Abstract
Latrophilin, a neuronal “adhesion G-protein-coupled receptor”, is the major brain receptor for α-latrotoxin, a black widow spider toxin which stimulates strong neuronal exocytosis in vertebrates. Latrophilin has an unusual structure consisting of two fragments that are produced by proteolytic cleavage of the parental molecule and behave independently in the plasma membrane. On binding an agonist, the fragments reassociate and send an intracellular signal. This signal, transduced by a heterotrimeric G-protein, causes release of calcium from intracellular stores and massive release of neurotransmitters. Latrophilin represents a phylogenetically conserved family of receptors, with orthologues found in all animals and up to three homologues present in most chordate species. From mammalian homologues, latrophilins 1 and 3 are expressed in neurons, while latrophilin 2 is ubiquitous. Latrophilin 1 controls synapse maturation and exocytosis, while latrophilin 2 is involved in breast cancer. Latrophilins may play different roles during development and in adult animals: thus, LAT-1 determines cell fate in early embryogenesis in Caenorhabditis elegans and controls neurotransmitter release in adult nematodes. This diversity suggests that the functions of latrophilins may be determined by their interactions with respective ligands. The finding of the ligand of latrophilin 1, the large postsynaptic protein lasso, is the first step in the quest for the physiological functions of latrophilins.
Introduction
Latrophilin was isolated in 19961,2. This was a result of extensive efforts of a number of laboratories trying to identify the functional receptor(s) of α-latrotoxin, a neurotoxin from black widow spider venom whose study had begun more than thirty years ago.
Alpha-latrotoxin causes exhaustive release of neurotransmitters from nerve terminals of vertebrates even in the absence of extracellular Ca2+.3,4 Due to its stimulating effect on all types of synapses and endocrine cells, the toxin has been widely used to study the mechanisms of regulated exocytosis (for a review see Ref. 5,6). When the toxin was found to require specific cell-surface receptors for its actions, several groups began looking for a high-affinity α-latrotoxin receptor7-9 which was envisaged to play a key role in exocytosis.
This protracted search eventually resulted in the isolation of latrophilin which has not only become one of the first members of the adhesion G protein-coupled receptors (GPCRs) to be identified, but also the only member with a known exogenous agonist (α-latrotoxin). Latrophilin research has greatly contributed to the general understanding of this peculiar receptor family. Thus, the idea of posttranslational cleavage of adhesion GPCRs, although first described for CD97,10 was developed into a conceptualised theory based on the studies of latrophilin.11 Latrophilin has also served as a model to propose the “split personality” hypothesis stating that the fragments of adhesion GPCRs can behave as independent proteins capable of ligand-induced reassociation and concomitant signalling.12,13 In addition, latrophilin is the first of the few adhesion GPCRs for which physical and functional association with G proteins has been directly demonstrated,12,14-16 justifying the name of the whole family.
The isolation of latrophilin
In 1990, by means of α-latrotoxin affinity chromatography in the presence of Ca2+, Petrenko et al. isolated several proteins from solubilised bovine brain.9 Two large components of this mixture were subsequently sequenced, cloned and termed neurexins17. Neurexins, a polymorphic family of neuronal cell-adhesion proteins, have later been shown to participate in synapse formation/stabilisation18,19 and to contribute to predisposition to autism.20 Although neurexin Iα was able to bind α-latrotoxin and mediate some of its toxic effects,21,22 its binding was Ca2+-dependent and its structural features did not suggest a signal transduction capability. This indicated the existence of another receptor that would bind α-latrotoxin in the absence of Ca2+ and have an ability to send intracellular signals.
In fact, another major protein was always present in the α-latrotoxin column eluate, but it was initially dismissed because, having the same molecular mass as α-latrotoxin (120-130 kDa), it was mistakenly considered to be the toxin leaching off the column. This protein actually was the N-terminal fragment (NTF) of latrophilin, a hitherto unknown brain protein which was simultaneously isolated by two groups in 1996, using α-latrotoxin affinity chromatography in the absence of calcium.1,2 This protein’s characteristics were consistent with the predicted receptor functions and, based on its high affinity for α-latrotoxin (~0.5 nM), one group termed it latrophilin,1,14 while the other called it CIRL (calcium-independent receptor of α-latrotoxin).2,11 This protein is also sometimes referred to as CL (CIRL/latrophilin) in the literature.23
The cDNA of latrophilin was cloned on the basis of peptide sequences from the 120 kDa protein (p120) isolated from bovine and rat brain.11,14 One long open reading frame was detected that encoded a protein consisting of 146614 or 147111 amino acid residues. Since the predicted size of the cloned protein (~162 kDa) was significantly larger than 120 kDa, the presence of the entire latrophilin sequence in the α-latrotoxin column eluate was investigated using antibodies.11 While an antibody against p120 recognised this band only, an antibody against the predicted C-terminal peptide failed to stain p120 but instead labelled some aggregated material at the top of the gel.11 This aggregate was resolved by supplementing SDS-polyacrylamide gels with 8 M urea and by not boiling the electrophoretic samples. The resulting C-terminal fragment (CTF) appeared as a fuzzy band of ~85 kDa and was termed p85.11 It has been subsequently shown, however, that CTF can be successfully analysed in standard SDS-gels if the samples are heated to 50 °C only;12,13 under these conditions the protein consistently migrates as a concise group of bands with an average molecular mass of ~69 kDa, exactly as predicted for this fragment (see below).
These results suggested that the translated protein was cleaved. This hypothesis was directly confirmed by N-terminal sequencing of CTF (p85) which demonstrated that the N-terminus of p85 corresponded to Thr-838 of the full-size protein.11 Thus, the cleavage at this position produced two fragments, or subunits. The predicted molecular mass of the α subunit was ~95 kDa (this corresponded to p120, which was known to be highly glycosylated).1,11 The predicted size of the β subunit was ~69 kDa,11 and its aberrant migration as p85 could be explained by the effect of urea.12 To avoid confusion and due to the partial independence of the two fragments, in the rest of this review we will refer to p120 (95 kDa fragment, α subunit) as NTF and to p85 (69 kDa fragment, β subunit) as CTF.
The latrophilin family
When the cDNA encoding rat and bovine latrophilin/CIRL was isolated and sequenced, three types of sequences were identified and found to represent a family of homologous mRNAs.23-25 One of these, encoding the protein purified by α-latrotoxin chromatography,1,2,11,14 was termed latrophilin 1 (CIRL1), while the other two sequences were assigned numbers (2 and 3) based on their homology to latrophilin 1. Consistent with its higher resemblance of latrophilin 1, latrophilin 2 also binds α-latrotoxin but with a much lower affinity,26 which does not allow its purification on α-latrotoxin columns, while latrophilin 3 does not bind the toxin appreciably.24 The genes encoding these mRNAs were termed lphn1, lphn2, and lphn3, the most widely used nomenclature. These genes are located, respectively, on chromosomes 19, 1 and 4 in humans and 8, 3 and 5 in mice. In a separate later study,27 three homologous genes encoding lectomedins (proteins containing lectin and olfactomedin domains) were identified by genome sequencing and termed lec1, lec2 and lec3. Sequence comparisons demonstrate that lec1 is identical to lphn2 and lec2 has the same sequence as lphn1, while lec3 corresponds to lphn3. Also, human “latrophilin 1” (lphh1)28 is in fact the human orthologue of rat and bovine latrophilin 2.
The latrophilin mRNAs have several sites of alternative splicing (two have been directly identified by cDNA sequencing in lphn1, five in lphn2 and four in lphn3).23 The exon boundaries in the three lphn genes are essentially the same, with a few exceptions, and many alternative splice sites coincide. The most notable are splice sites 5 and 7. The former alters the sequence of the third cytoplasmic loop between transmembrane regions (TMRs) 5 and 6 and is thus likely to affect G-protein coupling (see ‘Latrophilin as a GPCR’). The latter leads to the expression of different cytosolic domains in latrophilin 3. Alternative splicing at splice site 2 in latrophilin 1 truncates the protein immediately downstream of the N-terminal lectin-like domain (see ‘NTF’ below), producing a short protein which is probably secreted.
