Abstract
CmeABC, a multidrug efflux system in Campylobacter jejuni, plays an important role in the resistance to different antimicrobials and toxic compounds. Although this efflux system has been well characterized in C. jejuni and to a less extent in C. coli, it is unknown if CmeABC homologs are functional in other Campylobacter spp. In this study, the cmeABC homologs were identified and functionally characterized in five Campylobacter species including C. jejuni, C. coli, C. lari, C. upsaliensis, and C. fetus. Our results indicated that cmeABC is present in all five Campylobacter spp. and the genomic organization of this efflux operon is similar among the Campylobacter spp. Insertional mutagenesis of cmeB increased the susceptibilities of all the five Campylobacter spp. to structurally diverse antimicrobials. Together, these results indicated that the CmeABC efflux system is conserved at both the genomic and functional levels in all five Campylobacter spp. examined in this study, further highlighting the significant role of CmeABC in Campylobacter pathobiology.
Introduction
Multidrug efflux transporters have been recognized to play a major role in mechanisms responsible for intrinsic and acquired antibiotic resistances in many bacterial species (Poole, 2005). They are membrane transport proteins belonging to several superfamilies, including the ATP binding cassette superfamily, the resistance, nodulation, and cell division (RND) superfamily, the major facilitator superfamily, the small multidrug resistance family, and the multidrug and toxic compound extrusion family (Guillaume et al., 2004; Poole, 2005). The RND-type efflux pump is a tripartite efflux system consisting of an inner membrane transporter, a periplasmic membrane fusion protein, and an outer membrane channel protein (Poole, 2005). RND efflux systems extrude structurally diverse antimicrobials and toxic compounds directly out of bacterial cells (Piddock, 2006).
Campylobacter jejuni is a leading bacterial cause of human enteritis in developed countries (Allos, 2001). Other Campylobacter spp., such as C. coli, C. upsaliensis, C. lari, and C. fetus, have also been linked to gastrointestinal infections or other clinical diseases (Bourke et al., 1998; Martinot et al., 2001). Gastroenteritis caused by Campylobacter is usually self-limiting and does not require antibiotic therapy, but antibiotic treatment is needed in severe and prolonged cases, such as those occurring in immunoincompetent patients (Tee and Mijch, 1998). When clinical therapy is warranted, fluoroquinolone and macrolide antibiotics are the drugs often prescribed for the treatment of Campylobacter infections. Development of resistance to these antibiotics in Campylobacter reduces the effectiveness of antibiotic therapy and has emerged as a major public health problem worldwide (Luangtongkum et al., 2009).
Multiple mechanisms associated with antibiotic resistance have been identified in Campylobacter, but target mutations and drug efflux are most relevant to the resistance to fluoroquinolones and macrolides (Luangtongkum et al., 2009). These two mechanisms function together in conferring high-level resistance to the two classes of antibiotics. In C. jejuni an RND-type efflux pump, named CmeABC, was found to mediate the extrusion of structurally diverse antimicrobials and toxic compounds and contributes to the intrinsic and acquired resistances to various antimicrobials (Lin et al., 2002; Pumbwe and Piddock, 2002; Luo et al., 2003). CmeABC is also a major player in the efflux of bile acids and plays a critical role in facilitating Campylobacter colonization of the intestinal tract (Lin et al., 2003). Expression of cmeABC is subject to regulation by CmeR, a repressor encoded by a gene immediately upstream of cmeA (Lin et al., 2005a). CmeR binds directly to an inverted repeat in the promoter region of cmeABC and inhibits the transcription of this efflux operon. Bile salts induce the expression of cmeABC, by inhibiting the binding of CmeR to the promoter of cmeABC (Lin et al., 2005a, 2005b).
As discussed earlier, the genetic features and functions of CmeABC have been well defined in C. jejuni and to a less extent in C. coli (Cagliero et al., 2005). However, it is unknown if this efflux pump is also functional in the same fashion in other Campylobacter spp. The purposes of this study were to identify the orthologs of CmeABC and to characterize their function in different Campylobacter spp., including C. jejuni, C. coli, C. upsaliensis, C. lari, and C. fetus. We focused on these five species because they have been linked to clinical diseases in humans (Lastovica and Allos, 2008).
