1 mm EDTA or 35 μm CoCl2 (final [Co(II)] = 35–42 μm), respectively, under anaerobic conditions, and anisotropy, robs, was measured. In the presence of EDTA it is apparent that these data do not fit a 1:1 binding model, which is consistent with a large Δrobs implying binding of multiple dimers; the curve represents a sigmoidal fit, KKmtR = 0.6 μm. B, microtiter plate assays of Ni(II)-PAR formation (400 μm PAR), measured at 492 nm and zeroed against apo-PAR, as a function of [Ni(II)] in the absence (closed symbols) or presence (open symbols) of KmtR (2 μm).
Contrasting the Nickel-Cobalt Sensors NmtR and KmtR
We have previously shown (13) that NmtR is an ArsR-SmtB Ni(II)-Co(II) sensor in M. tuberculosis. The nmtR gene is located remote from kmtR at position 4195.440–4195.802 kb in the M. tuberculosis genome. Why does this organism possess two such sensors? A more parsimonious solution would be to possess one sensor that acts at all of the target promoters unless the two sensors differ in some other property, such as sensitivity or interactions with metal donors, perhaps. The responses of KmtR and NmtR to a range of concentrations of Ni(II) and Co(II), up to and including inhibitory doses, were compared (Fig. 8). Expression from the NmtR-regulated operator-promoter region (PnmtA) was low in cells grown in Middlebrook 7H9 medium with no metal supplement and was substantially increased in response to elevated concentrations of Co(II) and Ni(II). In contrast, expression from the KmtR-regulated promoter was highly elevated in cells grown with no metal supplement and was unaffected by elevated concentrations of Co(II), whereas a slight increase in expression was detected with non-inhibitory concentrations of Ni(II) (≤30 μm). This indicates that KmtR-mediated repression is alleviated in the complex medium while NmtR-mediated repression is not. With analogy to KmtR, NmtR was responsive to both Co(II) and Ni(II) in cells grown in Chelex-treated Sauton medium (data not shown). Hence the responses of KmtR and NmtR to Ni(II) and Co(II) differ under the two sets of different growth conditions.
FIGURE 8. Comparison of KmtR and NmtR metal responsiveness.
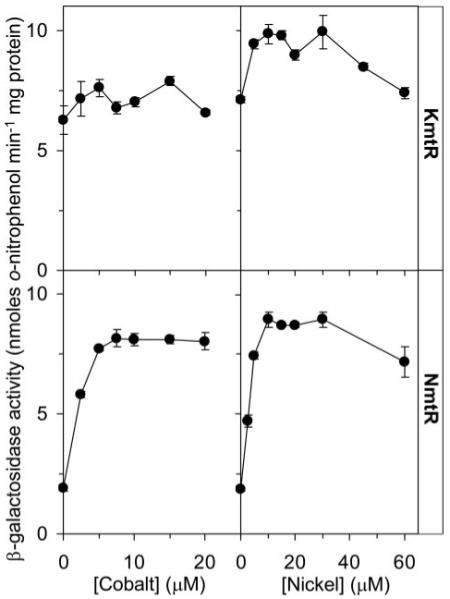
β-Galactosidase activity in M. smegmatis mc2155 cells containing KmtR-regulated (upper) or NmtR-regulated (lower) lacZ grown up to inhibitory [Ni(II)] or [Co(II)].
An explanation for the regulatory effects of growth conditions is that KmtR was more sensitive to Ni(II) and Co(II) than NmtR such that basal metal concentrations in complete medium allowed de-repression of KmtR target promoters. A tighter Co(II) and Ni(II) affinity of KmtR versus NmtR is one plausible basis for different sensitivities. NmtR was expressed and purified, and an Ni(II) binding isotherm was generated by monitoring tyrosine fluorescence (data not shown) replicating results reported previously (13) and showing saturation of 5 μm NmtR upon addition of 5 μm Ni(II). KmtR contains a single tryptophan residue, giving more intense fluorescence than NmtR and greater quenching upon Ni(II) binding, allowing the generation of a Ni(II) binding isotherm for KmtR (Fig. 9A). Solutions containing 5 μm KmtR also showed saturation upon addition of ~5 μm Ni(II), and therefore binding was too tight to allow measurement of KNi under these conditions; hence, the curve has not been modeled, but KNi must be tighter than 2.5 μm. By increasing λex to 295 nm, NmtR tyrosine fluorescence became negligible, whereas substantial KmtR tryptophan fluorescence was retained. A Ni(II)-dependent difference emission spectrum (Fig. 9B) showed that the magnitude of quenching of KmtR tryptophan fluorescence upon addition of 0.8 equivalents of Ni(II) was similar in the presence or absence of NmtR. Thus, KmtR had the greater affinity for Ni(II). Co(II) binding isotherms were similarly obtained (Fig. 9C) with the curve representing 1:1 (metal:monomer) model for Co(II)·KmtR. In a Co(II)-competition experiment KmtR showed similar quenching in the presence or absence of NmtR (Fig. 9D). Notably, and unlike Ni(II), when 0.8 equivalent of Co(II) was added to 5 μm KmtR, the protein was <80% saturated (Fig. 9C), and it remained possible that KCo for KmtR and NmtR was within an order of magnitude. Taken together, these data reveal that KmtR has tighter affinities for Ni(II) and for Co(II) than does NmtR.