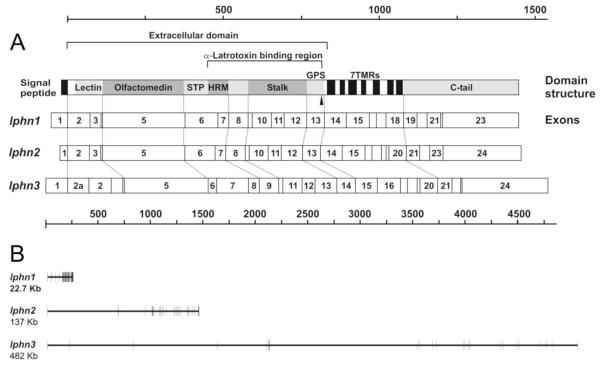
Variably spliced latrophilin gene orthologues are present in all animals, from coelenterates, nematodes and insects to tunicates and vertebrates,29,30 and have apparently evolved from a primordial adhesion GPCR gene. The nematodes possess two homologous genes, while one gene is found in arthropods. Coelenterates have three genes only weakly homologous to latrophilin and lacking many domains. Three proper latrophilin homologues are present in most chordates: from sea squirt to fish, platypus and humans; however, only two genes have been identified in the chicken genome. The family of three genes probably resulted from two rounds of gene duplication in early Chordata, with latrophilin 1 being the latest evolutionary acquisition. Given the early divergence of the three vertebrate lphn genes and their location on different chromosomes, it is not surprising that their introns differ vastly in size and sequence. However, the intron positions are highly conserved and almost precisely coincide with the borderlines between the domains of respective proteins (Fig. 1).
Figure 1.
The structure of latrophilin proteins and genes. A. The distribution of protein domains in latrophilin 1 (top diagram) is shown in comparison with the distribution of exons in the mRNAs of latrophilins 1-3. Only the translated exons are shown (the numbering starts at the first protein-coding exon). Arrowhead, the site of proteolytic cleavage. The scales show the size of the mature protein (top) and the mRNA (bottom). Exon 2a (alternative) in lphn3 is alternatively spliced. Note that many exons (or groups of exons) encode specific protein domains. B. Exon-intron structures and relative sizes of the three mouse latrophilin genes. Exons are depicted as vertical bars; introns, as horizontal lines. The size of each gene (including the translated exons and introns) is shown below each gene’s name. The gene structures shown are from the 129/SvJ mouse (some intron sizes differ between the 129/SvJ and CL57BL/6 mouse strains).
Interestingly, the latrophilin orthologues from such distant vertebrates as fish and humans are much more homologous than the three latrophilins within one organism, indicating that the three latrophilins possess different functions which are strictly preserved in the evolution of chordate animals. This is further supported by the different expression patterns of the three latrophilin homologues in various tissues.
Expression patterns of latrophilins
Northern blot analyses of different tissues have shown that latrophilins 1 and 3 are strictly brain specific, while latrophilin 2 is expressed in many tissues.14,23,24,26 Similar to neurexin Iα,17,31 very small levels of latrophilin 1 mRNA can be detected outside brain, in particular in kidneys and pancreas. It is possible that in the samples of these latter tissues latrophilin 1 mRNA is actually present in neurons from autonomic ganglia or in endocrine cells. Indeed latrophilin 1 mRNA has been found in many endocrine cells, e.g. pancreatic β-cells32 and chromaffin cells.33
Recently, a real-time PCR profiling of mRNA levels of all known adhesion GPCRs, including latrophilins, was conducted.34 Consistent with Northern blot studies carried out previously,14,23,24,26 it was shown in that study that latrophilin 1 (Lec2) and latrophilin 3 (Lec3) mRNAs are strongly enriched in the mouse brain and are essentially absent from, for example, mouse lung and liver. Again in line with the previous publications, latrophilin 2 (Lec1) mRNA was found in most mouse tissues. On the other hand, in the rat, the levels of latrophilin 1 and 3 mRNA in the liver and lung appeared as high as in the brain.34 This result is rather surprising because (1) it contradicts the multiple direct mRNA hybridisation experiments; (2) it is inconsistent with the close evolutionary relationship between mouse and rat and (3) latrophilin 1 cannot be detected in rat liver using α-latrotoxin chromatography or anti-latrophilin antibodies.1,24 It is possible that either the primers used in this work were able to amplify the latrophilin 2 message or that the samples of rat tissues used were fortuitously enriched with neuronal (e.g. autonomic ganglia) or endocrine cells.
It is also conceivable that even if some latrophilin 1 mRNA is present in non-neuronal tissues, it is not translated efficiently. From this point of view, tissue distribution of latrophilin protein should be studied directly. Indeed, our analysis of different rat tissues by Western blotting1 and especially immunohistochemistry (which permits unequivocal typifying of positive cells; paper in preparation) show that latrophilin 1 is absent from any non-neuronal cells and is present in very small amounts in adrenals, but not in liver, lung or kidneys. This result has been confirmed by immunoelectron microscopy demonstrating latrophilin 1 presence in synaptic junctions only (data not shown).
The structure of latrophilin
Latrophilin 1, due to its strong affinity for α-latrotoxin, is by far the most studied of the three latrophilins. However, the primary structures of these proteins are 48-63% identical, and there is no reason to believe that the processing and behaviour of latrophilins 2 and 3 should be grossly different. Therefore, the general protein architecture and behaviour as described below apply to all latrophilins and may be relevant for all adhesion GPCRs.
The primary structure of latrophilin comprises the following domains: an 851 residues-long extracellular domain; seven hydrophobic TMRs which, together with the intra- and extracellular loops, encompass 243 residues; and a cytoplasmic tail of 372 amino acids. Constitutive proteolysis within the extracellular domain (19 amino acids upstream of the first TMR) produces NTF (832 residues) and CTF (634 residues).
NTF
The extracellular domain, which gives rise to NTF, begins with a hydrophobic signal peptide. Immediately downstream lies a 108 residues-long cysteine-rich region homologous to galactose-binding lectin (GBL).11,14 GBL is present in most latrophilin orthologues found in animals from to nematodes (Caenorhabditis elegans) to vertebrates (but not in coelenterates).29 GBL is indeed able to bind D-galactose but shows much stronger preference for L-rhamnose35,36 and ouabain.35 The solution structure of this region, alternatively termed rhamnose-binding lectin, has been solved recently.35 This study argues, however, that carbohydrates are unlikely to be the endogenous ligands of this domain35 because (1) rhamnose is not normally found in animals; (2) the affinity of the monomeric lectin domain for rhamnose and especially other monosaccharides is insufficient for specific binding and (3) the residues critical for carbohydrate binding are not conserved in GBLs of latrophilin orthologues from different organisms. However, many GPCRs are dimeric, and it would be interesting to determine the affinity of GBL for D-galactose-containing glycans when latrophilin dimerises in response to agonist (α-latrotoxin) binding.12 In addition, the strong evolutionary conservation of GBL (amino acid sequence identity between latrophilins from nematodes, insects and mammals is 36-40%)29 suggests that GBL plays a very important role in the function of this receptor.
The GBL domain is followed by the region (260 amino acids) of homology to olfactomedin, a glycoprotein of the extracellular matrix of the olfactory neuroepithelium (for a review see Ref. 37). Olfactomedin domain is found in many different proteins all of which, apart from latrophilin, are secreted and most are expressed in the nervous system. Olfactomedin domain-containing proteins have been implicated in cell-cell interactions important for neurogenesis, neural crest formation, dorsoventral patterning and cell cycle regulation, while mutations in these proteins may be involved in various neurological diseases from glaucoma to psychiatric disorders. Interestingly, this domain is absent in all invertebrate orthologues of latrophilin and could be acquired during the early evolution of vertebrates.29
The 79 amino acid sequence downstream of the olfactomedin domain contains multiple serines, threonines and prolines (STP)25 but shows no significant homology to any known proteins, except some proline-rich bacterial proteins with low sequence complexity and unknown functions. The STP domain is found in insect and vertebrate orthologues of latrophilin, but not in C. elegans.
