Abstract
Bird migration is one of the most spectacular and best-studied phenomena in behavioural biology. Yet, while the patterns of variation in migratory behaviour and its ecological causes have been intensively studied, its genetic, physiological and neurological control remains poorly understood. The lack of knowledge of the molecular basis of migration is currently not only limiting our insight into the proximate control of migration, but also into its evolution. We investigated polymorphisms in the exons of six candidate genes for key behavioural traits potentially linked to migration, which had previously been identified in several bird species, and eight control loci in 14 populations of blackcaps (Sylvia atricapilla), representing the whole range of geographical variation in migration patterns found in this species, with the aim of identifying genes controlling variation in migration. We found a consistent association between a microsatellite polymorphism and migratory behaviour only at one candidate locus: the ADCYAP1 gene. This polymorphism explained about 2.6 per cent of the variation in migratory tendency among populations, and 2.7–3.5% of variation in migratory restlessness among individuals within two independent populations. In all tests, longer alleles were associated with higher migratory activity. The consistency of results among different populations and levels of analysis suggests that ADCYAP1 is one of the genes controlling the expression of migratory behaviour. Moreover, the multiple described functions of the gene product indicate that this gene might act at multiple levels modifying the shift between migratory and non-migratory states.
Keywords: ADCYAP1, PACAP, migratory restlessness, bird migration, neuropeptide
1. Introduction
Each year roughly 50 billion birds, involving about half of all avian species, perform some type of migratory movement [1]. This behaviour has fascinated lay people and scientists alike, and is probably one of the biological phenomenon with the longest research tradition [1,2]. Despite extensive research over decades, the molecular, physiological and endocrinological mechanisms underlying the regulation of migratory movements remain largely unknown [2–6]. In particular, it remains unclear whether endocrine changes are a cause or an effect of migratory processes [3]. There is a clear need to study the molecular genetic basis of migration, which may bridge the gap between genes and phenotype. Knowledge of genetic differences and differential gene expression between migratory and non-migratory individuals will help in solving the causality problem encountered in purely physiological studies. Studies on the molecular basis of migration may also help to improve our understanding of the evolutionary history involved in changes in migratory behaviour in response to past environmental shifts.
Before and during migration, migratory birds undergo a profound and synchronized shift in a set of physiological adaptations and behavioural traits. Migratory disposition, i.e. a state of readiness for prolonged flights, comprises hypertrophy of flight muscles, fat deposition in the adipose tissue, integrated changes in enzyme activities involved in the energy metabolism, hyperphagia, dietary changes and the development of migratory activity [4,7]. From a behavioural ecology perspective, the ability to shift circadian activity during migration represents a key element of avian migratory behaviour. Indeed, many species of diurnally active birds switch to additional nocturnal activity during the migratory season [4]. Such a change in the circadian activity patterns involves substantial physiological and hormonal shifts. Birds migrating at night maintain high levels of physical and cognitive functions such as prolonged flight, navigation performance and alertness against predators at times when they usually sleep [8]. It is suggested that components of the endogenous circadian clock control the nocturnal migratory behaviour [9–12]. Personality traits have also been discussed in the context of variation in migratory behaviour. For example, it has been suggested that initiation of migration behaviour and migration distance are related to individual competitive ability or dominance [13], which in turn may be linked to aggression and anxiety-related behaviour [14]. Furthermore, migratory and non-migratory birds may differ in exploratory behaviour [15–17].
Many components of migratory behaviour, such as the amount, timing and intensity of migratory activity, are under strong genetic control, at least in small night-migrating passerines [18–21]. Also cross-breeding experiments among groups of European blackcaps (Sylvia atricapilla) that differed in migratory behaviour indicated a strong genetic basis of this behaviour [22,23]. In the European blackcap, a model species for the study of avian migration, a new wintering area was established in only a few decades, involving evolutionary changes in migration distance and direction [24,25]. Moreover, a strong evolutionary reduction of migratory activity has been observed in a blackcap population, presumably in response to climate change [26]. These results suggest that the evolution of migratory behaviour in a resident population or of residency in a migratory population may be a common and rapid process [21]. Furthermore, quantitative genetic studies provide evidence for the genetic integration of migratory traits [20,27]. High genetic correlations among incidence, amount, intensity and timing of migratory activity in blackcaps suggest that these components of migratory behaviour are influenced by common genetic mechanisms [20,27]. As a consequence, we would expect that phenotypic variation of correlated migratory traits is linked to genetic variation at a single closely linked gene cluster or a few ‘regulatory genes’ with multiple pleiotropic effects [28].
Based on two behavioural elements of avian migration, nocturnality and exploratory behaviour, we selected nine exonic polymorphic loci in six candidate genes. The loci are reported microsatellites and single-nucleotide polymorphisms (SNPs) in the exons (coding and untranslated exonic regions (UTRs)) of candidate genes in birds that are known to be involved in the expression of circadian rhythms [29] or personality traits [30–32]. We made use of the profound knowledge on blackcap migration and of the large amount of geographical and within-population variation in migratory behaviour found in this species [21,33] to study the association between allelic variation at candidate loci and migratory behaviour at two levels: (i) among individuals within populations, and (ii) between populations that vary in the proportion of migrants and in migration distance. Here, we show that migratory restlessness is consistently associated with allele length at a 3′-UTR locus of the adenylate cyclase-activating polypeptide 1 (ADCYAP1) gene in two independent populations. Moreover, the same alleles are correlated with an estimated higher proportion of migratory individuals across 14 blackcap populations.