Materials and Methods
Bacterial strains and cultures
The bacterial strains of the five Campylobacter spp. used in this study are listed in Table 1. Campylobacter strains were grown in Mueller–Hinton (MH) broth or on MH agar plates at either 37°C (for C. upsaliensis, C. lari, and C. fetus) or 42°C (for C. jejuni and C. coli) under microaerobic conditions. For the culture of C. upsaliensis, C. lari, and C. fetus, the MH broth and agar plates were supplemented with either Campylobacter growth supplement (SR0232; Oxoid, Cambridge, UK) or 5% horse blood (Cleveland Scientific, Bath, OH). For the selection and culture of the cmeB insertional mutants in different Campylobacter spp., either 4 μg/mL of chloramphenicol or 20 μg/mL of kanamycin was added to the corresponding culture media depending on the antibiotic resistance cassette inserted in the mutants. Escherichia coli JM109 was routinely grown in Luria–Bertani (LB) medium. For the selection of E. coli transformants, the LB medium was supplemented with a final concentration of 30 μg/mL kanamycin or 20 μg/mL chloramphenicol, and 100 μg/mL ampicillin according to the selection marker(s) carried on the plasmid.
Table 1.
Bacterial Strains Used in This Study
| Name | Description | Source or reference |
|---|---|---|
| Escherichia coli stains and plasmid | ||
| JM109 | Cloning strain for blue and white colony screening | Promega |
| pGEM-T easy | Polymerase chain reaction cloning vector, ampr | Promega |
| Campylobacter strains | ||
| 700819 | C. jejuni strain NCTC 11168 | ATCC |
| S3b | C. jejuni, isolated from chicken | This laboratory |
| 993868 | C. coli, isolated from human | UIa |
| 33559 | C. coli, isolated from swine | ATCC |
| TF1-13 | C. coli, isolated from turkey | This laboratory |
| H-8 | C. coli, isolated from swine | This laboratory |
| 2744 | C. upsaliensis | NADCb |
| 3121 | C. lari, isolated from chicken | NADC |
| 3125 | C. lari, a environmental isolate from sediments | NADC |
| 5652 | C. fetus, isolated from bovine | NADC |
| 6953 | C. fetus, isolated from equine | NADC |
| S3b-B | S3b derivative; cmeB::cat | This study |
| 99B | 993868 derivative; cmeB::cat | This study |
| TF99B | TF1-13 derivative; cmeB:cat | This study |
| 44B | 2744 derivative; cmeB::cat | This study |
| 21B | 3121 derivative; cmeB::cat | This study |
| 25B | 3125 derivative; cmeB::cat | This study |
| 52B | 5652 derivative; cmeB::kan | This study |
| 53B | 6953 derivative; cmeB::kan | This study |
From N. Moyer, University of Iowa.
From I.V. Wesley, National Animal Disease Center, U.S. Department of Agriculture.
ATCC, American type culture collection.
DNA extraction and plasmid purification
Campylobacter genomic DNA was extracted using the Wizard® Genomic DNA Purification Kit (Promega, Madison, WI), following the manufacturer's instructions. E. coli plasmids were extracted from overnight cultures in LB broth supplemented with appropriate antibiotics using the Plasmid MiniPrep Kit (Qiagen, Valencia, CA), following the manufacturer's instructions.
cmeABC sequencing strategies
Polymerase chain reaction (PCR)-based sequencing in conjunction with direct sequencing of genomic DNA was used in determining the sequences of cmeABC in various Campylobacter spp. cmeABC-specific primers were designed from the genome sequences of C. jejuni 11168 and 81–176 for sequencing the cmeABC operon in C. jejuni and cmeB in C. coli. For the initial sequencing of the cmeABC operon in C. upsaliensis, C. lari, and C. fetus, for which the cmeABC DNA could not be amplified using primers designed from C. jejuni, a pair of degenerate primers (Table 2) for a fragment of the cmeB gene were designed and the amplified DNA was cloned into pGEM-T plasmid and subsequently sequenced. The known cmeB sequences of C. coli, C. upsaliensis, C. lari, and C. fetus were used to design outward primers, which were then used to obtain the complete cmeABC sequences in these Campylobacter spp. through direct sequencing of their genomic DNA. DNA sequencing was performed at the Iowa State University DNA Facilities.
Table 2.