The STP region is linked to a domain (~60 residues) characteristically found in Class II (or secretin family) peptide hormone GPCRs.38 It is variably called “hormone-binding domain”,38 “signature domain”24 or “hormone receptor motif” (HRM, adopted here).29 HRM contains two conserved tryptophan residues and three to four conserved cysteines, which may form internal disulphide bridges. This region appears in many but not all adhesion GPCRs; it is absent from the latrophilin orthologues from insects and in latrophilin 3 from the chicken, but is present in the C. elegans latrophilins.29
A unique 68 amino acid region downstream of HRM connects it to a glycosylated domain of 180 residues which is analogous (although only 20 % identical) to the “Stalk” region of another adhesion GPCR, EMR339 and its homologues. In EMR proteins, this region is essential for the cleavage of the ectodomain39,40 and is thought to be an autoproteolytic enzyme.41,42 Truncations of this domain in latrophilin render this receptor unable to bind its ligand, α-latrotoxin.25
The Stalk domain is attached to a short, highly conserved29 sequence containing four cysteine residues and termed GPCR proteolysis site (or GPS).11 To avoid confusion with the actual site of cleavage, we will be referring to this region as the “GPS motif”. It is difficult to define the borderlines of this motif because of the variable conservation of the sequences surrounding this region in different adhesion GPCRs. Therefore, we suggest to set the limits of the GPS motif according to the ends of the exon encoding this entire domain (see ‘The latrophilin family’ above). This would mean that the GPS motif in latrophilin is 57 amino acids-long (starting at Ala-788 and ending at Ile-844). Most importantly, this motif contains the site of posttranslational cleavage that divides latrophilin into the non-covalently bound NTF and CTF. The cleavage occurs between Leu-832 and Thr-833,11 which is 8 amino acids upstream of the C-terminal end of the GPS motif or 19 amino acids upstream of the first TMR. As a result of this cleavage, the GPS motif itself becomes unequally split between NTF and CTF.
Both the Stalk domain and the GPS motif are present in latrophilin orthologues from all animal taxa,29 as well as in all other adhesion GPCRs. Sequence identity between latrophilins from vertebrates, insects and worms is 16-33 % within the Stalk domain and 45-49 % within the GPS motif.29 These domains are even more conserved among the three latrophilin homologues found in any vertebrate animal, where sequence identity is 50-60 % between the Stalks and 72-82 % between the GPS motifs. It is possible that the Stalk and GPS motif form a single functional unit that is involved in the posttranslational cleavage of latrophilin.
CTF
CTF begins at the site of cleavage within the GPS motif (Thr-833).11,12 The most prominent feature of this fragment is the presence of seven TMRs that are highly homologous to those of the secretin family GPCRs (50-60 % sequence similarity and 30 % identity). Similar to peptide hormone GPCRs, extracellular loops 1 and 2 contain two cysteine residues which are believed to form an intramolecular disulphide bridge. In fact, CTF possesses many other features thought to be important for GPCRs, e.g. a negatively charged amino acid within the third TMR, proline residues in the fourth and fifth TMRs and potential sites of palmitoylation in the N-terminal part of the cytoplasmic tail.
The cytoplasmic tail is the least conserved domain among latrophilins. Thus, sequence identity in this region is 13-28% among latrophilin orthologues from worms, insects and vertebrates, and 35-49 % among the three latrophilin homologues found in each mammal. For comparison, the average sequence identity of the whole protein molecules among the three homologues is 49-63%, while within the TMRs (including loop regions) it is 69-80 %.
There are numerous potential phosphorylation sites for several types of protein kinases on the cytoplasmic C-terminal portion of CTF. In fact, CTF of latrophilin isolated from rat brain is phosphorylated on multiple positions, and this explains the behaviour of this fragment in SDS-electrophoresis (see Section ‘The isolation of latrophilin’). Phosphorylation does not normally occur in cultured cells expressing latrophilin, which suggests that this modification in the brain is a result of normal latrophilin function. The phosphorylation plays an important role in the interaction of latrophilin fragments: the phosphorylated forms of CTF bind NTF much stronger than the non-phosphorylated CTF (paper in preparation). This may have important implications for the behaviour of the two fragments after their reassociation and signalling.
Cleavage and unusual behaviour of latrophilin fragments
Latrophilin was the first adhesion GPCR for which the site of intramolecular posttranslational cleavage was identified by direct sequencing of CTF.11 Since then all adhesion GPCRs studied in respect of cleavage have been proven to undergo proteolysis at a strictly conserved position within the GPS motif. This autoproteolytic cleavage and the two-subunit structure probably represent a common feature of all the members of the adhesion GPCR family.11,43,44 Moreover, the cleavage site between NTF and CTF coincides with the tentative borderline between the “adhesion” and the “GPCR” halves of these chimerical receptors.43,44
The cleavage occurs constitutively in the endoplasmic reticulum and is required for latrophilin trafficking to the cell surface12,43 Full-size, non-cleaved adhesion-GPCRs are apparently short-lived and not normally detectable in live tissues. Although full size GRP56 (another member of the adhesion GPCR family) was reported to appear in large quantities in some mouse tissues,45 this was later disproved.46
As described above, the cleavage site is localised in the GPS motif, 19 amino acids upstream of TMR1. Thus NTF contains no TMRs, but it is not released into the medium and remains non-covalently associated with the membrane. Given that NTF and CTF have an ability to interact strongly with each other when isolated by α-latrotoxin chromatography and immunoprecipitation,11,12,43 NTF has been thought11,43 to attach to the cell surface through its interaction with CTF, a transmembrane protein. This is also supported by the fact that proteolysis at a second site (located between the GPS and TMR1) releases some NTF into the medium.47
However, this does not seem to be true for a large proportion of NTF and CTF. Mutagenesis of CTF showed that only eight43 or even seven12 residues in the C-terminal part of the GPS motif (after cleavage forming the N-terminus of CTF) are both necessary and sufficient for the cleavage. These seven amino acids are also sufficient for the interaction between NTF and CTF. Despite such a short sequence holding NTF and CTF together, NTF cannot be removed from the membranes by most chaotropic conditions: pH 2.5, pH 12, 4 M Mg2+ or 8 M urea (unpublished observations). Most detergents, while solubilising the membrane, do not affect the NTF-CTF complexes, and the latter can be isolated from solution. However, upon solubilisation of cells expressing latrophilin, a large percentage of each fragment remains free from the other. Furthermore, a weak detergent, perfluorooctanoic acid, which does not break up the membrane bilayer, removes a large amount of NTF from the plasma membrane, while leaving all CTF behind.13 Together, these data suggest that at least some proportion of NTF is anchored in the membrane independently of CTF, perhaps via hydrophobic amino acids or modification/s.12,13
This “split personality” hypothesis has been tested in a comprehensive series of experiments,12,13 which demonstrate that the two fragments do not always colocalise with each other on the cell surface and can even migrate in the membrane and internalise independently. When patched on live cells using antibodies, the two fragments behave as non-interacting free proteins.12 This corroborates the idea that NTF could have a hydrophobic anchor of its own.