FIGURE 9. KmtR has tighter affinity for nickel and cobalt than NmtR.

A, Ni(II) binding isotherm for KmtR (5 μm) monitored as tryptophan fluorescence (λex = 280 nm, λem = 440 nm). KNi is too tight to measure under these conditions. B, competition between KmtR (5 μm) and NmtR (5 μm) for Ni(II) (4 μm) monitored as tryptophan emission spectra following excitation at 295 nm. Only KmtR contains a tryptophan residue. Ni(II)-dependent difference spectra are shown for KmtR (5 μm) alone (open circles) and KmtR in the presence of equimolar NmtR (closed circles). The lower curve (triangles) shows the negligible Ni(II)-dependent difference spectrum for tyrosine residues of NmtR (5 μm) alone at this excitation wavelength. C, Co(II) binding isotherm for KmtR monitored as tryptophan fluorescence (λex = 280 nm, λem = 440 nm). The curve represents a 1:1 ligand binding model, which under these conditions (including 1 mm DTT) gives KKmtR = 6.9 μm. KNmtR under these conditions is estimated to be weaker than KKmtR and weaker than we reported previously in the absence of DTT (13). D, competition between KmtR (5 μm) and NmtR (5 μm) for Co(II) (4 μm) monitored as tryptophan emission spectra following excitation at 295 nm. Co(II)-dependent difference spectra are shown for KmtR (5 μm) alone (open circles) and KmtR (5 μm) in the presence of equimolar NmtR (closed circles). The lower curve (triangles) shows the negligible Co(II)-dependent difference spectrum for tyrosine residues of NmtR (5 μm) alone at this excitation wavelength. Conditions are 20 mm HEPES, 1 mm DTT, 50 mm NaCl (pH 7.5), 22 °C.
Identification of Residues Essential for Nickel and Cobalt Recognition in Vivo
Some difference in the effector recognition site of KmtR compared with NmtR might alter its sensitivity to Ni(II) and Co(II) or otherwise change its access to these metals within the cytosol. NmtR senses Co(II) and Ni(II) by forming six-coordinate complexes at an α5C site (13, 40). However, KmtR lacks residues corresponding to a typical α5C site or any other previously defined ArsR-SmtB family metal-binding motif (Fig. 1). The next challenge was therefore to identify the inducer recognition site of KmtR. Metal sensing by many ArsR-SmtB family members involves Cys. KmtR possesses a single Cys residue within the deduced α1 helix. Ser substitution of Cys16 did not impair either KmtR-mediated repression or Ni(II) and Co(II) recognition in mycobacterial cells grown in Chelex-treated Sauton medium (Fig. 10).
FIGURE 10. Metal sensing site of KmtR.
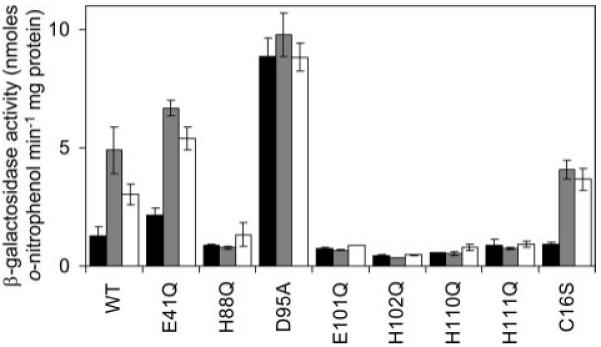
β-Galactosidase activity was measured in M. smegmatis containing wild-type KmtR (WT) and various derivatives with indicated codon substitutions. Cells were grown in Chelex-treated Sauton medium with no metal supplement (black) or maximum permissive [Ni(II)] (gray) or [Co(II)] (white).