Key Polymerase Chain Reaction Primers Used in This Study
| Target amplified | ||
|---|---|---|
| Degenerate primers for cmeB of C. upsaliensis, C. lari, and C. fetus | ||
| cmeB1213F | GGDATHGTYGTAGATGATGC | cmeB |
| cmeB2817R | GCRAAYTCWACRATYAARATHGCATT | cmeB |
| Mutagenesis primers | ||
| chlU | ACGGATCCAAAGAGTGACCGCCGAGA (BamHI)a | cat cassette |
| chlL | ACTCTAGACAGTGCGACAAACTGGGA (XbaI) | cat cassette |
| kanU | ATGGATCCTGCAAGGAACAGTGAAT (BamHI) | kanr cassette |
| kanL | ATTCTAGAGCGATGAAGTGCGTAAG (XbaI) | kanr cassette |
| 99UBF | CAATCCGTTAGTACAATGAG | C. coli cmeB |
| 99UBR | ACGGATCCCATATTATCTGCGCCATTGA (BamHI) | C. coli cmeB |
| 99LBF | GGTCTAGAGGAGTGGATCAGCTTATCAA (XbaI) | C. coli cmeB |
| 99LBR | GTATTCTCCTACTGCTCCTA | C. coli cmeB |
| 44BUF | CAGCTTGTAAGCGATTATGA | C. upsaliensis cmeB |
| 44BUR | ACGGATCCAGCGATAACTTCTAGTATGC (BamHI) | C. upsaliensis cmeB |
| 44BLF | ACTCTAGAAGCTAGTATGGCTCTAATCG (XbaI) | C. upsaliensis cmeB |
| 44BLR | CTAAGGTAGAAGCAGCTATC | C. upsaliensis cmeB |
| LarUF | TCTAGCTTGTGCTTCGCTTA | C. lari cmeB |
| LarUR | TTGGATCCGAAGAGCTTGAGGTTGAATC (BamHI) | C. lari cmeB |
| LarLF | AATCTAGAGGCCATTGGTATTGTGGTTG (XbaI) | C. lari cmeB |
| LarLR | GTGCAGGATCACCTTGAACT | C. lari cmeB |
| FetUF | TATGCCAGTAACTGTTATGC | C. fetus cmeB |
| FetUR | ACGGATCCTAGTATCGGCTATAGTCTGT (BamHI) | C. fetus cmeB |
| FetLF | ACTCTAGATGTTCGCACTTATACTAGCC (XbaI) | C. fetus cmeB |
| FetLR | TGATACTACCGAGTCTAACG | C. fetus cmeB |
Restriction sites are underlined and labeled in parentheses.
Polymerase chain reaction
PCR was performed in a reaction volume of 50 μL containing 200 nM each of the deoxynucleoside triphosphates, 2.0 mM of MgCl2, 200 nM each of the primers, 50–100 ng of template DNA, and 2.5 units of the Taq polymerase (Promega) or the Pfu Turbo polymerase (Stratagene, La Jolla, CA). PCR cycling conditions were determined according to the estimated annealing temperatures of the primers and the length of the amplified products. PCR products for DNA sequencing and cloning were purified using a commercial PCR purification kit (Qiagen).
Mutagenesis of the CmeABC efflux pump
The cmeB mutant was generated for each Campylobacter spp. through the insertion of a chloramphenicol (cat) or kanamycin (kanr) resistance cassette in the cmeB gene based on the previously published method with minor modifications (Akiba et al., 2006). PCR primers used for the mutagenesis are listed in Table 2. In the first step of mutagenesis, a suicide vector carrying either a cat or kanr cassette flanked by 0.5–1.2 kb of the cmeB fragments was constructed. Briefly, the cat or kanr cassette was amplified using the Pfu polymerase (Stratagene) from pUOA18 (cat) or pMW10 plasmids (kanr) and purified with a PCR purification kit (Qiagen). The antibiotic cassette was inserted into the cmeB sequence by direct ligation or an inverse PCR method. For the direct ligation method, the amplified cmeB fragments and the antibiotic cassette were treated with restriction enzymes and then ligated by T4 DNA ligase (Promega). The ligated DNA with cmeB DNA flanking the antibiotic cassette was purified by agarose gel electrophoresis. The DNA band of the expected size was excised and ligated to pGEM-T (Promega) through A–T cloning. For the inverse PCR method, the cmeB DNA was amplified by the Taq polymerase and cloned into pGEM-T, which was then transformed into E. coli JM109 cells. The purified pGEM-T constructs were used as templates in the inverse PCR using the Pfu polymerase and a pair of cmeB inverse PCR primers. The amplified DNA was ligated to the antibiotic cassette by T4 DNA ligase, resulting in a suicide vector carrying the antibiotic cassette flanked by the cmeB sequences. The suicide vectors were transformed into Campylobacter cells via electroporation according to the protocol described previously (Guerry et al., 1994). After overnight incubation on a nonselective MH agar plate, the electroporated cells were plated onto selective MH agar plate with appropriate antibiotics and incubated for 3–5 days microaerobically at 42°C. Single colonies of transformants were obtained and the cmeB insertional mutations were confirmed by PCR.