Under certain conditions, e.g. the binding of agonists (see ‘Extracellular ligands’ below) and also upon membrane solubilisation with detergents, the free latrophilin fragments can reassociate.12 Treatment of latrophilin-expressing cells with α-latrotoxin or its mutant LTXN4C results in the formation of large ternary complexes (α-latrotoxin-NTF-CTF) on the plasma membrane. This leads to intracellular signal transduction to intracellular Ca2+ stores (described in ‘Latrophilin as a GPCR’). The mechanism of this reassociation apparently involves dimerisation of NTF domains, which increases the affinity between NTF and CTF.12,13
Most intriguingly, the ligand-induced reassociation of latrophilin fragments does not always occur within the same cleaved receptor molecule, as the ligand-bound NTF can interact with CTF from another latrophilin molecule.13 Moreover, due to the high conservation of GPS motifs in all adhesion GPCRs, NTF of latrophilin can even bind to CTF from another member of this receptor family. Such criss-cross association of NTFs and CTFs produced by the cleavage of different receptors creates functional complexes capable of intracellular signalling and has the potential of greatly diversifying the transduced signal.13
This “split personality” architecture of receptors, consisting of two independent modules that associate interchangeably on binding their ligands, is rather enigmatic but not entirely unprecedented. Several other signalling systems require co-receptors. For example, there are two receptors for the Wnt signalling proteins: Frizzled (a GPCR) and low-density-lipoprotein receptor-related protein.48 Normally such co-receptors are phylogenetically unrelated proteins, both of which bind the same ligand molecule. However, in the case of latrophilin and possibly other adhesion GPCRs, both “co-receptors” are the complementary fragments of the same (or structurally related) parental proteins. In addition, at least when α-latrotoxin is used, the ligand apparently only interacts with NTF, which then serves as an activated ligand of CTF.12,13 Pleiotropy of downstream effects in this case is achieved not by one ligand activating two different receptors (as in the Wnt pathway), but by the ligand-bound NTF activating CTF from one or another adhesion GPCR. This mechanism may have important implications for the biology of all adhesion GPCRs.
Latrophilin as a GPCR
Latrophilin was classified as a GPCR14 on the basis of its high sequence homology with the TMRs of the secretin family GPCRs and the features within CTF that are important for G protein coupling (see Section ‘CTF’ above). However, this cannot be considered a proof that this protein signals through G proteins. This aspect of receptor function was studied in some detail and, as a result, latrophilin has become the first receptor of the adhesion GPCR group for which specific binding to G proteins, namely Gα0 and Gαq/11, was demonstrated.14-16 This interaction is strong because it persists through two consecutive affinity chromatographies on different adsorbents that bind NTF only.14,15 (It needs to be pointed out that G proteins can only interact with CTF, and their isolation on NTF-binding columns is only possible due to NTF and CTF forming strong complexes). Moreover, the interaction between CTF and G proteins is dynamic and depends on the ability of G proteins to cleave GTP.15 Thus, G proteins copurify abundantly with latrophilin only when GDP and EGTA are added to the solubilisation buffer, i.e. under the conditions when the GTPase activity of G proteins is inhibited and they normally interact with respective GPCRs. The addition of GTP and Mg2+ to purification buffer supports the GTPase activity and causes the dissociation of G proteins, resulting, as expected, in their loss from the column eluate.15 Thus, excess of GTP is able to reverse the interaction of latrophilin with its requisite G protein(s), suggesting that this association is not only physical but also functional.
Intracellular signalling mechanisms of latrophilin have so far been studied using its exogenous agonist, α-latrotoxin. The signalling induced by α-latrotoxin is also consistent with the activation of G protein pathways. In particular, non-neuronal cells expressing latrophilin respond by stimulation of adenylate cyclise and phospholipase C (PLC) and release of intracellular calcium.14 Similarly, the toxin triggers activation of PLC and increase in cytosolic Ca2+ in PC12 cells49 and synaptosomes.50 However, studies of signal transduction from α-latrotoxin have been complicated by the fact that the toxin binds to at least two disparate receptors (neurexin and latrophilin) and also forms Ca2+-permeable transmembrane pores.5,6 The effect of Ca2+ influx through the toxin pore may obscure any physiological signalling. A breakthrough has been achieved with the creation in Tom Südhof’s laboratory of a mutant α-latrotoxin, LTXN4C.51 This mutant lacks the ability to form pores33 but still stimulates neurotransmitter secretion in hippocampal slices, neuronal cultures, neuromuscular junctions and synaptosomes,6,52-55 indicating that its effect is based on stimulation of a receptor. The subsequent receptor transduction pathway involves a G protein coupled to activation of PLC, production of inositol-trisphosphate and release of Ca2+ from intracellular stores.53 To determine which receptor is involved in this signalling, neuroblastoma cells expressing latrophilin or neurexin were stimulated with LTXN4C. It was demonstrated that only the cells expressing latrophilin (but not neurexin or latrophilin mutant with a single TMR) reacted by activating PLC and producing cytosolic Ca2+ waves.12 These results unequivocally indicate that latrophilin – via its CTF – can send intracellular signals to PLC and intracellular Ca2+ stores.
Until recently, however, it has been difficult to prove that the same signalling cascade (LTXN4C – latrophilin – G protein – PLC – Ca2+ stores)12,13 also underlies the LTXN4C-induced neurotransmitter release in nerve terminals.33,53 This is because LTXN4C also binds two other neuronal receptors (neurexin Iα and PTPσ) which might contribute to or mediate its effect in neurons. In addition, LTXN4C action in nerve terminals requires extracellular Ca2+,33,53 possibly suggesting that the toxin might induce influx of Ca2+e rather than release of Ca2+i. Indeed, when large LTXN4C binds to any receptor, it might interact with ion channels and thus cause exocytosis irrespective of latrophilin. These arguments have been remarkably answered by our recent finding that a recombinant single-chain antibody A1 against latrophilin 1, isolated from a phage display library, can cause burst-like neurotransmitter exocytosis similar to that induced by LTXN4C (paper in preparation). This ultimately proves that stimulation of neuronal latrophilin by agonists sends an exocytotic signal via a G protein pathway.
Neuronal studies also demonstrate that the main signals sent by latrophilin are relatively fast and reach a maximum within several seconds or minutes. Furthermore, this signalling retains all its characteristics even in synaptosomes or neuromuscular junctions, subcellular systems consisting of severed nerve terminals and lacking the neuronal somata. This leads to an important conclusion that, consistent with Ca2+ signalling, latrophilin acts locally, within presynaptic nerve terminals and does not necessarily send signals to the cell body and the nucleus.52,55
Of course, it is also possible that latrophilin can link to other signalling pathways, especially considering the promiscuous reassociation of its NTF with CTF’s from other adhesion GPCRs. Therefore, in-depth studies of both G protein-coupled and any alternative mechanisms are required.
Ligands and interacting partners of latrophilins
Extracellular ligands
α-Latrotoxin
The main exogenous ligand of latrophilin 1 is α-latrotoxin. The toxin stimulates its receptor and thus can be classed as an agonist. The interaction of α-latrotoxin with latrophilin was tested using various truncated constructs of latrophilin.25 This study demonstrated that a very large fragment of NTF containing HRM, Stalk and GPS (390 amino acids) may be necessary to bind α–latrotoxin strongly, suggesting a multi-point interaction between toxin and latrophilin, with low-affinity binding at each point. In particular, HRM, a putative ligand-binding region of hormone receptors, alone was unable to mediate toxin binding. It may be possible that some peptide within the toxin molecule mimics a natural ligand of HRM but interacts with this domain only weakly. The association of toxin with NTF at additional sites may provide sufficient retention of the hormone-mimetic toxin peptide in contact with HRM, leading to receptor activation.
Black widow spider venom also contains another component (possibly ε-latroinsectotoxin) that kills C. elegans worms on injection.56 Knockout and RNAi studies have shown that the toxic effects of the venom is mediated by the LAT-1 orthologue of latrophilins in C. elegans, but not by LAT-2.56
Cyclooctadepsipeptides
Latrophilin orthologues from the parasitic nematode Haemonchus contortus (HC110-R) and C. elegans (LAT-1) were thought to bind the anthelmintic cyclical depsipeptides, PF1022A (a natural secondary metabolite of the fungus Mycelia sterilia) and its semisynthetic derivative emodepside.57 Electrophysiological studies revealed that emodepside inhibited pharyngeal pumping of the nematodes in a concentration-dependent manner.58 In C. elegans LAT-1 knockout mutants emodepside had a decreased paralysing effect on the pharyngeal muscle.59 These studies suggested that cyclodepsipeptides were exogenous agonists of the latrophilin-like proteins in nematodes, leading to the release of an unidentified inhibitory transmitter which acted postsynaptically to relax both pharyngeal and somatic body wall muscle, causing flaccid paralysis of the nematode.