KmtR is located within a sub-group of group 8 of the tree diagram (Fig. 1) formed by ten proteins, all from Actinobacteria, which lack typical metal binding patterns. It is noted that one of these sequences, from Corynebacterium glutamicum ATC13032, appears to be represented twice (EMBL-EBI accession numbers Q6M276 and Q8NM04) due to different start codon assignments. An alignment of the nine remaining sequences (supplementary Fig. 2) reveals a conserved Glu (Glu41 in KmtR) in the deduced α3 helix region and a number of conserved residues in the deduced α5 helix (His88, Asp95, Glu101, and His102) and carboxyl-terminal region (His110 and His111) that could function as metal-binding ligands. Substitution of Glu41 with Gln causes slightly elevated expression from Pcdf in mycobacterial cells grown in Chelex-treated Sauton medium with no metal supplement, consistent with some loss of repressor function (Fig. 10). However, expression was highly elevated in cells exposed to Ni(II) and Co(II) demonstrating that inducer responsiveness by KmtR either did not involve α3/α3N ligands or Glu41 was not an essential α3/α3N ligand. Substitution of His88, Glu101, His102, His110, and His111 with Gln also created functional repressors that mediated low expression of lacZ from Pcdf in cells grown with no metal supplements (Fig. 10). Most significantly, β-galactosidase activity was not elevated at Ni(II) and Co(II) concentrations that cause loss of repression by wild-type KmtR and also by the Glu41 and Cys16 substituted mutants (Fig. 10). These results demonstrate that at least His88, Glu101, His102, His110, and His111 are obligatory for Ni(II) and Co(II) recognition. Asp95 (essential for repression (Fig. 10)) is a potential sixth ligand. Thus the KmtR metal-sensing ligands are located around predicted helix α5 but differ from the two sites at α5 helices identified previously (α5 and α5C), and hence are designated α5-3.
DISCUSSION
A second DNA-binding (Figs. 2 and 3), Co(II)- and Ni(II)-sensing ArsR-SmtB transcriptional repressor (Fig. 6), KmtR, has been characterized from the intracellular pathogen M. tuberculosis. The residues required for KmtR metal responsiveness (Fig. 10) are distinct from NmtR and define a new metal-sensing motif (Fig. 11). More metal sensors can now be predicted from data base entries. Purified KmtR formed specific complexes in vitro with 32- and 34-bp regions of the cdf and kmtR operator-promoters, respectively, which contain a conserved 13-4-13 region of dyad symmetry. Inducing metals bound to KmtR in vitro (Figs. 7B and 9) and impaired DNA binding in vitro (Fig. 7A). Gene profiling experiments reveal elevated cdf transcript abundance in a kmtR mutant of M. tuberculosis. Furthermore, expression of β-galactosidase activity from the cdf and kmtR operator-promoter regions was repressed in mycobacterial cells containing functional kmtR grown in minimal medium but elevated when a stop codon was introduced within the kmtR open reading frame (Fig. 6). Elevated expression from KmtR-responsive promoters was observed in cells grown in complete medium (Fig. 8). In contrast, for the related sensor NmtR, repression and Ni(II)-Co(II) responsiveness were retained in complete medium (Fig. 8). Thus, KmtR and NmtR respond under different surplus cobalt and nickel conditions implying exquisitely subtle adaptation to excess amounts of these metals in the cytosol.
FIGURE 11. Sensory sites in ArsR-SmtB family members.