2. Material and methods
(a). Samples
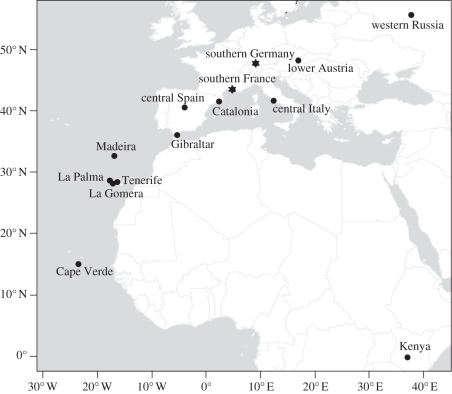
Thirteen European/African blackcap populations representing the entire range of geographical variation in migration patterns, from Cape Verde to western Russia, have been sampled in the years 1989–1996 (figure 1). We also included a sample of birds captured in Kenya in the year 2000. The geographical coordinates and sampling information for each population are listed in electronic supplementary material, table S1. All birds investigated were sampled randomly within a restricted geographical area, which we defined as population. Birds held in captivity (Madeira, southern France, southern Germany, western Russia) were collected as nestlings from the populations in the wild. All other birds—except those from central Italy and Kenya—were captured with mistnets in the wild during or after the reproductive season, but before the start of migration. The samples of central Italy and Kenya were captured with mistnets during winter.
Figure 1.
Map of the sampled blackcap populations. The two populations, which were used for the intrapopulational analyses are indicated with stars.
Blood samples (ca 50 µl) were obtained by puncturing the brachial vein. An isotonic NaCl–EDTA buffer (0.85% NaCl) was used to prevent blood cell lysis and coagulation. For the genetic analyses, the erythrocyte fraction was used as a source of DNA.
(b). Determination of migratory status of individuals and populations
Migratory behaviour of individuals from the southern France and southern Germany populations was quantified by measuring nocturnal migratory activity in registration cages under identical, standardized conditions. These measures of migratory activity have been analysed and published previously (southern France: [27,34] and southern Germany: [18,26]). The amount of migratory activity was measured in inexperienced hand-reared birds as the total number of 30 min intervals with activity during the autumn migration period. This variable is correlated to the distance the bird would migrate in the wild [4].
The migratory status of each population was classified using all available information on the migratory behaviour of individuals from that population, including capture–recapture data, direct observations and laboratory measures of migratory restlessness (Cape Verde, Tenerife, Madeira, southern France, southern Germany, lower Austria and western Russia). Integrating this information, we classified blackcaps from Cape Verde [23] and Gibraltar [35,36] as being completely resident, blackcaps from Tenerife [24], Madeira [37] and, presumably, La Gomera and La Palma as residents, but showing some residual migratory restlessness in the laboratory. Blackcaps breeding in Catalonia (G. Gargallo 1992, personal communication) and southern France [27,34] are partially migratory. Birds breeding in the uplands north of Madrid, central Spain, are migratory but, presumably, migrate only short distances to the south of the Iberian Peninsula [36]. The blackcap populations breeding in southern Germany and lower Austria and the winter sample of central Italy are presumably completely migratory with largely intermediate migration distances [38,39]. Finally, individuals from western Russia and the winter sample of Kenya are classified as distinct long-distance migrants (all birds covering distances greater than 3500 km). This pattern is in general agreement with a leap-frog migration pattern [1].
(c). Genotyping
We selected all known exonic di- and trinucleotide microsatellite loci in candidate genes for circadian behaviour: CLOCK, ADCYAP1, CREB1 and NPAS2 (for selection strategy, primer, PCR and scoring details see [29]). We also genotyped one exonic trinucleotide microsatellite and four exonic SNPs of the major candidate genes for ‘exploratory behaviour’ or ‘anxiety-related behaviour’: DRD4 and SERT (for details, see electronic supplementary material, table S2). This set represents a complete list of all currently known microsatellites in exons of candidate genes for circadian behaviour and personality in birds [29,31]. We focused on exonic microsatellites because they are mostly conserved across species, thus promoting primer transferability. As there was no microsatellite in the DRD4 candidate gene, we identified blackcap-specific SNPs by sequencing eight blackcap individuals at exon 3 of the DRD4 gene. This limited set of easy accessible polymorphisms with exonic location has been selected because of their a priori chance to directly influence gene product structure and gene expression.
For comparative association analyses, we used eight anonymous di- and tetranucleotide microsatellite loci (Syl1, Syl2, Syl4, Syl5, Syl6, Syl9, Ppi2 and Pca8), which are presumably neutral (for genotyping details see [40] and electronic supplementary material, table S2). All birds were sexed using Griffiths et al.'s [41] P2 and P8 primers. An ABI 3130 sequencer was used for the microsatellite fragment analysis and the ABI SnaPshot protocol for SNP genotyping.
(d). Data analyses
We tested associations between individual migratory restlessness and genotypes using mixed-effects regression models. Because the samples of southern France and southern Germany comprised some sibships (i.e. nestlings collected from the same nest), we minimized the effect of pseudoreplication by including sibship as a random factor in the models (R package lme4 [42,43]). Sibship was always significant (p < 0.002; tested with R package RLRsim [44]) and the variance owing to sibship was estimated as 43 and 29 per cent of the total variance in southern France and southern Germany, respectively, reflecting the high heritability in this trait [18,26]. We also tested the interaction between sex and genotype, but this was not significant in both populations. This term was, therefore, excluded from the final model. All p-value and standardized regression coefficient estimates are based on 10 000 Markov chain Monte Carlo (MCMC) samples (R package languageR [45]). Individual microsatellite genotypes were coded as mean allele lengths averaged over the two alleles and SNP genotypes were coded according to the allele dose model (copy number of one of the two alleles) in the within-population analyses. These standard genotype coding models are powerful with one degree of freedom, and are known to capture most allelic effects in association studies [46]. Note, however, that these models implicitly assume that alleles are codominant and that allele length is linearly related to variation in the phenotype.
We analysed the relationship between the migratory status and the genetic composition of populations using partial Mantel tests with 10 000 permutations on appropriate distance matrices accounting for geographical distances ([47], R package ecodist [48]). In samples where sibships were included (i.e. Madeira, southern France, southern Germany and western Russia), we randomly selected one individual per sibship. The genetic differentiation at each locus and for all population pairs was calculated as multi-allelic FST-values according to Weir & Cockerham [49] using Genepop [50]. Distances in migratory status were calculated after coding pure resident populations as ‘0’, resident populations with some migratory restlessness as ‘0.5’, partial migratory populations as ‘1’, completely migratory populations migrating short-distances as ‘1.5’, intermediate-distance migratory populations as ‘2’ and distinct long-distance migratory populations as ‘2.5’ (electronic supplementary material, table S1). Surface geographical distances were calculated using the R package gmt [51]. Genetic variance partitioning among groups of differing migration status was performed in a hierarchical analysis of molecular variance (AMOVA) framework with 10 000 permutations using Arlequin [52].