However, the expression of depsiphilin, a LAT-1 orthologue from the canine hookworm Ancylostoma caninum, did not correlate with emodepside sensitivity.60 Also, in C. elegans LAT-1 knockout worms only pharyngeal pumping was resistant to the inhibitory effect of emodepside, while locomotion was blocked by the drug even in the double mutant lat-1, lat-2 worms.61 Ultimately, emodepside has been found to target directly a Ca2+-activated potassium channel, SLO-1, which is expressed in both neurons and muscles. One pathway involving neuronal SLO-1 and controlled by LAT-1, is responsible for pharynx pumping. The second pathway, based on both neuronal and muscle SLO-1 that is independent of LAT-1 or LAT-2, is responsible for locomotion.61
Putative small endogenous ligands
The 54 kDa N-terminal fragment of latrophilin-like receptor HC110-R from H. contortus has been tested for its ability to bind different FMRFamide-like neuropeptides.62 Three of these peptides (AF1, AF10 and PF2) exhibited low-affinity interaction with the receptor with dissociation constants of 11 μM, 52 μM and 583 μM, respectively. These data suggest that AF1, AF10, and PF2 might represent natural ligands of HC110-R and might be involved in controlling pharyngeal pumping in nematodes.
Endogenous adhesion ligand
The structure of latrophilin, with its large adhesion-like N-terminal domain, and the finding of large protein ligands for other adhesion GPCRs63,64 suggest that latrophilin may be capable of interacting with ligands on adjacent cells or in the extracellular matrix. Therefore, in our attempts to isolate an endogenous ligand of latrophilin, we used NTF. Several variants of recombinant NTF were expressed, purified and used to synthesise an affinity adsorbent. Out of these constructs, only one resulted in an active column, which allowed us to isolate a protein from solubilised rat brain that specifically binds latrophilin (paper in preparation). This protein, termed lasso (latrophilin-associated synaptic surface organiser), is a large glycoprotein expressed on the postsynaptic membrane. The binding of lasso to NTF of latrophilin involves multiple sites in each molecule and causes specific association of cells expressing these proteins. Moreover, the interaction between latrophilin and lasso is required for synapse formation and maturation.
Intracellular partners
Most importantly for its signalling function, latrophilin has been found to bind two types of α-subunits of heterotrimeric G proteins14-16 (described in detail in ‘Latrophilin as a GPCR’).
Close to the C-terminus of CTF, there is a proline-rich region,14 which could bind SH3 domains of proteins involved in signalling.
In addition, CTF may stably or transiently interact with structurally important proteins. In a yeast two-hybrid system, the C-terminal cytoplasmic tail of latrophilin was able to bind Shank, a proline-rich postsynaptic scaffolding protein.65,66 The significance of this interaction is unclear: Shank contains a PDZ domain that binds proteins with a consensus sequence Ser/Thr-X-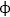 , where
, where 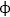 is a bulky hydrophobic amino acid. However, this sequence is present in all three latrophilin homologues, and consistently both latrophilins 1 and 2 were isolated in this artificial system.66 It is possible that the ubiquitous latrophilin 2 is the physiological target of Shank in the postsynaptic density, while latrophilin 1 is normally found in the presynaptic terminals (discussed in Section ‘Expression patterns of latrophilins’).
is a bulky hydrophobic amino acid. However, this sequence is present in all three latrophilin homologues, and consistently both latrophilins 1 and 2 were isolated in this artificial system.66 It is possible that the ubiquitous latrophilin 2 is the physiological target of Shank in the postsynaptic density, while latrophilin 1 is normally found in the presynaptic terminals (discussed in Section ‘Expression patterns of latrophilins’).
Finally, CTF of latrophilin has been shown to interact with TRIP8b, tetratricopeptide repeat-containing Rab8b-interacting protein,67,68 a cytosolic protein that binds clathrin and the adaptor protein AP2. This indicates that latrophilin may play a role in receptor-mediated endocytosis or trafficking of neuronal channels.69
Latrophilin gene knockouts
Mouse
To study the physiological role of latrophilin 1, its gene has been knocked out in mice by deleting exon 270 or the distal part of exon 1 plus the proximal part of exon 2 (unpublished data). In our laboratory the first lphn1-/- mouse only appeared after about 40 rounds of mating heterozygous animals, suggesting that the lphn1 deletion is actually embryonically lethal. This is supported by our consistent finding of dead embryos in pregnant heterozygous female mice. However, both knockout approaches eventually resulted in live and fertile lphn1-/- mice, indicating that the lack of latrophilin 1 can be effectively compensated for by a mutation or upregulation of another gene that either occurs spontaneously or is introduced by C57BL/6 backcrossing. Both compensated mutant strains display lack of maternal behaviour (pup nursing, nest building, etc.). In our colony, these maternal nurturing defects reciprocally correlate with the dose of lphn1 gene. In addition, our knockout mice show increased aggression. It should be noted that a very similar phenotype has been reported for mice lacking Gαq71 or PLC-β1,72 the proteins known to be involved in the downstream signalling cascade of latrophilin. Despite the mild phenotypic manifestation of latrophilin 1 deletion, which is apparently due to compensatory changes in the genetic background, further behavioural studies are needed to throw more light on the functions of latrophilin 1 in those cells and brain regions where the genetic compensation is less pronounced. In addition, it would be especially revealing to determine the nature of the compensatory mutation/s.
At the biochemical level, knockout mice demonstrated a decreased binding of α-latrotoxin and a great decrease in toxin-evoked glutamate release from nerve terminals, both in the presence and absence of Ca2+,70 indicating that latrophilin is the major receptor for α-latrotoxin. However, this study employed the wild-type toxin, whose ability to form Ca2+-permeable pores complicated the results and made it impossible to detect an inhibition of latrophilin signalling in knockout mice. An in-depth exploration of the role of latrophilin in nerve terminals must be conducted using the non-pore-forming mutant LTXN4C or other tools.
C. elegans
The orthologues of mammalian latrophilins in the nematode C. elegans are encoded by two genes: lat-1 and lat-2. The LAT proteins are 25-28 % identical to all latrophilins, but not particularly related to any one latrophilin.
The results obtained from lat-1 knockout in C. elegans strongly support the hypothesis that LAT-1 is presynaptic in adult nematodes and that its stimulation, similar to the mammalian latrophilin pathway,12 signals via activation of Gαq protein and phospholipase C-β1, leading to the mobilisation of diacylglycerol (DAG). DAG then activates UNC-13, an important protein that regulates the tethering of presynaptic vesicles to the plasma membrane, and synaptobrevin, a vesicular protein that binds vesicles to the plasma membrane. This is thought to result in transmitter release.59,73
In addition, loss-of-function mutations in lat-1 (but not in lat-2) have indicated a different role for LAT-1 in C. elegans development.74 The lack of this protein results in defects in anterior-posterior polarity, leading to arrest of larval development and suggesting that LAT-1, in parallel with the wnt pathway, controls the polarity of cell division and cell migration during nematode embryogenesis. Both the extracellular N-terminal region (including the GBL/RBL domain) and the C-terminal domain are required for this mechanism. This indicates that in the process of early worm development LAT-1 acts by transforming the interaction of NTF with adjacent cells into intracellular signals. These signals are probably different from those sent by the protein in terminally differentiated cells of the adult worm.