Minimal inhibitory concentration tests on Campylobacter strains and mutants
The broth microdilution method was used to measure the minimal inhibitory concentration (MIC) of various antimicrobials as described previously (Lin et al., 2002). C. jejuni 11168 was used as an in-house quality control. Fresh Campylobacter cultures were grown for 24–48 hours in MH (for C. jejuni and C. coli) or MH with Campylobacter growth supplement (for C. upsaliensis, C. lari, and C. fetus) and used as inocula. The supplement (SR0232; Oxiod) contains no antibiotics, but sodium pyruvate, sodium metabisulphite, and ferrous sulphate, and was used to stimulate the growth of the slow-growing Campylobacter spp. Twofold dilution series of antibiotics or chemicals in 50 μL of the same bacterial culture media were made in 96-well plates. Fifty microliters of Campylobacter culture containing 5 × 105 colony forming unit (CFU) cells/mL were added to the 50 μL of diluted antibiotics or chemicals in the 96-well plates and mixed well by gentle shaking before incubation. The plates were incubated at 37°C (C. upsaliensis, C. lari, and C. fetus) or 42°C (C. jejuni and C. coli) for 48 hours under microaerobic conditions. The MIC test was repeated at least three times.
Sequence alignment
The obtained DNA sequences were assembled and analyzed by Vector NIT software (Invitrogen, Carlsbad, CA). The CmeABC sequences were aligned by the ClustalW software and phylogenetic trees were constructed by the neighbor-joining method.
Sequence accession number
The GenBank accession numbers of the CmeABC sequences used in this study are FJ797669, FJ797670, FJ797671, FJ797672, FJ797673, FJ797674, FJ797675, FJ797676, FJ797677, and FJ797678.
Results and Discussion
Comparison of the cmeABC operon in different Campylobacter spp.
Five species of Campylobacter were examined in this study, including C. jejuni, C. coli, C. upsaliensis, C. lari, and C. fetus. The genomic organization of the CmeABC operon was conserved among all examined Campylobacter spp., with one nucleotide overlapping between cmeA and cmeB and eight nucleotides overlapping between cmeB and cmeC. The cmeR gene, which encodes a local repressor of CmeABC in C. jejuni (Lin et al., 2005a), was also identified in all of the Campylobacter spp. and it is located immediately upstream of cmeABC. Cj0364, encoding a hypothetical protein with an unknown function, is immediately downstream of cmeABC in C. jejuni and this is also the case in C. coli and C. upsaliensis. These findings are consistent with the results of a previous study conducted in C. coli (Corcoran et al., 2005). However, in C. lari and C. fetus, cmeABC is followed by an open reading frame that is distinct from Cj0364 (data not shown).
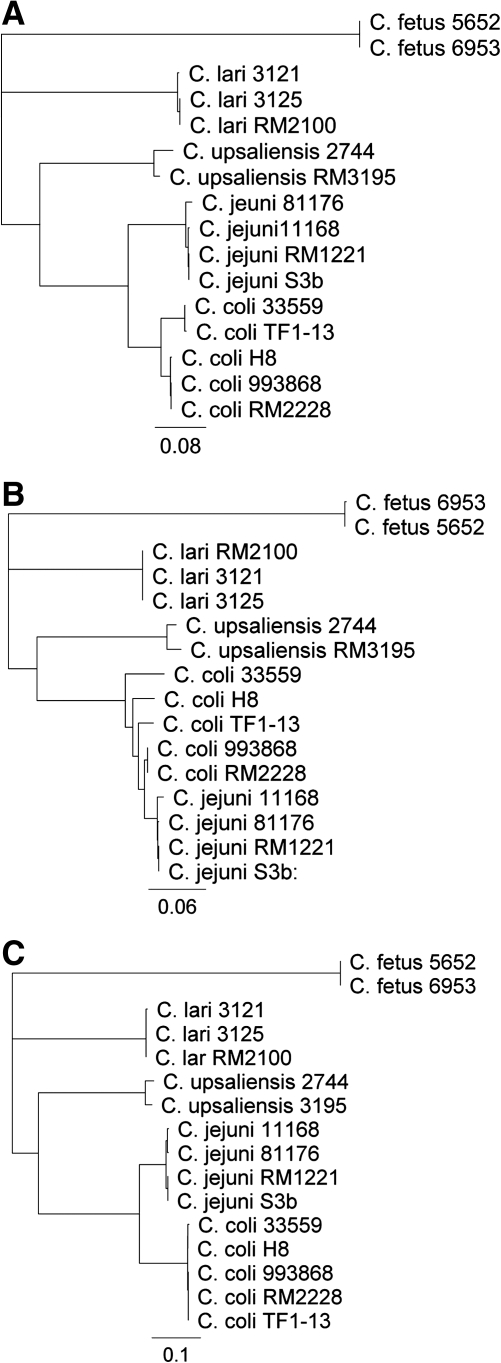
Sequence analysis showed that cmeB and cmeC are the most and least conserved, respectively, in the operon among the five Campylobacter species. Based on the amino acid sequences of CmeABC, the phylogenetic relatedness of the Campylobacter species in relation to C. jejuni was in the order of C. coli, C. upsaliensis, C. lari, and C. fetus (Fig. 1), which is in agreement with the finding from a previous report by comparing 12 conserved protein sequences (Fouts et al., 2005), but different from the phylogenetic tree constructed from the 16S rRNA sequences (Vandamme, 2000). As the number of strains of each species sequenced in this study was limited, we also included the CmeABC sequences available in GenBank for alignment, which showed that within each Campylobacter species, the CmeABC sequences are highly conserved, with amino acid identities ranging from 95% to 100% (data not shown). This finding is consistent with a previous report in which the alignment of partial cmeC sequences from 131 C. jejuni and C. coli strains indicated the conserved nature of CmeC sequences within each species (Fakhr and Logue, 2007). However, another study reported a variant CmeB in C. jejuni that was only 80% identical to that of C. jejuni 81–176 (Cagliero et al., 2006).