3. Results
(a). Within-population tests
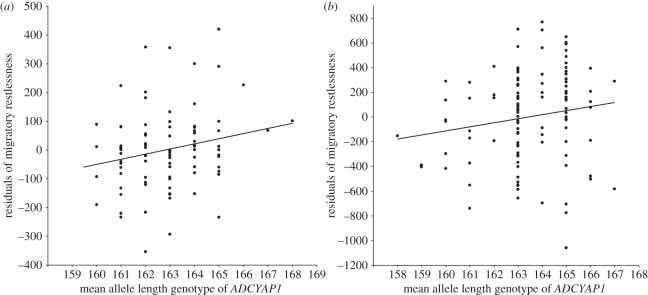
In both populations with data on individual migratory activity (southern France and southern Germany; figure 1), migratory restlessness was associated with the genotypes of the ADCYAP1 locus (table 1). Individual mean allele length at ADCYAP1 correlated positively with migratory restlessness in both populations (figure 2). The mean genotypes explained 2.7 and 3.5 per cent of the variance in migratory restlessness in southern France and southern Germany, respectively. Note that the test in the southern France population failed nominal significance (p = 0.056). We consider this a type II statistical error, given the smaller sample size, and the consistency of allelic effects in terms of strength and direction in both populations and in the among-population test (see below). We also explored two alternative genotype coding models at the ADCYAP1 locus by using the shorter (or longer) allele in each individual as a measure of genotype. Whereas the ‘longer allele’ model was always non-significant (p > 0.22), the ‘shorter allele’ genotypes were significantly associated with migratory restlessness in both southern France (p = 0.048; standardized regression coefficient = 0.21) and southern Germany (p = 0.022; standardized regression coefficient = 0.21). This could indicate that the shorter alleles are more effective in influencing migratory restlessness in these populations than longer alleles (but see among-population results). We also found significant associations with migratory restlessness at two control microsatellite loci (table 1), but these were not consistent across the within- and among-population tests.
Table 1.
Results (p-values) of three independent association tests between variation in migratory behaviour and genetic variation at 17 polymorphic loci. n.a., not applicable owing to monomorphic locus. p-values < 0.05 are in bold; 0.05 < p-value < 0.1 in bold italic.
| gene/locus name | southern Francea (n = 87)c | southern Germanya (n = 119)c | all populationsb |
|---|---|---|---|
| CLOCK | 0.11 (0.16) | 0.76 (0.03) | 0.59 (−0.04) |
| ADCYAP1 | 0.056 (0.24) | 0.025 (0.20) | 0.0038 (0.45) |
| CREB1 | 0.74 (−0.04) | 0.12 (0.14) | 0.80 (−0.15) |
| NPAS2 | n.a. | 0.76 (0.03) | n.a. |
| SERT | 0.67 (0.05) | n.a. | n.a. |
| DRD4_366 | 0.34 (−0.04) | 0.24 (−0.09) | 0.83 (−0.17) |
| DRD4_524 | 0.39 (−0.02) | n.a. | n.a. |
| DRD4_815 | 0.59 (0.01) | n.a. | n.a. |
| DRD4_890 | 0.50 (−0.02) | 0.91 (0.01) | 0.70 (−0.08) |
| Syl1 | 0.31 (0.10) | 0.91 (0.01) | 0.82 (−0.15) |
| Syl2 | 0.046 (0.22) | 0.27 (−0.11) | 0.11 (0.23) |
| Syl4 | 0.61 (0.05) | 0.30 (0.09) | 0.75 (−0.11) |
| Syl5 | 0.85 (−0.04) | 0.40 (−0.07) | 0.77 (−0.11) |
| Syl6 | 0.10 (0.19) | 0.60 (−0.05) | 0.21 (0.15) |
| Syl9 | 0.74 (0.03) | 0.21 (−0.12) | 0.16 (0.19) |
| Ppi2 | 0.020 (−0.27) | 0.063 (−0.17) | 0.83 (−0.15) |
| Pca8 | 0.088 (−0.19) | 0.70 (−0.03) | 0.44 (0.004) |
aIndividual-based association between migratory restlessness and mean allele length genotype (microsatellites) or allele dose genotype (SNPs) using mixed-effects models with sibship as random factor (in brackets MCMC estimate of standardized regression coefficient).
bPopulation-based correlation between migration status distances and genetic differentiation (FSTs) among all summer population samples using a partial Mantel test conditional on geographical distances (in brackets Mantel correlation coefficient).
cActual sample sizes. The smaller sample sizes in comparison to the ones presented in electronic supplementary material, table S1 are explained by the fact that migratory restlessness was not measured in all birds for which DNA samples were available.
Figure 2.
Mean allele length genotypes at the ADCYAP1 locus plotted against residuals of the null mixed-effects model on migratory restlessness with sibship as random factor and no fixed effect (genotype). (a) Southern France and (b) southern Germany.
(b). Among-population tests
We found a strong general correlation between the between-population differentiation in migration status and genetic differentiation at the ADCYAP1 locus, but not at any other locus (table 1). To account for geography-based genetic similarity between breeding populations owing to common colonization histories and gene flow, the analysis included surface geographical distances between sample sites, and excluded the winter samples of central Italy and Kenya, for which the breeding areas of the birds could not be determined. The results of this analysis were robust against slightly different estimates in migration status for populations with little migration data. The genotypic variance at the ADCYAP1 locus explained 2.6 per cent of the variance among groups of populations differing in migration status (AMOVA). It is thus similar to the variance in migratory restlessness among individuals explained by this polymorphism. Note, however, that this estimate might be inflated because the AMOVA does not account for genetic similarity owing to geographical proximity of the populations.