Latrophilins in disease
To our knowledge, genetic links between the lphn1 gene and an inheritable disease have not been established yet. This may suggest – in line with our knockout results above – that most mutations in latrophilin 1, as well as its ablation, are embryonically lethal. Indirect evidence suggests that latrophilin 1 may be associated with such mental disorders as schizophrenia and bipolar disorder. Thus, chronic administration of risperidone, an antipsychotic drug often used to treat schizophrenia, led to an upregulation of lphn1 in rats.75 Also, the lack of latrophilin in mice, despite the compensatory changes, led to behaviours consistent with schizophrenia phenotypes.76 Schizophrenia is a complex neuropsychiatric disease, and multiple genes and environmental factors can contribute to its manifestation, making further research into latrophilin gene/s even more important.
On the other hand, mutations in the human gene lphh1 encoding the ubiquitous latrophilin 2 have been associated with breast cancer.28 Analyses of tumour cell lines showed that lphh1 expression was variable. Also, gene product variability was higher in the tumour than in normal breast tissue.
Conclusions
Taken together these data suggest that the ancient physiological role of latrophilins in animals is to convert cell contacts into intracellular signals. However, the members of this family have distinct distributions and functions, from early patterning during embryogenesis to controlling release of neurotransmitters in neurons. The identification of specific ligands that bind different latrophilin homologues, or each latrophilin during different stages of animal development, will bring about a new level of understanding of these unusual receptors.
Reference List
- 1.Davletov BA, Shamotienko OG, Lelianova VG, et al. Isolation and biochemical characterization of a Ca2+-independent α-latrotoxin-binding protein. J Biol Chem. 1996;271:23239–23245. doi: 10.1074/jbc.271.38.23239. [DOI] [PubMed] [Google Scholar]
- 2.Krasnoperov VG, Beavis R, Chepurny OG, et al. The calcium-independent receptor of α-latrotoxin is not a neurexin. Biochem Biophys Res Commun. 1996;227:868–875. doi: 10.1006/bbrc.1996.1598. [DOI] [PubMed] [Google Scholar]
- 3.Clark AW, Mauro A, Longenecker HE, Jr, et al. Effects of black widow spider venom on the frog neuromuscular junction. Effects on the fine structure of the frog neuromuscular junction. Nature. 1970;225:703–705. doi: 10.1038/225703a0. [DOI] [PubMed] [Google Scholar]
- 4.Longenecker HE, Hurlbut WP, Mauro A, et al. Effects of black widow spider venom on the frog neuromuscular junction. Effects on end-plate potential, miniature end-plate potential and nerve terminal spike. Nature. 1970;225:701–703. doi: 10.1038/225701a0. [DOI] [PubMed] [Google Scholar]
- 5.Ushkaryov YA, Rohou A, Sugita S. α-Latrotoxin and its receptors. In: Sudhof TC, Starke K, Boehm S, editors. Pharmacology of neurotransmitter release, Handb Exp Pharmacol. Vol. 184. Springer-Verlag; Berlin Heidelberg: 2008. pp. 171–206. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Silva JP, Suckling J, Ushkaryov Y. Penelope’s web: using α-latrotoxin to untangle the mysteries of exocytosis. J Neurochem. 2009;111:275–290. doi: 10.1111/j.1471-4159.2009.06329.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Tzeng MC, Siekevitz P. The binding interaction between α-latrotoxin from black widow spider venom and a dog cerebral cortex synaptosomal membrane preparation. J Neurochem. 1979;33:263–274. doi: 10.1111/j.1471-4159.1979.tb11728.x. [DOI] [PubMed] [Google Scholar]
- 8.Scheer H, Meldolesi J. Purification of the putative α-latrotoxin receptor from bovine synaptosomal membranes in an active binding form. EMBO J. 1985;4:323–327. doi: 10.1002/j.1460-2075.1985.tb03632.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Petrenko AG, Kovalenko VA, Shamotienko OG, et al. Isolation and properties of the α-latrotoxin receptor. EMBO J. 1990;9:2023–2027. doi: 10.1002/j.1460-2075.1990.tb08331.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Gray JX, Haino M, Roth MJ, et al. CD97 is a processed, seven-transmembrane, heterodimeric receptor associated with inflammation. J Immunol. 1996;157:5438–5447. [PubMed] [Google Scholar]
- 11.Krasnoperov VG, Bittner MA, Beavis R, et al. α-Latrotoxin stimulates exocytosis by the interaction with a neuronal G-protein-coupled receptor. Neuron. 1997;18:925–937. doi: 10.1016/s0896-6273(00)80332-3. [DOI] [PubMed] [Google Scholar]
- 12.Volynski KE, Silva JP, Lelianova VG, et al. Latrophilin fragments behave as independent proteins that associate and signal on binding of LTXN4C. EMBO J. 2004;23:4423–4433. doi: 10.1038/sj.emboj.7600443. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Silva JP, Lelianova V, Hopkins C, et al. Functional cross-interaction of the fragments produced by the cleavage of distinct adhesion G-protein-coupled receptors. J Biol Chem. 2009;284:6495–6506. doi: 10.1074/jbc.M806979200. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Lelianova VG, Davletov BA, Sterling A, et al. α-Latrotoxin receptor, latrophilin, is a novel member of the secretin family of G protein-coupled receptors. J Biol Chem. 1997;272:21504–21508. doi: 10.1074/jbc.272.34.21504. [DOI] [PubMed] [Google Scholar]
- 15.Rahman MA, Ashton AC, Meunier FA, et al. Norepinephrine exocytosis stimulated by α-latrotoxin requires both external and stored Ca2+ and is mediated by latrophilin, G proteins and phospholipase C. Phil Trans R Soc Lond B. 1999;354:379–386. doi: 10.1098/rstb.1999.0390. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Serova OV, Popova NV, Deev IE, et al. Identification of proteins in complexes with α-latrotoxin receptors. Bioorg Khim. 2008;34:747–753. doi: 10.1134/s1068162008060046. [DOI] [PubMed] [Google Scholar]
- 17.Ushkaryov YA, Petrenko AG, Geppert M, et al. Neurexins: synaptic cell surface proteins related to the α-latrotoxin receptor and laminin. Science. 1992;257:50–56. doi: 10.1126/science.1621094. [DOI] [PubMed] [Google Scholar]
- 18.Scheiffele P, Fan J, Choih J, et al. Neuroligin expressed in nonneuronal cells triggers presynaptic development in contacting axons. Cell. 2000;101:657–669. doi: 10.1016/s0092-8674(00)80877-6. [DOI] [PubMed] [Google Scholar]
- 19.Varoqueaux F, Aramuni G, Rawson RL, et al. Neuroligins determine synapse maturation and function. Neuron. 2006;51:741–754. doi: 10.1016/j.neuron.2006.09.003. [DOI] [PubMed] [Google Scholar]
- 20.Szatmari P, Paterson AD, Zwaigenbaum L, et al. Mapping autism risk loci using genetic linkage and chromosomal rearrangements. Nat Genet. 2007;39:319–328. doi: 10.1038/ng1985. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Geppert M, Khvotchev M, Krasnoperov V, et al. Neurexin Iα is a major α-latrotoxin receptor that cooperates in α-latrotoxin action. J Biol Chem. 1998;273:1705–1710. doi: 10.1074/jbc.273.3.1705. [DOI] [PubMed] [Google Scholar]