FIG. 1.
Dendrograms constructed with the amino acid sequences of CmeA (A), CmeB (B), and CmeC (C) from different Campylobacter species. The rooted trees were constructed with the neighbor-joining method based on ClustalW alignments of the CmeA, CmeB, and CmeC amino sequences of Camplylobacter strains from this study (C. jejuni S3b; C. coli 993868, 33559, TF1-13, and H-8; C. upsaliensis 2744; C. lari 3121and 3125; C. fetus 5652 and 6953) and representative genome sequences deposited in GenBank (C. jejuni 11168, 81176, and RM1221; C. coli RM2228; C. upsaliensis RM3195; and C. lari RM2100).
The cmeR sequence is also conserved among the different Campylobacter spp. The inverted repeat, which serves as a binding site for CmeR (Lin et al., 2005a), was also identified in the intergenic region between cmeR and cmeA for all Campylobacter spp. (data not shown). These findings suggest that the CmeABC efflux system and its regulator CmeR are conserved in Campylobacter spp. and this system is important for Campylobacter pathobiology.
CmeABC contributes to antibiotic resistance in different Campylobacter spp.
In this study, the cmeB homologs in different Campylobacter spp. were inactivated successfully by insertional mutagenesis. The mutants and their corresponding wild-type strains were compared for susceptibility to various antibiotics and the results are listed in Table 3. The mutagenesis of CmeB resulted in a substantial increase in the susceptibility of all Campylobacter spp. to the majority of antibiotics and toxic compounds examined in this study.
Table 3.
Minimal Inhibitory Concentrations (μg/ml) of Various Antimicrobials in Different Campylobacter Spp. and Their Isogenic cmeB Mutants
| Antimicrobials | C. jejuni 11168 | C. jejuni S3B | C. coli 993868 | C. coli TF1-13 | C. upsaliensis 2744 | C. lari 3121 | C. fetus 5652 |
|---|---|---|---|---|---|---|---|
| Ciprofloxacin | 0.25/0.031 (8) | 0.5/0.063 (8) | 0.25/0.031 (8) | 0.25/0.031 (8) | 0.5/0.063 (8) | 32/2 (16) | 1/0.0625 (16) |
| Erythromycin | 1/0.063 (16) | 1/0.031 (32) | 1/0.031 (32) | 256/4 (64) | 0.125/0.087 (16) | 2/0.031 (64) | 1/0.25 (4) |
| Ampicillin | 8/2 (4) | 16/1 (16) | 256/128 (2) | 16/8 (2) | 1/0.25 (4) | 2/0.125 (16) | 8/0.25 (4) |
| Cephalothin | 256/8 (32) | 256/4 (64) | 2048/32 (64) | 2048/8 (256) | 2/0.25 (8) | 128/0.5 (256) | 16/0.0625 (256) |
| Cefoperazone | 256/16 (16) | 1024/16 (64) | 2048/128 (16) | 1024/32 (32) | 16/0.063 (256) | 256/0.125 (2048) | 256/0.125 (2048) |
| Cefotaxime | 8/0.063 (128) | 16/0.063 (256) | 16/0.25 (64) | 16/0.063 (256) | 1/0.063 (16) | 32/0.063 (512) | 8/0.016 (512) |
| Rifampin | 256/1 (256) | 256/0.5 (512) | 64/0.0625 (1024) | 128/0.25 (512) | 8/0.063 (128) | 64/0.5 (128) | 128/128 (1) |
| Gentamicin | 2/1 (2) | 2/1 (2) | 1/0.5 (2) | 1/0.5 (2) | 8/8 (1) | 16/16 (1) | 32/32 (1) |
| Tetracycline | 0.125/0.063 (2) | 64/8 (8) | 0.5/0.125 (4) | 128/8 (16) | 1/0.125 (8) | 8/0.5 (16) | 8/0.5 (16) |
| Polymyxin B | 8/4 (2) | 4/2 (2) | 2/4 (0.5) | 2/4 (0.5) | 2/4 (0.5) | 4/8 (0.5) | >512/>512 (–) |
| Ethidium bromide | 1/0.125 (8) | 0.5/0.0625 (8) | 8/0.5 (16) | 4/0.25 (16) | 8/0.25 (32) | 4/1 (4) | 16/1 (16) |
| Cholic acid | 8192/128 (64) | 8192/128 (64) | 4096/256 (16) | 8192/256 (32) | 2048/32 (64) | 8192/32 (256) | 8192/128 (64) |
| Sodium dodecyl sulphate | 16/4 (4) | 32/4 (8) | 16/4 (4) | 32/8 (4) | 64/8 (8) | 256/8 (32) | 256/16 (16) |
Values indicate minimal inhibitory concentrations of the wild-type/its isogenic cmeB mutant. Values in bold are fold changes.