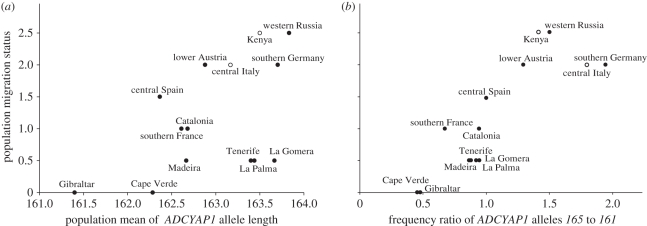
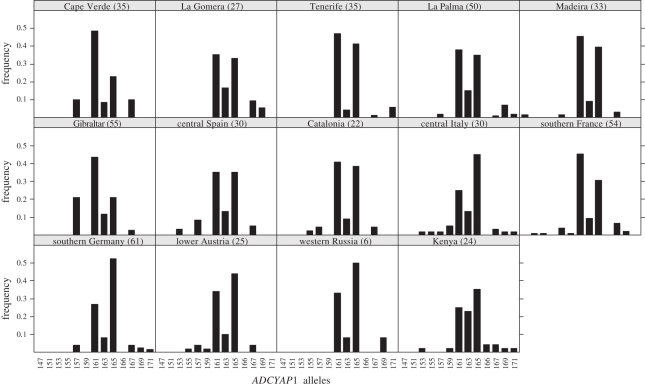
The distance approach used above does not provide information on the direction of the relationship between migration status and allele length. To explore the direction of the relationship, we directly tested the correlation between population migration status and population mean allele length and found a significant positive correlation (Spearman rank correlation: ρ = 0.57, p = 0.034; figure 4a). We also tested this relationship with alternative population summary statistics, such as median allele length (electronic supplementary material, figure S1), mean of the shorter (or longer) allele in each individual (electronic supplementary material, figure S1), proportion of allele 161 or shorter (Spearman rank correlation: ρ = −0.78, p = 0.0011) and proportion of allele 165 or longer (Spearman rank correlation: ρ = 0.62, p = 0.017). The allele frequency distribution at the ADCYAP1 locus showed a bimodal pattern in all populations (figure 3). The most frequent alleles (161 and 165) are two mutational steps apart, assuming a stepwise mutation model, and show considerable frequency variation among populations. As this pattern is indicative of an old balanced polymorphism, we also tested the frequency ratio of these two major alleles as a potential predictor for population migration status. Migration status was significantly linked to this frequency ratio (165 to 161) (Spearman rank correlation: ρ = 0.87, p = 0.000054), whereby the longer allele was more prevalent in more migratory populations (figure 4b).
Figure 4.
The association between population migration status and (a) population mean of ADCYAP1 allele length (Spearman rank correlation: ρ = 0.57, p = 0.034) or (b) the frequency ratio between ADCYAP1 alleles 165 and 161 (Spearman rank correlation: ρ = 0.87, p = 0.000054). Here, the winter samples of Kenya and central Italy are included in the analyses.
Figure 3.
Allele frequency distributions of the ADCYAP1 locus in 14 blackcap populations (sample sizes in brackets).
All but one of the seven—non-independent—tests of the among-population effect were significant, indicating the robustness of the association between population migration status and allele length at the ADCYAP1 polymorphism. The combinatorial probability of finding three significant effects at the same locus in the same direction in three independent studies (two within- and one between-population study) on 14 loci each (here, the four DRD4 loci are treated as one locus) by chance equals 0.00017.
4. Discussion
In this study, we investigated the effect of nine known exonic polymorphisms in six candidate genes for behavioural traits on the expression of migratory behaviour in a migratory bird species. Three independent tests indicate that long alleles at a microsatellite in the 3′-UTR of the ADCYAP1 gene are associated with high migratory activity in blackcaps, either measured as migratory restlessness of individuals in the laboratory or assessed as the proportion of migrants and migration distance in natural populations. Both the within-population analyses and the among-population comparison indicate that genotypic variation at this gene explains about 3 per cent of phenotypic variation in ‘migratoriness’, which amounts to a maximum of 6–8% of the additive genetic variance in this trait, assuming a mean heritability of this trait of 0.43 [26]. This represents a relatively large single-gene effect on a complex behavioural trait when compared with reported genetic effects on other complex traits [53,54]. There are only few other gene variants reported to influence a behavioural trait in a wild bird population with a similar strength, for instance, an exonic DRD4 SNP explaining about 5 per cent of the exploratory behaviour in great tits [31,32]. The large proportion of unexplained additive genetic variance indicates that many additional still unknown loci contribute to the expression of migratory behaviour. In general, complex traits are expected to show a genetic architecture with a high number of contributing genes with epistatic effects and gene by environment interactions [53,55].
The ADCYAP1 polymorphism is located in the 3′-UTR of the gene, which is known to comprise important regulatory elements of post-transcriptional processes [56–58]. It has been suggested that the insertion of simple sequence repeats in 3′-UTR regulatory elements and the structural variation at the 3′-UTR mediated by microsatellite variation can interact with and modify the 3′-UTR regulatory functions [59,60]. The polymorphism could also be in linkage disequilibrium with a different functional polymorphism in the gene region influencing peptide structure or transcription level. The allelic association, however, needs to be consistent across the populations to explain the observed patterns. Only direct expression studies in different tissues can reveal the link between genotypes and levels of the different splice variants and/or isoforms at the ADCYAP1 gene [61]. General functionality of the ADCYAP1 polymorphism is indicated by conservation across avian and mammalian species. A similar dinucleotide sequence repeat in the 3′-UTR with different levels of motif purity and polymorphism (where tested) has been found in 40 bird species, human, mouse and rat ([29,62]; our unpublished data; UCSC genome browser at http://genome.ucsc.edu/). Obviously, only further work on other species will show to what extent the association between the polymorphism and the expression of avian migration can be generalized.