- 22.Sugita S, Khvochtev M, Südhof TC. Neurexins are functional α-latrotoxin receptors. Neuron. 1999;22:489–496. doi: 10.1016/s0896-6273(00)80704-7. [DOI] [PubMed] [Google Scholar]
- 23.Sugita S, Ichtchenko K, Khvotchev M, et al. α-Latrotoxin receptor CIRL/latrophilin 1 (CL1) defines an unusual family of ubiquitous G-protein-linked receptors. G-protein coupling not required for triggering exocytosis. J Biol Chem. 1998;273:32715–32724. doi: 10.1074/jbc.273.49.32715. [DOI] [PubMed] [Google Scholar]
- 24.Matsushita H, Lelianova VG, Ushkaryov YA. The latrophilin family: multiply spliced G protein-coupled receptors with differential tissue distribution. FEBS Lett. 1999;443:348–352. doi: 10.1016/s0014-5793(99)00005-8. [DOI] [PubMed] [Google Scholar]
- 25.Krasnoperov V, Bittner MA, Holz RW, et al. Structural requirements for α-latrotoxin binding and α-latrotoxin-stimulated secretion. A study with calcium-independent receptor of α-latrotoxin (CIRL) deletion mutants. J Biol Chem. 1999;274:3590–3596. doi: 10.1074/jbc.274.6.3590. [DOI] [PubMed] [Google Scholar]
- 26.Ichtchenko K, Bittner MA, Krasnoperov V, et al. A novel ubiquitously expressed α-latrotoxin receptor is a member of the CIRL family of G-protein-coupled receptors. J Biol Chem. 1999;274:5491–5498. doi: 10.1074/jbc.274.9.5491. [DOI] [PubMed] [Google Scholar]
- 27.Lander ES, Linton LM, Birren B, et al. Initial sequencing and analysis of the human genome. Nature. 2001;409:860–921. doi: 10.1038/35057062. [DOI] [PubMed] [Google Scholar]
- 28.White GR, Varley JM, Heighway J. Isolation and characterization of a human homologue of the latrophilin gene from a region of 1p31.1 implicated in breast cancer. Oncogene. 1998;17:3513–3519. doi: 10.1038/sj.onc.1202487. [DOI] [PubMed] [Google Scholar]
- 29.Rohou A, Nield J, Ushkaryov YA. Insecticidal toxins from black widow spider venom. Toxicon. 2007;49:531–549. doi: 10.1016/j.toxicon.2006.11.021. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Nordstrom KJ, Lagerstrom MC, Waller LM, et al. The Secretin GPCRs descended from the family of Adhesion GPCRs. Mol Biol Evol. 2009;26:71–84. doi: 10.1093/molbev/msn228. [DOI] [PubMed] [Google Scholar]
- 31.Occhi G, Rampazzo A, Beffagna G, et al. Identification and characterization of heart-specific splicing of human neurexin 3 mRNA (NRXN3) Biochem Biophys Res Commun. 2002;298:151–155. doi: 10.1016/s0006-291x(02)02403-8. [DOI] [PubMed] [Google Scholar]
- 32.Lang J, Ushkaryov Y, Grasso A, et al. Ca2+-independent insulin exocytosis induced by α-latrotoxin requires latrophilin, a G protein-coupled receptor. EMBO J. 1998;17:648–657. doi: 10.1093/emboj/17.3.648. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Volynski KE, Capogna M, Ashton AC, et al. Mutant α-latrotoxin (LTXN4C) does not form pores and causes secretion by receptor stimulation. This action does not require neurexins. J Biol Chem. 2003;278:31058–31066. doi: 10.1074/jbc.M210395200. [DOI] [PubMed] [Google Scholar]
- 34.Haitina T, Olsson F, Stephansson O, et al. Expression profile of the entire family of Adhesion G protein-coupled receptors in mouse and rat. BMC Neurosci. 2008;9:43. doi: 10.1186/1471-2202-9-43. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Vakonakis I, Langenhan T, Promel S, et al. Solution structure and sugar-binding mechanism of mouse latrophilin-1 RBL: a 7TM receptor-attached lectin-like domain. Structure. 2008;16:944–953. doi: 10.1016/j.str.2008.02.020. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Terada T, Watanabe Y, Tateno H, et al. Structural characterization of a rhamnose-binding glycoprotein (lectin) from Spanish mackerel (Scomberomorous niphonius) eggs. Biochim Biophys Acta. 2007;1770:617–629. doi: 10.1016/j.bbagen.2006.11.003. [DOI] [PubMed] [Google Scholar]
- 37.Tomarev SI, Nakaya N. Olfactomedin domain-containing proteins: possible mechanisms of action and functions in normal development and pathology. Mol Neurobiol. 2009;40:122–138. doi: 10.1007/s12035-009-8076-x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Harmar AJ. Family-B G-protein-coupled receptors. Genome Biol. 2001;2:3013.1–3013.10. doi: 10.1186/gb-2001-2-12-reviews3013. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Stacey M, Lin HH, Hilyard KL, et al. Human epidermal growth factor (EGF) module-containing mucin-like hormone receptor 3 is a new member of the EGF-TM7 family that recognizes a ligand on human macrophages and activated neutrophils. J Biol Chem. 2001;276:18863–18870. doi: 10.1074/jbc.M101147200. [DOI] [PubMed] [Google Scholar]
- 40.Chang GW, Stacey M, Kwakkenbos MJ, et al. Proteolytic cleavage of the EMR2 receptor requires both the extracellular stalk and the GPS motif. FEBS Lett. 2003;547:145–150. doi: 10.1016/s0014-5793(03)00695-1. [DOI] [PubMed] [Google Scholar]
- 41.Lin HH, Chang GW, Davies JQ, et al. Autocatalytic cleavage of the EMR2 receptor occurs at a conserved G protein-coupled receptor proteolytic site motif. J Biol Chem. 2004;279:31823–31832. doi: 10.1074/jbc.M402974200. [DOI] [PubMed] [Google Scholar]
- 42.Hsiao CC, Cheng KF, Chen HY, et al. Site-specific N-glycosylation regulates the GPS autoproteolysis of CD97. FEBS Lett. 2009 doi: 10.1016/j.febslet.2009.09.001. [DOI] [PubMed] [Google Scholar]
- 43.Krasnoperov V, Lu Y, Buryanovsky L, et al. Post-translational proteolytic processing of the calcium-independent receptor of α-latrotoxin (CIRL), a natural chimera of the cell adhesion protein and the G protein-coupled receptor. Role of the G protein-coupled receptor proteolysis site (GPS) motif. J Biol Chem. 2002;277:46518–46526. doi: 10.1074/jbc.M206415200. [DOI] [PubMed] [Google Scholar]
- 44.Fredriksson R, Lagerstrom MC, Lundin LG, et al. The G-protein-coupled receptors in the human genome form five main families. Phylogenetic analysis, paralogon groups, and fingerprints. Mol Pharmacol. 2003;63:1256–1272. doi: 10.1124/mol.63.6.1256. [DOI] [PubMed] [Google Scholar]
- 45.Iguchi T, Sakata K, Yoshizaki K, et al. Orphan G protein-coupled receptor GPR56 regulates neural progenitor cell migration via a G alpha 12/13 and Rho pathway. J Biol Chem. 2008;283:14469–14478. doi: 10.1074/jbc.M708919200. [DOI] [PubMed] [Google Scholar]
- 46.Huang Y, Fan J, Yang J, et al. Characterization of GPR56 protein and its suppressed expression in human pancreatic cancer cells. Mol Cell Biochem. 2008;308:133–139. doi: 10.1007/s11010-007-9621-4. [DOI] [PubMed] [Google Scholar]
- 47.Krasnoperov V, Deyev I, Serova O, et al. Dissociation of CIRL subunits as a result of two-step proteolysis. Biochemistry. 2009 doi: 10.1021/bi802163p. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Kikuchi A, Yamamoto H, Kishida S. Multiplicity of the interactions of Wnt proteins and their receptors. Cell Signal. 2007;19:659–671. doi: 10.1016/j.cellsig.2006.11.001. [DOI] [PubMed] [Google Scholar]
- 49.Vicentini LM, Meldolesi J. α Latrotoxin of black widow spider venom binds to a specific receptor coupled to phosphoinositide breakdown in PC12 cells. Biochem Biophys Res Commun. 1984;121:538–544. doi: 10.1016/0006-291x(84)90215-8. [DOI] [PubMed] [Google Scholar]