There were 8- to 16- and 4- to 64-fold changes in the MICs of ciprofloxacin and erythromycin, respectively (Table 3). It was known from previous studies that CmeABC and target mutations, such as the 23S rRNA and gyrA mutations, work synergistically to confer high-level resistance to macrolide and fluoroquinolone antibiotics in C. jejuni and C. coli (Luo et al., 2003; Cagliero et al., 2005; Mamelli et al., 2005). The high-level resistance of C. coli TF1-13 to erythromycin and C. lari 3121 to ciprofloxacin observed in this study was likely owing to the presence of resistance-conferring mutations in these two isolates. Regardless, mutation of cmeB in these two isolates resulted in a significant reduction in the MICs of ciprofloxacin or erythromycin (Table 3), further indicating the importance of CmeABC in conferring resistance to the antibiotics. The cmeB mutation also led to 2- to 16-fold reduction in the MICs of tetracycline. The resistance to gentamicin was slightly affected by the cmeB mutation in C. jejuni and C. coli (twofold change in the MIC), but not in C. upsaliensis, C. lari, and C. fetus. The cmeB mutation also affected the susceptibility to ampicillin in different Campylobacter spp., with 2- to16-fold changes in the MIC observed.
All Campylobacter spp. examined in this study were highly resistant to cephalothin and cefoperazone and moderately resistant to cefotaxime (Table 3). Consistent and drastic decreases in MICs of cephalosporin antibiotics (8- to 2048-fold) were observed in the cmeB mutants of different Campylobacter species. Previously, the high-level intrinsic resistance of Campylobacter to cephalosporins was attributed to the low binding affinity of the antibiotics to the bacterial penicillin-binding protein or the low permeability of the cell membrane (Lachance et al., 1991). The results from this study suggest that the efflux mediated by CmeABC also plays a significant role in the resistance to these cephalosporin antibiotics in different Campylobacter spp.
Campylobacter is intrinsically resistant to rifampin, and mutation of CmeABC was shown to cause a dramatic decrease in the MIC of rifampin in C. jejuni. Similar results were observed in this study with C. coli, C. upsaliensis, and C. lari; however, no changes in the MIC of rifampin occurred in the cmeB mutant of C. fetus (Table 3). This finding suggests that CmeABC is important for the intrinsic resistance to rifampin in C. jejuni, C. coli, C. upsaliensis, and C. lari, but does not play a role in rifampin resistance in C. fetus. Rifampin resistance in other bacteria is conferred by mutations in the DNA-dependent RNA polymerase (RpoB) (Musser, 1995), but in some Campylobacter spp. the resistance appears to be mainly mediated by efflux. Because the cmeB mutation did not affect the MIC of rifampin in C. fetus, the resistance in this species is likely due to a different mechanism.
The cmeB mutation also significantly increased the susceptibility to cholic acid in all five species of Campylobacter, indicating that this efflux pump plays a major role in the adaptation to bile-containing environments such as the intestinal tract (Table 3). The resistance to sodium dodecyl sulphate and ethidium bromide was also decreased (Table 3). These results are consistent with previous findings with CmeABC in C. jejuni and indicate that CmeABC is important for the resistance to antimicrobial compounds in different Campylobacter spp.
Several antibiotics, such as rifampin, cephalothin, and cefoperazone, have been incorporated into commercial Campylobacter selective media for the isolation of Campylobacter spp. from clinical samples (Karmali et al., 1981; Goossens et al., 1986; Ng et al., 1988; Burnens and Nicolet, 1992). The results in this study indicate that CmeABC is associated with high-level intrinsic resistance to these antibiotics and our data also provide a molecular explanation for the use of these antibiotics as Campylobacter selective agents. On the other hand, the results from this study also revealed that C. upsaliensis appears to be intrinsically more susceptible to the often used selective antibiotics, which highlights the need to lower the concentration of antibiotics in selective media to successfully recover C. upsaliensis from clinical samples. It has been reported that the use of filtration has a significantly better recovery rate of C. upsaliensis in clinical samples than using selective medium (Lastovica and Le Roux, 2001). Further work in understanding the antimicrobial resistance mechanisms will facilitate the selection of antibiotics for clinical treatment of campylobacteriosis and the formulation of diagnostic media for various Campylobacter spp.