The ADCYAP1 gene encodes the pituitary adenylate cyclase-activating polypeptide (PACAP), which is one of the most studied neuropeptides (more than 3000 papers deal directly with PACAP) ([61], Web of Science at http://apps.isiknowledge.com). The peptide and its receptors are widely distributed in the brain and in various peripheral organs [61,63]. In comparison with the products of our other tested candidate loci, PACAP has a broad spectrum of biological functions with profound influence on physiology and behaviour. Most of the reported effects exerted by PACAP are indeed strongly linked to the physiological and behavioural shifts described for avian migration. For example, exposure of the chicken pineal gland to PACAP induces a transitory increase in melatonin secretion, but does not cause phase shift of the melatonin rhythm [64–66]. PACAP has also been shown to directly influence clock gene expression [67] and affect signalling pathways that integrate the molecular clock in the functionality of circadian rhythms in a dose- and phase-dependent manner [68]. Further studies are needed to determine whether the ADCYAP1 polymorphism exerts its effect on nocturnal migratory restlessness via a phase-shift of the endogenous oscillator(s) or via a modulation of the downstream processes of the molecular clock. PACAP has also a strong modifying effect on the energy metabolism. Intracerebroventricularly administered PACAP appeared to stimulate catabolic effects on energy metabolism in chicken [69]: it increased body temperature, metabolic rate and lipid utilization. Moreover, increased PACAP concentrations in the chicken brain inhibited feeding [70], which was later shown to be mediated by corticosterone release [71]. All these shifts in metabolism and feeding behaviour have been described as the characteristics of birds preparing for or performing long-distance migratory flights [4]. Given the multiple pleiotropic functions of PACAP, ADCYAP1 has at least the potential to modify multiple physiological and behavioural changes during the migratory period. Similar pleiotropic regulators have been postulated to explain expression covariation in large gene sets for different life-history transitions, including a migration state transition, in the Atlantic salmon [72].
Comparative studies suggest that migratory activity and/or residency have rapidly and independently evolved in different bird lineages as a response to environmental changes, and recently to global warming [21,26,73]. The existence of a strong positive correlation between the frequency of migrants in a population and the average migratory activity of individuals has led to the threshold model hypothesis [27]. This model links the continuous trait of migratory restlessness to the phenotypic dichotomy between migrants and non-migrants. We hypothesize that adaptive allele frequency shifts at the ADCYAP1 locus could modulate population migratory activity and, as a consequence, the frequency of migratory and resident individuals in a population.
Acknowledgements
We are grateful to Peter Berthold for initiating genetic studies on bird migration and critical discussions, which have been fundamental to this study. We thank Wolfgang Fiedler and Leonida Fusani for providing blackcap samples from the Cape Verde Islands, central Italy and Kenya, and the numerous field assistants that helped catching blackcaps.
This work was supported by the Max Planck Society. F.P. was financially supported by grants from the Spanish Ministry of Science and Innovation (MICINN; RYC-2007-01861 and CGL2009-12397). The funders had no role in study design, data collection and analysis, decision to publish, or preparation of the article.
References
- 1.Berthold P. 2001. Bird migration: a general survey, 2nd edn Oxford, UK: Oxford University Press [Google Scholar]
- 2.Newton I. 2008. The migration ecology of birds. London, UK: Academic Press [Google Scholar]
- 3.Wingfield J. C., Schwabl H., Mattocks P. W., Jr 1990. Endocrine mechanisms of migration. In Bird migration: physiology and ecophysiology (ed. Gwinner E.), pp. 232–256 Berlin, Germany: Springer [Google Scholar]
- 4.Berthold P. 1996. Control of bird migration. London, UK: Chapman and Hall [Google Scholar]
- 5.Silverin B. 2003. Behavioural and hormonal dynamics in a partial migrant—the willow tit. In Avian migration (eds Berthold P., Gwinner E., Sonnenschein E.), pp. 127–140 Berlin, Germany: Springer [Google Scholar]
- 6.Wingfield J. C. 2003. Avian migration: regulation of facultative-type movements. In Avian migration (eds Berthold P., Gwinner E., Sonnenschein E.), pp. 113–125 Berlin, Germany: Springer [Google Scholar]
- 7.Ramenofsky M., Wingfield J. C. 2007. Regulation of migration. Bioscience 57, 135–143 10.1641/B570208 (doi:10.1641/B570208) [DOI] [Google Scholar]
- 8.Rattenborg N. C., Mandt B. H., Obermeyer W. H., Winsauer P. J., Huber R., Wikelski M., Benca R. M. 2004. Migratory sleeplessness in the white-crowned sparrow (Zonotrichia leucophrys gambelii). PLoS Biol. 2, 924–936 10.1371/journal.pbio.0020212 (doi:10.1371/journal.pbio.0020212) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Gwinner E., Helm B. 2003. Circannual and circadian contributions to the timing of avian migration. In Avian migration (eds Berthold P., Gwinner E., Sonnenschein E.), pp. 81–95 Berlin, Germany: Springer [Google Scholar]