- 50.Davletov BA, Meunier FA, Ashton AC, et al. Vesicle exocytosis stimulated by α-latrotoxin is mediated by latrophilin and requires both external and stored Ca2+ EMBO J. 1998;17:3909–3920. doi: 10.1093/emboj/17.14.3909. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Ichtchenko K, Khvotchev M, Kiyatkin N, et al. α-Latrotoxin action probed with recombinant toxin: receptors recruit α-latrotoxin but do not transduce an exocytotic signal. EMBO J. 1998;17:6188–6199. doi: 10.1093/emboj/17.21.6188. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Ashton AC, Volynski KE, Lelianova VG, et al. α-Latrotoxin, acting via two Ca2+-dependent pathways, triggers exocytosis of two pools of synaptic vesicles. J Biol Chem. 2001;276:44695–44703. doi: 10.1074/jbc.M108088200. [DOI] [PubMed] [Google Scholar]
- 53.Capogna M, Volynski KE, Emptage NJ, et al. The α-latrotoxin mutant LTXN4C enhances spontaneous and evoked transmitter release in CA3 pyramidal neurons. J Neurosci. 2003;23:4044–4053. doi: 10.1523/JNEUROSCI.23-10-04044.2003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Deák F, Liu X, Khvochtev M, et al. α-Latrotoxin stimulates a novel pathway of Ca2+-dependent synaptic exocytosis independent of the classical synaptic fusion machinery. J Neurosci. 2009;29:8639–8648. doi: 10.1523/JNEUROSCI.0898-09.2009. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55.Lelyanova VG, Thomson D, Ribchester RR, et al. Activation of α-latrotoxin receptors in neuromuscular synapses leads to a prolonged splash acetylcholine release. Bull Exp Biol Med. 2009;147:701–703. doi: 10.1007/s10517-009-0600-5. [DOI] [PubMed] [Google Scholar]
- 56.Mee CJ, Tomlinson SR, Perestenko PV, et al. Latrophilin is required for toxicity of black widow spider venom in Caenorhabditis elegans. Biochem J. 2004;378:185–191. doi: 10.1042/BJ20031213. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57.Saeger B, Schmitt-Wrede HP, Dehnhardt M, et al. Latrophilin-like receptor from the parasitic nematode Haemonchus contortus as target for the anthelmintic depsipeptide PF1022A. FASEB J. 2001 doi: 10.1096/fj.00-0664fje. [DOI] [PubMed] [Google Scholar]
- 58.Harder A, Schmitt-Wrede HP, Krucken J, et al. Cyclooctadepsipeptides-an anthelmintically active class of compounds exhibiting a novel mode of action. Int J Antimicrob Agents. 2003;22:318–331. doi: 10.1016/s0924-8579(03)00219-x. [DOI] [PubMed] [Google Scholar]
- 59.Willson J, Amliwala K, Davis A, et al. Latrotoxin receptor signaling engages the UNC-13-dependent vesicle-priming pathway in C. elegans. Curr Biol. 2004;14:1374–1379. doi: 10.1016/j.cub.2004.07.056. [DOI] [PubMed] [Google Scholar]
- 60.Kruger N, Harder A, von Samson-Himmelstjerna G. The putative cyclooctadepsipeptide receptor depsiphilin of the canine hookworm Ancylostoma caninum. Parasitol Res. 2009;105(Suppl 1):S91–100. doi: 10.1007/s00436-009-1500-3. [DOI] [PubMed] [Google Scholar]
- 61.Guest M, Bull K, Walker RJ, et al. The calcium-activated potassium channel, SLO-1, is required for the action of the novel cyclo-octadepsipeptide anthelmintic, emodepside, in Caenorhabditis elegans. Int J Parasitol. 2007;37:1577–1588. doi: 10.1016/j.ijpara.2007.05.006. [DOI] [PubMed] [Google Scholar]
- 62.Muhlfeld S, Schmitt-Wrede HP, Harder A, et al. FMRFamide-like neuropeptides as putative ligands of the latrophilin-like HC110-R from Haemonchus contortus. Mol Biochem Parasitol. 2009;164:162–164. doi: 10.1016/j.molbiopara.2008.12.003. [DOI] [PubMed] [Google Scholar]
- 63.Mikesch JH, Buerger H, Simon R, et al. Decay-accelerating factor (CD55): a versatile acting molecule in human malignancies. Biochim Biophys Acta. 2006;1766:42–52. doi: 10.1016/j.bbcan.2006.04.001. [DOI] [PubMed] [Google Scholar]
- 64.Yona S, Lin HH, Siu WO, et al. Adhesion-GPCRs: emerging roles for novel receptors. Trends Biochem Sci. 2008;33:491–500. doi: 10.1016/j.tibs.2008.07.005. [DOI] [PubMed] [Google Scholar]
- 65.Tobaben S, Sudhof TC, Stahl B. The G protein-coupled receptor CL1 interacts directly with proteins of the shank family. J Biol Chem. 2000;275:36204–36210. doi: 10.1074/jbc.M006448200. [DOI] [PubMed] [Google Scholar]
- 66.Kreienkamp HJ, Zitzer H, Gundelfinger ED, et al. The calcium-independent receptor for α-latrotoxin from human and rodent brains interacts with members of the ProSAP/SSTRIP/Shank family of multidomain proteins. J Biol Chem. 2000;275:32387–32390. doi: 10.1074/jbc.C000490200. [DOI] [PubMed] [Google Scholar]
- 67.Popova NV, Plotnikov A, Deev IE, et al. Interaction of calcium-independent latrotoxin receptor with intracellular adapter protein TRIP8b. Dokl Biochem Biophys. 2007;414:149–151. doi: 10.1134/s1607672907030155. [DOI] [PubMed] [Google Scholar]
- 68.Popova NV, Plotnikov AN, Ziganshin RK, et al. Analysis of proteins interacting with TRIP8b adapter. Biochemistry (Mosc ) 2008;73:644–651. doi: 10.1134/s0006297908060035. [DOI] [PubMed] [Google Scholar]
- 69.Santoro B, Piskorowski RA, Pian P, et al. TRIP8b splice variants form a family of auxiliary subunits that regulate gating and trafficking of HCN channels in the brain. Neuron. 2009;62:802–813. doi: 10.1016/j.neuron.2009.05.009. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 70.Tobaben S, Sudhof TC, Stahl B. Genetic analysis of α-latrotoxin receptors reveals functional interdependence of CIRL/Latrophilin 1 and neurexin Iα. J Biol Chem. 2002;277:6359–6365. doi: 10.1074/jbc.M111231200. [DOI] [PubMed] [Google Scholar]
- 71.Wettschureck N, Moers A, Hamalainen T, et al. Heterotrimeric G proteins of the Gq/11 family are crucial for the induction of maternal behavior in mice. Mol Cell Biol. 2004;24:8048–8054. doi: 10.1128/MCB.24.18.8048-8054.2004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 72.Bohm D, Schwegler H, Kotthaus L, et al. Disruption of PLC-beta 1-mediated signal transduction in mutant mice causes age-dependent hippocampal mossy fiber sprouting and neurodegeneration. Mol Cell Neurosci. 2002;21:584–601. doi: 10.1006/mcne.2002.1199. [DOI] [PubMed] [Google Scholar]
- 73.Harder A, Holden-Dye L, Walker R, et al. Mechanisms of action of emodepside. Parasitol Res. 2005;97(Suppl 1):S1–10. doi: 10.1007/s00436-005-1438-z. [DOI] [PubMed] [Google Scholar]
- 74.Langenhan T, Promel S, Mestek L, et al. Latrophilin signaling links anterior-posterior tissue polarity and oriented cell divisions in the C. elegans embryo. Dev Cell. 2009;17:494–504. doi: 10.1016/j.devcel.2009.08.008. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 75.Chen ML, Chen CH. Microarray analysis of differentially expressed genes in rat frontal cortex under chronic risperidone treatment. Neuropsychopharmacology. 2005;30:268–277. doi: 10.1038/sj.npp.1300612. [DOI] [PubMed] [Google Scholar]
- 76.Kellendonk C, Simpson EH, Kandel ER. Modeling cognitive endophenotypes of schizophrenia in mice. Trends Neurosci. 2009;32:347–358. doi: 10.1016/j.tins.2009.02.003. [DOI] [PMC free article] [PubMed] [Google Scholar]