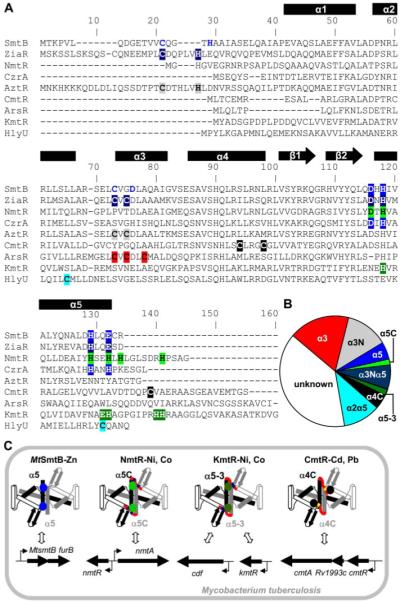
A, alignment of representative sequences for the different metal-sensing sites, involving ligands at an α5 site (blue) in cyanobacterial SmtB and CzrA (24-26); α3N and α5 (dark blue) in ZiaR (16); α5C (light green) in NmtR (13); α3N (gray) in AztR (19); α4C (black) in CmtR (14); α3 (red) in ArsR (27); and α5-3 (dark green) in KmtR. The hypothetical α2α5 site (pale blue) is also shown for HlyU. The α3N site in SmtB (residues in blue) is not required for metal responsiveness. B, chart showing the abundance of the various metal-sensing sites among the 554 ArsR-SmtB family representatives. Proteins designated α3Nα5 (ZiaR-like) possess both α5 and α3N sites, although in some cases (e.g. cyanobacterial SmtB and CadC) only one site may be required for metal-mediated allostery. C, representation of the four characterized ArsR-SmtB sensors in M. tuberculosis with effector binding sites for Zn(II) at α5 in dimeric MtSmtB (blue), for Ni(II) and Co(II) at α5C (light green) in NmtR and α5-3 (dark green) in KmtR, and for Cd(II) and Pb(II) at α4C (black) in CmtR. α-helices (boxes), β-strands (arrows), DNA-binding helix-turn-helix region (open boxes), and carboxyl-terminal extension (red) are indicated, the latter being absent from MtSmtB. Binding of their effectors inhibits DNA binding and alleviates repression of their target genes (horizontal arrows). Permutations in the effector binding sites allows detection of the different metals within the cytosol, whereas differences in the sensing ligands at α5C and α5-3 in NmtR and KmtR, respectively, may alter their affinities for Co(II) and Ni(II) allowing these proteins to respond under different surplus Co(II) and Ni(II) conditions.
KmtR had tighter affinities for nickel and cobalt than NmtR (Fig. 9). Hence, the different sensitivities of KmtR and NmtR in complete medium are most likely explained by KmtR, but not NmtR, sensing basal concentrations of nickel and/or cobalt leading to de-repression of KmtR target promoters. Similarly, elevated expression from PMtsmtB in complete medium most likely corresponds to MtSmtB sensing increased basal zinc levels in this medium. Notably, the level of nickel in both minimal and complete media was estimated to be ~50-fold lower than the level required for half-maximal expression from a KmtR-regulated promoter in minimal medium (KNi = 2.5 μm), whereas the cobalt content in complete medium was within an order of magnitude (KCo = 0.1 μm). Hence, accumulation of cobalt, and possibly nickel, in the context of elevated levels of other trace metals and altered expression of other genes is most likely to be triggering loss of KmtR-mediated repression in the complete medium. We conclude that differences in the effector binding sites at α5C and α5-3 in NmtR and KmtR (Fig. 11) allow these proteins to differentially regulate gene expression under different surplus cobalt and nickel concentrations such that as cytosolic levels increase KmtR detects these metals and confers metal export via a CDF-family transporter, whereas, only when a higher threshold of these metals is reached does NmtR detect these metals and confer export via the P1-type ATPase NmtA.
In humans, M. tuberculosis infects macrophages and lives inside their phagosomes. In this environment its survival is dependent on the activity of nickel-dependent urease, with urea hydrolysis contributing to nitrogen availability and environmental pH modulation (41). Furthermore, production of ammonia in this reaction can block phagosome-lysosome fusion providing further protection from host killing mechanisms (42). Both 7H9 (which contains nitrogen in the form of 3.8 mm ammonium sulfate and 3.4 mm glutamic acid) and Sauton (which contains 15 mm asparagine) media are considered nitrogen-rich (41), although urease activity of M. tuberculosis is easily measured in both (41) implying a requirement for urea hydrolysis under both sets of growth conditions. It is, however, tempting to speculate that a difference in urease activity in cells grown in the different media modifies availability of these ions to the surplus nickel sensors. Differing interaction with a nickel-metallochaperone for urease could be a factor. Cobalt is also required by this organism for the biosynthesis of vitamin B12 (43). Acquisition of cobalt for vitamin B12 synthesis may also contribute to M. tuberculosis survival in macrophages. Vitamin B12-binding protein is a component of neutrophil-specific granules, which can be acquired by macrophages, due to phagocytosis of apoptotic neutrophils, to assist in killing M. tuberculosis (44).