- 10.Bartell P. A., Gwinner E. 2005. A separate circadian oscillator controls nocturnal migratory restlessness in the songbird Sylvia borin. J. Biol. Rhythms 20, 538–549 10.1177/0748730405281826 (doi:10.1177/0748730405281826) [DOI] [PubMed] [Google Scholar]
- 11.Kumar V., Rani S., Singh B. P. 2006. Biological clocks help reduce the physiological conflicts in avian migrants. J. Ornithol. 147, 281–286 10.1007/s10336-006-0055-7 (doi:10.1007/s10336-006-0055-7) [DOI] [Google Scholar]
- 12.Rani S., Malik S., Trivedi A. K., Singh S., Kumar V. 2006. A circadian clock regulates migratory restlessness in the blackheaded bunting, Emberiza melanocephala. Curr. Sci. 91, 1093–1096 [Google Scholar]
- 13.Helm B., Piersma T., van der Jeugd H. 2006. Sociable schedules: interplay between avian seasonal and social behaviour. Anim. Behav. 72, 245–262 10.1016/j.anbehav.2005.12.007 (doi:10.1016/j.anbehav.2005.12.007) [DOI] [Google Scholar]
- 14.Reale D., Reader S. M., Sol D., McDougall P. T., Dingemanse N. J. 2007. Integrating animal temperament within ecology and evolution. Biol. Rev. 82, 291–318 10.1111/j.1469-185X.2007.00010.x (doi:10.1111/j.1469-185X.2007.00010.x) [DOI] [PubMed] [Google Scholar]
- 15.Mettke-Hofmann C., Greenberg R. 2005. Behavioral and cognitive adaptation to long-distance migration. In Birds of two worlds (eds Greenberg R., Marra P. P.), pp. 114–123 Baltimore, MD: Johns Hopkins University Press [Google Scholar]
- 16.Mettke-Hofmann C., Ebert C., Schmidt T., Steiger S., Stieb S. 2005. Personality traits in resident and migratory warbler species. Behaviour 142, 1357–1375 10.1163/156853905774539427 (doi:10.1163/156853905774539427) [DOI] [Google Scholar]
- 17.van Noordwijk A. J., et al. 2006. A framework for the study of genetic variation in migratory behaviour. J. Ornithol. 147, 221–233 10.1007/s10336-005-0047-z (doi:10.1007/s10336-005-0047-z) [DOI] [Google Scholar]
- 18.Berthold P., Pulido F. 1994. Heritability of migratory activity in a natural bird population. Proc. R. Soc. Lond. B 257, 311–315 10.1098/rspb.1994.0131 (doi:10.1098/rspb.1994.0131) [DOI] [Google Scholar]
- 19.Pulido F., Berthold P., Mohr G., Querner U. 2001. Heritability of the timing of autumn migration in a natural bird population. Proc. R. Soc. Lond. B 268, 953–959 10.1098/rspb.2001.1602 (doi:10.1098/rspb.2001.1602) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Pulido F., Berthold P. 2003. Quantitative genetic analysis of migratory behaviour. In Avian migration (eds Berthold P., Gwinner E., Sonnenschein E.), pp. 53–77 Berlin, Germany: Springer [Google Scholar]
- 21.Pulido F. 2007. The genetics and evolution of avian migration. Bioscience 57, 165–174 10.1641/B570211 (doi:10.1641/B570211) [DOI] [Google Scholar]
- 22.Berthold P., Querner U. 1981. Genetic basis of migratory behavior in European warblers. Science 212, 77–79 10.1126/science.212.4490.77 (doi:10.1126/science.212.4490.77) [DOI] [PubMed] [Google Scholar]
- 23.Berthold P., Wiltschko W., Miltenberger H., Querner U. 1990. Genetic transmission of migratory behavior into a nonmigratory bird population. Experientia 46, 107–108 10.1007/BF01955432 (doi:10.1007/BF01955432) [DOI] [Google Scholar]
- 24.Berthold P., Helbig A. J., Mohr G., Querner U. 1992. Rapid microevolution of migratory behaviour in a wild bird species. Nature 360, 668–670 10.1038/360668a0 (doi:10.1038/360668a0) [DOI] [Google Scholar]
- 25.Rolshausen G., Segelbacher G., Hobson K. A., Schaefer H. M. 2009. Contemporary evolution of reproductive isolation and phenotypic divergence in sympatry along a migratory divide. Curr. Biol. 19, 2097–2101 10.1016/j.cub.2009.10.061 (doi:10.1016/j.cub.2009.10.061) [DOI] [PubMed] [Google Scholar]
- 26.Pulido F., Berthold P. 2010. Current selection for lower migratory activity will drive the evolution of residency in a migratory bird population. Proc. Natl Acad. Sci. USA 107, 7341–7346 10.1073/pnas.0910361107 (doi:10.1073/pnas.0910361107) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Pulido F., Berthold P., van Noordwijk A. J. 1996. Frequency of migrants and migratory activity are genetically correlated in a bird population: evolutionary implications. Proc. Natl Acad. Sci. USA 93, 14 642–14 647 10.1073/pnas.93.25.14642 (doi:10.1073/pnas.93.25.14642) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Bell A. M., Aubin-Horth N. 2010. What can whole genome expression data tell us about the ecology and evolution of personality? Phil. Trans. R. Soc. B 365, 4001–4012 10.1098/rstb.2010.0185 (doi:10.1098/rstb.2010.0185) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Steinmeyer C., Mueller J. C., Kempenaers B. 2009. Search for informative polymorphisms in candidate genes: clock genes and circadian behaviour in blue tits. Genetica 136, 109–117 10.1007/s10709-008-9318-y (doi:10.1007/s10709-008-9318-y) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Savitz J. B., Ramesar R. S. 2004. Genetic variants implicated in personality: a review of the more promising candidates. Am. J. Med. Genet. (Part B) 131B, 20–32 10.1002/ajmg.b.20155 (doi:10.1002/ajmg.b.20155) [DOI] [PubMed] [Google Scholar]