Acknowledgments
The authors thank Irene V. Wesley at the USDA ARS National Animal Disease Center and Nelson Moyer at the University of Iowa for providing some of the Campylobacter isolates used in this study. This work was supported by the National Institutes of Health (grant no. 5RO1DK063008).
Disclosure Statement
No competing financial interests exist.
References
- Akiba M. Lin J. Barton YW. Zhang Q. Interaction of CmeABC and CmeDEF in conferring antimicrobial resistance and maintaining cell viability in Campylobacter jejuni. J Antimicrob Chemother. 2006;57:52–60. doi: 10.1093/jac/dki419. [DOI] [PubMed] [Google Scholar]
- Allos BM. Campylobacter jejuni infections: update on emerging issues and trends. Clin Infect Dis. 2001;32:1201–1206. doi: 10.1086/319760. [DOI] [PubMed] [Google Scholar]
- Bourke B. Chan VL. Sherman P. Campylobacter upsaliensis: waiting in the wings. Clin Microbiol Rev. 1998;11:440–449. doi: 10.1128/cmr.11.3.440. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Burnens AP. Nicolet J. Detection of Campylobacter upsaliensis in diarrheic dogs and cats, using a selective medium with cefoperazone. Am J Vet Res. 1992;53:48–51. [PubMed] [Google Scholar]
- Cagliero C. Cloix L. Cloeckaert A. Payot S. High genetic variation in the multidrug transporter cmeB gene in Campylobacter jejuni and Campylobacter coli. J Antimicrob Chemother. 2006;58:168–172. doi: 10.1093/jac/dkl212. [DOI] [PubMed] [Google Scholar]
- Cagliero C. Mouline C. Payot S. Cloeckaert A. Involvement of the CmeABC efflux pump in the macrolide resistance of Campylobacter coli. J Antimicrob Chemother. 2005;56:948–950. doi: 10.1093/jac/dki292. [DOI] [PubMed] [Google Scholar]
- Corcoran D. Quinn T. Cotter L. O'Halloran F. Fanning S. Characterization of a cmeABC operon in a quinolone-resistant Campylobacter coli isolate of Irish origin. Microb Drug Resist. 2005;11:303–308. doi: 10.1089/mdr.2005.11.303. [DOI] [PubMed] [Google Scholar]
- Fakhr MK. Logue CM. Sequence variation in the outer membrane protein-encoding gene cmeC, conferring multidrug resistance among Campylobacter jejuni and Campylobacter coli strains isolated from different hosts. J Clin Microbiol. 2007;45:3381–3383. doi: 10.1128/JCM.01208-07. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fouts DE. Mongodin EF. Mandrell RE. Miller WG. Rasko DA. Ravel J. Brinkac LM. DeBoy RT. Parker CT. Daugherty SC. Dodson RJ. Durkin AS. Madupu R. Sullivan SA. Shetty JU. Ayodeji MA. Shvartsbeyn A. Schatz MC. Badger JH. Fraser CM. Nelson KE. Major structural differences and novel potential virulence mechanisms from the genomes of multiple Campylobacter species. PLoS Biol. 2005;3:e15. doi: 10.1371/journal.pbio.0030015. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Goossens H. De Boeck M. Coignau H. Vlaes L. Van den Borre C. Butzler JP. Modified selective medium for isolation of Campylobacter spp. from feces: comparison with Preston medium, a blood-free medium, and a filtration system. J Clin Microbiol. 1986;24:840–843. doi: 10.1128/jcm.24.5.840-843.1986. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Guerry P. Yao R. Alm RA. Burr DH. Trust TJ. Systems of experimental genetics for Campylobacter species. Methods Enzymol. 1994;235:474–481. doi: 10.1016/0076-6879(94)35163-5. [DOI] [PubMed] [Google Scholar]
- Guillaume G. Ledent V. Moens W. Collard JM. Phylogeny of efflux-mediated tetracycline resistance genes and related proteins revisited. Microb Drug Resist. 2004;10:11–26. doi: 10.1089/107662904323047754. [DOI] [PubMed] [Google Scholar]
- Karmali MA. De Grandis S. Fleming PC. Antimicrobial susceptibility of Campylobacter jejuni with special reference to resistance patterns of Canadian isolates. Antimicrob Agents Chemother. 1981;19:593–597. doi: 10.1128/aac.19.4.593. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lachance N. Gaudreau C. Lamothe F. Lariviere LA. Role of the beta-lactamase of Campylobacter jejuni in resistance to beta-lactam agents. Antimicrob Agents Chemother. 1991;35:813–818. doi: 10.1128/aac.35.5.813. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lastovica AJ. Allos BM. Clinical significance of Campylobacter and related species other than Campylobacter jejuni and Campylobacter coli. In: Nachamkin I, editor; Szymanski CM, editor; Blaser MJ, editor. Campylobacter. 3rd. Washington, DC: ASM Press; 2008. pp. 123–149. [Google Scholar]