Metal discrimination by KmtR and MtSmtB is completely inverted; KmtR repression is alleviated by Ni(II) and Co(II) but not Zn(II) (Fig. 6), whereas MtSmtB detects Zn(II) but fails to respond to Co(II) or Ni(II) (Fig. 5). Intrinsic features of these proteins must allow discrimination of these metals in the mycobacterial cytosol. In common with Zn(II)-sensing SmtB from Synechococcus PCC 7942, MtSmtB possesses residues expected to form a tetrahedral α5 site (Asp116, His118, His129, and Glu132). KmtR can bind Zn(II) in vitro (Fig. 4C), but the sensory ligands located around helix α5 differ from SmtB or any other sites previously identified (α5or α5C). Six ligands are available in the metal coordination sphere of NmtR (13, 40), which is ideal for Ni(II) and Co(II) that prefer a higher coordination number. Five ligands (His88, Glu101, His102, His110, and His111) are critical for inducer recognition by KmtR, whereas a sixth (Asp95) is required for repressor function and hence cannot be excluded as a potential ligand (Fig. 10). Present predictions therefore support five- or six-coordinate Co(II) and Ni(II) liganding derived from the α5-helix and carboxyl-terminal region of KmtR. By analogy to discrimination against Zn(II) by NmtR (40), Zn(II) is expected to only bind a sub-set of KmtR ligands and inefficiently trigger allostery.
KmtR is the fourth M. tuberculosis ArsR-SmtB metal sensor to be characterized. The possession of multiple related metal sensors in a single organism provides advantages for exploring the rules governing metal selectivity in vivo. So far, each characterized sensor from M. tuberculosis uses a distinct effector-binding site; α4C CmtR, α5 MtSmtB, α5C NmtR and α5-3 KmtR (Fig. 11C). Eight further M. tuberculosis sensors are present within the Pfam data base HTH_5 family, although three (Rv1460, Rv0324 and Rv1674c) were excluded in the CLANS-derived ensemble. Of the remaining five sequences, Rv2640c, Rv2034, Rv0081 and Rv0576 are among the unknowns in groups 8, 6, 5 and 7, respectively lacking defined metal sensing sites. Rv2642 is in group 3 possessing an α3N site (CadC-like) and hence predicted to sense Zn(II), Cd(II), and/or Pb(II) which is also consistent with its location adjacent to a cadmium inducible gene, cadI (45) and hence likely target.
In addition to the ArsR-SmtB sensors, M. tuberculosis possesses representatives from other families of metal sensing transcriptional regulators, including the DtxR family (IdeR and SirR (9)), MerR family (two uncharacterized representatives), Fur-family (FurA and FurB/Zur (11, 46)), and CsoR (47). Notably this organism lacks homologues of the nickel sensors NikR and RcnA that regulate nickel uptake in E. coli (48), despite M. tuberculosis possessing a deduced nickel importer (NicT/NixA) to supply urease.
A KmtR sub-group of group 8 consists of ten proteins all from Actinobacteria (supplementary Table S1). The newly defined KmtR metal-sensing site (α5-3) is conserved among these sequences but absent from all other groups. The non-mycobacterial kmtR-like genes are adjacent to genes encoding deduced CDF family divalent cation transporters or a major facilitator superfamily (MFS_1) transporter (supplementary Table S1). The mycobacterial kmtR-like sequences, including M. tuberculosis kmtR, are remote from cdf sequences but adjacent to deduced truncated transposases and tRNA genes. Chromosomal genes associated with mobility-related elements are often linked with environmental adaptation and, in the case of pathogens, virulence (49). Known ArsR-SmtB metal-sensing sites and their abundances are summarized in Fig. 11. Sequence group 5 (Fig. 1) contains, almost exclusively, no predicted metal binding pattern. The largest subgroup includes HlyU from Vibrio cholerae, which positively regulates expression of hemolysin (HlyA) (50), although the binding of HlyU to the hlyA operator-promoter region has not been demonstrated, and SoxR from Pseudaminobacter salicylatoxidans, which negatively regulates expression of the sulfur oxidation (sox) operon (51). Notably, these proteins possess two Cys residues in helices α2 and α5 (Fig. 11A), which are shared by other sequences within the same sub-group (supplementary Fig. S3). During this study, we expressed and purified HlyU and a second member of this sub-group from Erwinia carotovora, designated EcaR, which formed specific complexes with DNA fragments containing regions of dyad symmetry found in the hlyU and ecaR operator-promoter regions, respectively (supplementary Fig. S4). These complexes were not impaired by metal ions consistent with the conserved Cys in helices α2 and α5 most likely predicting non-metal sensors.