- 31.Fidler A. E., van Oers K., Drent P. J., Kuhn S., Mueller J. C., Kempenaers B. 2007. Drd4 gene polymorphisms are associated with personality variation in a passerine bird. Proc. R. Soc. B 274, 1685–1691 10.1098/rspb.2007.0337 (doi:10.1098/rspb.2007.0337) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Korsten P., et al. 2010. Association between DRD4 gene polymorphism and personality variation in great tits: a test across four wild populations. Mol Ecol 19, 832–843 10.1111/j.1365-294X.2009.04518.x (doi:10.1111/j.1365-294X.2009.04518.x) [DOI] [PubMed] [Google Scholar]
- 33.Perez-Tris J., Bensch S., Carbonell R., Helbig A. J., Telleria J. L. 2004. Historical diversification of migration patterns in a passerine bird. Evolution 58, 1819–1832 10.1554/03-731 (doi:10.1554/03-731) [DOI] [PubMed] [Google Scholar]
- 34.Berthold P., Mohr G., Querner U. 1990. Control and evolutionary potential of obligate partial migration: results of a two-way selective breeding experiment with the Blackcap (Sylvia atricapilla). J. Ornithol. 131, 33–45 10.1007/BF01644896 (doi:10.1007/BF01644896) [DOI] [Google Scholar]
- 35.Finlayson C. 1992. Birds of the strait of Gibraltar. London, UK: Poyser [Google Scholar]
- 36.Pérez-Tris J., Tellería J. L. 2002. Regional variation in seasonality affects migratory behaviour and life-history traits of two Mediterranean passerines. Acta Oecol. 23, 13–21 10.1016/S1146-609X(01)01129-8 (doi:10.1016/S1146-609X(01)01129-8) [DOI] [Google Scholar]
- 37.Bletz H., Weindler P., Wiltschko R., Wiltschko W., Berthold P. 1996. The magnetic field as reference for the innate migratory direction in blackcaps, Sylvia atricapilla. Naturwissenschaften 83, 430–432 10.1007/BF01142071 (doi:10.1007/BF01142071) [DOI] [Google Scholar]
- 38.Helbig A. J. 1992. SE- and SW-migrating blackcap (Sylvia atricapilla) populations in central Europe: orientation of birds in the contact zone. J. Evol. Biol. 4, 657–670 10.1046/j.1420-9101.1991.4040657.x (doi:10.1046/j.1420-9101.1991.4040657.x) [DOI] [Google Scholar]
- 39.Spina F., Volponi S. 2008. Atlante della Migrazione degli Uccelli in Italia II. Passeriformi. Ministero dell'Ambiente e della Tutela del Territorio e del Mare, Istituto Superiore per la Protezione e la Ricerca Ambientale (ISPRA), Roma [Google Scholar]
- 40.Segelbacher G., Rolshausen G., Weis-Dootz T., Serrano D., Schaefer H. M. 2008. Isolation of 10 tetranucleotide microsatellite loci in the blackcap (Sylvia atricapilla). Mol. Ecol. Resour. 8, 1108–1110 10.1111/j.1755-0998.2008.02171.x (doi:10.1111/j.1755-0998.2008.02171.x) [DOI] [PubMed] [Google Scholar]
- 41.Griffiths R., Double M. C., Orr K., Dawson R. J. G. 1998. A DNA test to sex most birds. Mol. Ecol. 7, 1071–1075 10.1046/j.1365-294x.1998.00389.x (doi:10.1046/j.1365-294x.1998.00389.x) [DOI] [PubMed] [Google Scholar]
- 42.Bates D., Maechler M., Dai B. lme4: Linear mixed-effects models using S4 classes. 2008. R package v. 0.999375-28. See http://cran.r-project.org/web/packages/lme4/index.html . [Google Scholar]
- 43.R Development Core Team 2009. R: a language and environment for statistical computing. Vienna, Austria: R Foundation for Statistical Computing; See http://www.R-project.org [Google Scholar]
- 44.Scheipl F., Greven S., Kuechenhoff H. 2008. Size and power of tests for a zero random effect variance or polynomial regression in additive and linear mixed models. Comput. Stat. Data Anal. 52, 3283–3299 10.1016/j.csda.2007.10.022 (doi:10.1016/j.csda.2007.10.022) [DOI] [Google Scholar]
- 45.Baayen R. H. 2008. languageR: data sets and functions with ‘analyzing linguistic data: a practical introduction to statistics’. R package v. 0.953. See http://cran.r-project.org/web/packages/languageR/index.html [Google Scholar]
- 46.Ioannidis J. P. A., Thomas G., Daly M. J. 2009. Validating, augmenting and refining genome-wide association signals. Nat. Rev. Genet. 10, 318–329 10.1038/nrg2544 (doi:10.1038/nrg2544) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Smouse P. E., Long J. C., Sokal R. R. 1986. Multiple regression and correlation extensions of the Mantel test of matrix correspondence. Syst. Zool. 35, 627–632 10.2307/2413122 (doi:10.2307/2413122) [DOI] [Google Scholar]
- 48.Goslee S. C., Urban D. L. 2007. The ecodist package for dissimilarity-based analysis of ecological data. J. Stat. Softw. 22, 1–19 [Google Scholar]
- 49.Weir B. S., Cockerham C. C. 1984. Estimating F-statistics for the analysis of population structure. Evolution 38, 1358–1370 10.2307/2408641 (doi:10.2307/2408641) [DOI] [PubMed] [Google Scholar]
- 50.Rousset F. 2008. GENEPOP '007: a complete re-implementation of the GENEPOP software for Windows and Linux. Mol. Ecol. Resour. 8, 103–106 10.1111/j.1471-8286.2007.01931.x (doi:10.1111/j.1471-8286.2007.01931.x) [DOI] [PubMed] [Google Scholar]
- 51.Magnusson A. gmt: Interface between GMT map-making software and R. 2009. R package v. 1.1–3. See http://cran.r-project.org/web/packages/gmt/index.html . [Google Scholar]
- 52.Excoffier L., Smouse P., Quattro J. 1992. Analysis of molecular variance inferred from metric distances among DNA haplotypes: application to human mitochondrial DNA restriction data. Genetics 131, 479–491 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Flint J., Mackay T. F. C. 2009. Genetic architecture of quantitative traits in mice, flies, and humans. Genome Res. 19, 723–733 10.1101/gr.086660.108 (doi:10.1101/gr.086660.108) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Frazer K. A., Murray S. S., Schork N. J., Topol E. J. 2009. Human genetic variation and its contribution to complex traits. Nat. Rev. Genet. 10, 241–251 10.1038/nrg2554 (doi:10.1038/nrg2554) [DOI] [PubMed] [Google Scholar]