- Lastovica AJ. Le Roux E. Efficient isolation of Campylobacter upsaliensis from stools. J Clin Microbiol. 2001;39:4222–4223. doi: 10.1128/JCM.39.11.4222-4223.2001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lin J. Akiba M. Sahin O. Zhang Q. CmeR functions as a transcriptional repressor for the multidrug efflux pump CmeABC in Campylobacter jejuni. Antimicrob Agents Chemother. 2005a;49:1067–1075. doi: 10.1128/AAC.49.3.1067-1075.2005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lin J. Cagliero C. Guo B. Barton YW. Maurel MC. Payot S. Zhang Q. Bile salts modulate expression of the CmeABC multidrug efflux pump in Campylobacter jejuni. J Bacteriol. 2005b;187:7417–7424. doi: 10.1128/JB.187.21.7417-7424.2005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lin J. Michel LO. Zhang Q. CmeABC functions as a multidrug efflux system in Campylobacter jejuni. Antimicrob Agents Chemother. 2002;46:2124–2131. doi: 10.1128/AAC.46.7.2124-2131.2002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lin J. Sahin O. Michel LO. Zhang Q. Critical role of multidrug efflux pump CmeABC in bile resistance and in vivo colonization of Campylobacter jejuni. Infect Immun. 2003;71:4250–4259. doi: 10.1128/IAI.71.8.4250-4259.2003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Luangtongkum T. Jeon B. Han J. Plummer P. Logue CM. Zhang Q. Antibiotic resistance in Campylobacter: emergence, transmission, and persistence. Future Microbiol. 2009;4:189–200. doi: 10.2217/17460913.4.2.189. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Luo N. Sahin O. Lin J. Michel LO. Zhang Q. In vivo selection of Campylobacter isolates with high levels of fluoroquinolone resistance associated with gyrA mutations and the function of the CmeABC efflux pump. Antimicrob Agents Chemother. 2003;47:390–394. doi: 10.1128/AAC.47.1.390-394.2003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mamelli L. Prouzet-Mauleon V. Pages JM. Megraud F. Bolla JM. Molecular basis of macrolide resistance in Campylobacter: role of efflux pumps and target mutations. J Antimicrob Chemother. 2005;56:491–497. doi: 10.1093/jac/dki253. [DOI] [PubMed] [Google Scholar]
- Martinot M. Jaulhac B. Moog R. De Martino S. Kehrli P. Monteil H. Piemont Y. Campylobacter lari bacteremia. Clin Microbiol Infect. 2001;7:96–97. doi: 10.1046/j.1469-0691.2001.00212.x. [DOI] [PubMed] [Google Scholar]
- Musser JM. Antimicrobial agent resistance in mycobacteria: molecular genetic insights. Clin Microbiol Rev. 1995;8:496–514. doi: 10.1128/cmr.8.4.496. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ng LK. Taylor DE. Stiles ME. Characterization of freshly isolated Campylobacter coli strains and suitability of selective media for their growth. J Clin Microbiol. 1988;26:518–523. doi: 10.1128/jcm.26.3.518-523.1988. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Piddock LJ. Clinically relevant chromosomally encoded multidrug resistance efflux pumps in bacteria. Clin Microbiol Rev. 2006;19:382–402. doi: 10.1128/CMR.19.2.382-402.2006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Poole K. Efflux-mediated antimicrobial resistance. J Antimicrob Chemother. 2005;56:20–51. doi: 10.1093/jac/dki171. [DOI] [PubMed] [Google Scholar]
- Pumbwe L. Piddock LJ. Identification and molecular characterisation of CmeB, a Campylobacter jejuni multidrug efflux pump. FEMS Microbiol Lett. 2002;206:185–189. doi: 10.1111/j.1574-6968.2002.tb11007.x. [DOI] [PubMed] [Google Scholar]
- Tee W. Mijch A. Campylobacter jejuni bacteremia in human immunodeficiency virus (HIV)-infected and non-HIV-infected patients: comparison of clinical features and review. Clin Infect Dis. 1998;26:91–96. doi: 10.1086/516263. [DOI] [PubMed] [Google Scholar]
- Vandamme P. Taxonomy of the family Campylobacteraceae. In: Nachamkin I, editor; Blaser MJ, editor. Campylobacter. 2nd. Washington, DC: ASM Press; 2000. pp. 3–26. [Google Scholar]