For ~70% of the close relatives of ArsR-SmtB sensors, the capacity to detect metals can now be predicted based upon the presence of one or other of the identified metal-sensing (or non-sensing) motifs (Fig. 11B). To date, there has been complete correspondence between these motifs and the ability to sense specific metals in all ArsR-SmtB family proteins that have been investigated by experiment. Conversely, protein sequence similarity is a poor predictor of metal sensing or metal specificity in this family of proteins. ArsR-SmtB proteins are separated into eight groups, based on sequence similarity, which do not correlate with defined motifs or established organism phyla (Fig. 1). Extensive convergent evolution and concerted evolution may have occurred. This is relevant to many current missassignments of ArsR-SmtB and, probably other, metal sensors in bacterial sequence databases. We do not yet know whether KmtR and/or one or more of the other ArsR-SmtB metal sensors in M. tuberculosis contributes to the ability of M. tuberculosis to survive within the human host. A role for KmtR in M. tuberculosis virulence needs to be specifically tested. Within host cells, M. tuberculosis must protect against metal-mediated toxicity and must supply essential metal ions to metal-requiring proteins needed for survival within this environment. One could foresee that the possession of multiple sensors for a range of different metal ions provides an advantage for this pathogen within its intracellular niche allowing rapid responses to host mediated metal fluxes. Elucidation of the target genes and metal specificities of the remaining M. tuberculosis ArsR-SmtB sensors with no defined motifs may provide clues as to the survival strategies employed by this organism and aid the design of drugs that disrupt these mechanisms. Any therapeutic approach that assists host clearance of M. tuberculosis is a priority.
Supplementary Material
Acknowledgments
We acknowledge the Bacterial Microarray Group at St. George’s, University of London for supply of the microarrays and advice, and Kate Gould who provided microarray training.
Footnotes
The on-line version of this article (available at http://www.jbc.org) contains supplemental Figs. S1–S4 and Table S1.
This work was supported by grants from the Biotechnology and Biological Sciences Research Council (BBSRC) Plant and Microbial Sciences committee and Agri-Food committee and BBSRC studentships (to D. R. C. and K. E. C.). The Wellcome Trust for funding the multicollaborative microbial pathogen microarray facility under its Functional Genomics Resources Initiative.
- BCG
- Bacille Calmette-Guérin
- DTT
- dithiothreitol
- PAR
- 4-(2-pyridylazo)resorcinol
This article must therefore be hereby marked “advertisement” in accordance with 18 U.S.C. Section 1734 solely to indicate this fact
REFERENCES
- 1.Ginsberg AM, Spigelman M. Nat. Med. 2007;13:290–294. doi: 10.1038/nm0307-290. [DOI] [PubMed] [Google Scholar]
- 2.Warner DF, Mizrahi V. Nat. Med. 2007;13:282–284. doi: 10.1038/nm0307-282. [DOI] [PubMed] [Google Scholar]
- 3.Malik S, Abel L, Tooker H, Poon A, Simkin L, Girard M, Adams GJ, Starke JR, Smith KC, Graviss EA, Musser JM, Schurr E. Proc. Natl. Acad. Sci. U. S. A. 2005;102:12183–12188. doi: 10.1073/pnas.0503368102. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Hoal EG, Lewis LA, Jamieson SE, Tanzer F, Rossouw M, Victor T, Hillerman R, Beyers N, Blackwell JM, Van Helden PD. Int. J. Tuberc. Lung Dis. 2004;8:1464–1471. [PubMed] [Google Scholar]