- 55.Dingemanse N. J., Kazem A. J. N., Reale D., Wright J. 2010. Behavioural reaction norms: animal personality meets individual plasticity. Trends Ecol. Evol. 25, 81–89 [DOI] [PubMed] [Google Scholar]
- 56.Hirokawa N., Takemura R. 2005. Molecular motors and mechanisms of directional transport in neurons. Nat. Rev. Neurosci. 6, 201–214 10.1038/nrn1624 (doi:10.1038/nrn1624) [DOI] [PubMed] [Google Scholar]
- 57.Bartel D. P. 2009. MicroRNAs: target recognition and regulatory functions. Cell 136, 215–233 10.1016/j.cell.2009.01.002 (doi:10.1016/j.cell.2009.01.002) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58.Chatterjee S., Pal J. K. 2009. Role of 5′- and 3′-untranslated regions of mRNAs in human diseases. Biol. Cell 101, 251–262 10.1042/BC20080104 (doi:10.1042/BC20080104) [DOI] [PubMed] [Google Scholar]
- 59.Li Y.-C., Korol A. B., Fahima T., Nevo E. 2004. Microsatellites within genes: structure, function, and evolution. Mol. Biol. Evol. 21, 991–1007 10.1093/molbev/msh073 (doi:10.1093/molbev/msh073) [DOI] [PubMed] [Google Scholar]
- 60.Riley D. E., Krieger J. N. 2009. UTR dinucleotide simple sequence repeat evolution exhibits recurring patterns including regulatory sequence motif replacements. Gene 429, 80–86 10.1016/j.gene.2008.09.030 (doi:10.1016/j.gene.2008.09.030) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 61.Vaudry D., et al. 2009. Pituitary adenylate cyclase-activating polypeptide and its receptors: 20 years after the discovery. Pharmacol. Rev. 61, 283–357 10.1124/pr.109.001370 (doi:10.1124/pr.109.001370) [DOI] [PubMed] [Google Scholar]
- 62.Olano-Marin J., Dawson D. A., Girg A., Hansson B., Ljungqvist M., Kempenaers B., Mueller J. C. 2010. A genome-wide set of 106 microsatellite markers for the blue tit (Cyanistes caeruleus). Mol. Ecol. Res. 10, 516–532 10.1111/j.1755-0998.2009.02777.x (doi:10.1111/j.1755-0998.2009.02777.x) [DOI] [PubMed] [Google Scholar]
- 63.Nowak J. Z., Zawilska J. B. 2003. PACAP in avians: origin, occurrence, and receptors—pharmacological and functional considerations. Curr. Pharm. Des. 9, 67–481 10.2174/1381612033391586 (doi:10.2174/1381612033391586) [DOI] [PubMed] [Google Scholar]
- 64.Nakahara K., Abe Y., Murakami T., Shiota K., Murakami N. 2002. Pituitary adenylate cyclase-activating polypeptide (PACAP) is involved in melatonin release via the specific receptor PACAP-r1, but not in the circadian oscillator, in chick pineal cells. Brain Res. 939, 19–25 10.1016/S0006-8993(02)02538-6 (doi:10.1016/S0006-8993(02)02538-6) [DOI] [PubMed] [Google Scholar]
- 65.Csernus V., Jozsa R., Reglodi D., Hollosy T., Somogyvari-Vigh A., Arimura A. 2004. The effect of PACAP on rhythmic melatonin release of avian pineals. Gen. Comp. Endocrinol. 135, 62–69 10.1016/S0016-6480(03)00284-3 (doi:10.1016/S0016-6480(03)00284-3) [DOI] [PubMed] [Google Scholar]
- 66.Faluhelyi N., Reglodi D., Csernus V. 2005. Development of the circadian melatonin rhythm and its responsiveness to PACAP in the embryonic chicken pineal gland. Ann. NY Acad. Sci. 1040, 305–309 10.1196/annals.1327.048 (doi:10.1196/annals.1327.048) [DOI] [PubMed] [Google Scholar]
- 67.Nagy A. D., Csernus V. J. 2007. The role of PACAP in the control of circadian expression of clock genes in the chicken pineal gland. Peptides 28, 1767–1774 10.1016/j.peptides.2007.07.013 (doi:10.1016/j.peptides.2007.07.013) [DOI] [PubMed] [Google Scholar]
- 68.Racz B., et al. 2008. Effects of PACAP on the circadian changes of signalling pathways in chicken pinealocytes. J. Mol. Neurosci. 36, 220–226 10.1007/s12031-008-9112-4 (doi:10.1007/s12031-008-9112-4) [DOI] [PubMed] [Google Scholar]
- 69.Tachibana T., Oikawa D., Adachi N., Boswell T., Furuse M. 2007. Central administration of vasoactive intestinal peptide and pituitary adenylate cyclase-activating polypeptide differentially regulates energy metabolism in chicks. Comp. Biochem. Physiol. (Part A) 147, 156–164 10.1016/j.cbpa.2006.12.043 (doi:10.1016/j.cbpa.2006.12.043) [DOI] [PubMed] [Google Scholar]
- 70.Tachibana T., Saitoa S., Tomonaga S., Takagi T., Saito E.-S., Boswell T., Furuse M. 2003. Intracerebroventricular injection of vasoactive intestinal peptide and pituitary adenylate cyclase-activating polypeptide inhibits feeding in chicks. Neurosci. Lett. 339, 203–206 10.1016/S0304-3940(03)00017-X (doi:10.1016/S0304-3940(03)00017-X) [DOI] [PubMed] [Google Scholar]
- 71.Tachibana T., Saito E. S., Takahashi H., Saito S., Tomonaga S., Boswell T., Furuse M. 2004. Anorexigenic effects of pituitary adenylate cyclase-activating polypeptide and vasoactive intestinal peptide in the chick brain are mediated by corticotrophin-releasing factor. Regul. Pept. 120, 99–105 10.1016/j.regpep.2004.02.016 (doi:10.1016/j.regpep.2004.02.016) [DOI] [PubMed] [Google Scholar]
- 72.Aubin-Horth N., Letcher B. H., Hofmann H. A. 2009. Gene-expression signatures of Atlantic salmon's plastic life cycle. Gen. Comp. Endocrinol. 163, 278–284 10.1016/j.ygcen.2009.04.021 (doi:10.1016/j.ygcen.2009.04.021) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 73.Piersma T., Perez-Tris J., Mouritsen H., Bauchinger U., Bairlein F. 2005. Is there a ‘migratory syndrome’ common to all migrant birds? Ann. NY Acad. Sci. 1046, 282–293 10.1196/annals.1343.026 (doi:10.1196/annals.1343.026) [DOI] [PubMed] [Google Scholar]