- 5.Zhang W, Shao L, Weng X, Hu Z, Jin A, Chen S, Pang M, Chen ZW. Clin. Infect. Dis. 2005;40:1232–1236. doi: 10.1086/428726. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Jabado N, Jankowski A, Dougaparsad S, Picard V, Grinstein S, Gros P. J. Exp. Med. 2000;192:1237–1248. doi: 10.1084/jem.192.9.1237. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Goswami T, Bhattacharjee A, Babal P, Searle S, Moore E, Li M, Blackwell JM. Biochem. J. 2001;354:511–519. doi: 10.1042/0264-6021:3540511. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Rodriguez GM, Smith I. J. Bacteriol. 2006;188:424–430. doi: 10.1128/JB.188.2.424-430.2006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Manabe YC, Saviola BJ, Sun L, Murphy JR, Bishai WR. Proc. Natl. Acad. Sci. U. S. A. 1999;96:12844–12848. doi: 10.1073/pnas.96.22.12844. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Agranoff D, Krishna S. Front. Biosci. 2004;9:2996–3006. doi: 10.2741/1454. [DOI] [PubMed] [Google Scholar]
- 11.Lucarelli D, Russo S, Garman E, Milano A, Meyer-Klaucke W, Pohl E. J. Biol. Chem. 2007;282:9914–9922. doi: 10.1074/jbc.M609974200. [DOI] [PubMed] [Google Scholar]
- 12.Tottey S, Harvie DR, Robinson NJ. Acc. Chem. Res. 2005;38:775–783. doi: 10.1021/ar0300118. [DOI] [PubMed] [Google Scholar]
- 13.Cavet JS, Meng W, Pennella MA, Appelhoff RJ, Giedroc DP, Robinson NJ. J. Biol. Chem. 2002;277:38441–38448. doi: 10.1074/jbc.M207677200. [DOI] [PubMed] [Google Scholar]
- 14.Cavet JS, Graham AI, Meng W, Robinson NJ. J. Biol. Chem. 2003;278:44560–44566. doi: 10.1074/jbc.M307877200. [DOI] [PubMed] [Google Scholar]
- 15.Canneva F, Branzoni M, Riccardi G, Provvedi R, Milano A. J. Bacteriol. 2005;187:5837–5840. doi: 10.1128/JB.187.16.5837-5840.2005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Thelwell C, Robinson NJ, Turner-Cavet JS. Proc. Natl. Acad. Sci. U. S. A. 1998;95:10728–10733. doi: 10.1073/pnas.95.18.10728. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Xu C, Shi W, Rosen BP. J. Biol. Chem. 1996;271:2427–2432. doi: 10.1074/jbc.271.5.2427. [DOI] [PubMed] [Google Scholar]
- 18.Ye J, Kandegedara A, Martin P, Rosen BP. J. Bacteriol. 2005;187:4214–4221. doi: 10.1128/JB.187.12.4214-4221.2005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Liu T, Golden JW, Giedroc DP. Biochemistry. 2005;44:8673–8683. doi: 10.1021/bi050450+. [DOI] [PubMed] [Google Scholar]
- 20.Singh VK, Xiong A, Usgaard TR, Chakrabarti S, Deora R, Misra TK, Jayaswal RK. Mol. Microbiol. 1999;33:200–207. doi: 10.1046/j.1365-2958.1999.01466.x. [DOI] [PubMed] [Google Scholar]
- 21.Kuroda M, Hayashi H, Ohta T. Microbiol. Immunol. 1999;43:115–125. doi: 10.1111/j.1348-0421.1999.tb02382.x. [DOI] [PubMed] [Google Scholar]
- 22.Liu T, Nakashima S, Hirose K, Shibasaka M, Katsuhara M, Ezaki B, Giedroc DP, Kasamo K. J. Biol. Chem. 2004;279:17810–17818. doi: 10.1074/jbc.M310560200. [DOI] [PubMed] [Google Scholar]
- 23.Cook WJ, Kar SR, Taylor KB, Hall LM. J. Mol. Biol. 1998;275:337–347. doi: 10.1006/jmbi.1997.1443. [DOI] [PubMed] [Google Scholar]
- 24.Turner JS, Glands PD, Samson ACR, Robinson NJ. Nucleic Acids Res. 1996;24:3714–3721. doi: 10.1093/nar/24.19.3714. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.VanZile ML, Chen X, Giedroc DP. Biochemistry. 2002;41:9776–9786. doi: 10.1021/bi020178t. [DOI] [PubMed] [Google Scholar]
- 26.Eicken C, Pennella MA, Chen X, Koshlap KM, VanZile ML, Sacchettini JC, Giedroc DP. J. Mol. Biol. 2003;333:683–695. doi: 10.1016/j.jmb.2003.09.007. [DOI] [PubMed] [Google Scholar]
- 27.Shi W, Dong J, Scott RA, Ksenzenko MY, Rosen BP. J. Biol. Chem. 1996;271:9291–9297. doi: 10.1074/jbc.271.16.9291. [DOI] [PubMed] [Google Scholar]
- 28.Busenlehner LS, Pennella MA, Giedroc DP. FEMS Microbiol. Rev. 2003;27:131–143. doi: 10.1016/S0168-6445(03)00054-8. [DOI] [PubMed] [Google Scholar]
- 29.Busenlehner LS, Weng TC, Penner-Hahn JE, Giedroc DP. J. Mol. Biol. 2003;319:685–701. doi: 10.1016/S0022-2836(02)00299-1. [DOI] [PubMed] [Google Scholar]
- 30.Harvie DR, Andreini C, Cavallaro G, Meng W, Connolly BA, Yoshida K, Fujita Y, Harwood CR, Radford DS, Tottey S, Cavet JS, Robinson NJ. Mol. Microbiol. 2006;59:1341–1356. doi: 10.1111/j.1365-2958.2006.05029.x. [DOI] [PubMed] [Google Scholar]
- 31.Sambrook J, Fritsch EF, Maniatis T. Molecular Cloning: A Laboratory Manual. 2nd Ed. Cold Spring Harbor Laboratory Press; Cold Spring Harbor, NY: 1989. [Google Scholar]
- 32.Timm J, Lim EM, Gicquel B. J. Bacteriol. 1994;176:6749–6753. doi: 10.1128/jb.176.21.6749-6753.1994. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Parish T, Stoker NG. Microbiology. 2000;146:1969–1975. doi: 10.1099/00221287-146-8-1969. [DOI] [PubMed] [Google Scholar]
- 34.Stewart GR, Wernisch L, Stabler R, Mangan JA, Hinds J, Laing KG, Young DB, Butcher PD. Microbiology. 2002;148:3129–3138. doi: 10.1099/00221287-148-10-3129. [DOI] [PubMed] [Google Scholar]
- 35.Frickey T, Lupas A. Bioinformatics. 2004;20:3702–3704. doi: 10.1093/bioinformatics/bth444. [DOI] [PubMed] [Google Scholar]
- 36.Felsenstein J. Cladistics. 1989;5:164–166. [Google Scholar]
- 37.VanZile ML, Cosper NJ, Scott RA, Giedroc DP. Biochemistry. 2000;39:11818–11829. doi: 10.1021/bi001140o. [DOI] [PubMed] [Google Scholar]
- 38.Milano A, Branzoni M, Canneva F, Profumo A, Riccardi G. Res. Microbiol. 2004;155:192–200. doi: 10.1016/j.resmic.2003.11.009. [DOI] [PubMed] [Google Scholar]
- 39.McCall KA, Fierke CA. Anal. Biochem. 2000;284:307–315. doi: 10.1006/abio.2000.4706. [DOI] [PubMed] [Google Scholar]
- 40.Pennella MA, Shokes JE, Cosper NJ, Scott RA, Giedroc DP. Proc. Natl. Acad. Sci. U. S. A. 2003;100:3713–3718. doi: 10.1073/pnas.0636943100. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Clemens DL, Lee BY, Horwitz MA. J. Bacteriol. 1995;177:5644–5652. doi: 10.1128/jb.177.19.5644-5652.1995. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Gordon AH, Hart PD, Young MR. Nature. 1980;286:79–80. doi: 10.1038/286079a0. [DOI] [PubMed] [Google Scholar]
- 43.Karasseva V, Weiszfeiler JG, Lengyel Z. Zentralbl. Bakteriol. 1977;239:514–520. [PubMed] [Google Scholar]
- 44.Tan BH, Meinken C, Bastian M, Bruns H, Legaspi A, Ochoa MT, Krutzik SR, Bloom BR, Ganz T, Modlin RL, Stenger S. J. Immunol. 2006;177:1864–1871. doi: 10.4049/jimmunol.177.3.1864. [DOI] [PubMed] [Google Scholar]
- 45.Hotter GS, Wilson T, Collins DM. FEMS Microbiol. Lett. 2001;200:151–155. doi: 10.1111/j.1574-6968.2001.tb10707.x. [DOI] [PubMed] [Google Scholar]
- 46.Zahrt TC, Song J, Siple J, Deretic V. Mol. Microbiol. 2001;39:1174–1185. doi: 10.1111/j.1365-2958.2001.02321.x. [DOI] [PubMed] [Google Scholar]
- 47.Liu T, Ramesh A, Ma Z, Ward SK, Zhang L, George GN, Talaat AM, Sacchettini JC, Giedroc DP. Nat. Chem. Biol. 2007;3:60–68. doi: 10.1038/nchembio844. [DOI] [PubMed] [Google Scholar]
- 48.Iwig JS, Rowe JL, Chivers PT. Mol. Microbiol. 2006;62:252–262. doi: 10.1111/j.1365-2958.2006.05369.x. [DOI] [PubMed] [Google Scholar]
- 49.Gal-Mor O, Finlay BB. Cell. Microbiol. 2006;8:1707–1719. doi: 10.1111/j.1462-5822.2006.00794.x. [DOI] [PubMed] [Google Scholar]
- 50.Williams SG, Attridge SR, Manning PA. Mol. Microbiol. 1993;9:751–760. doi: 10.1111/j.1365-2958.1993.tb01735.x. [DOI] [PubMed] [Google Scholar]
- 51.Mandal S, Chatterjee S, Dam B, Roy P, Gupta KD. Microbiology. 2007;153:80–91. doi: 10.1099/mic.0.29197-0. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.


