Abstract
Females of many taxa often copulate with multiple males and incite sperm competition. On the premise that males of high genetic quality are more successful in sperm competition, it has been suggested that females may benefit from polyandry by accruing ‘good genes’ for their offspring. Laboratory studies have shown that multiple mating can increase female fitness through enhanced embryo viability, and have exposed how polyandry influences the evolution of the ejaculate. However, such studies often do not allow for both female mate choice and male–male competition to operate simultaneously. Here, I took house mice (Mus domesticus) from selection lines that had been evolving with (polygamous) and without (monogamous) sperm competition for 16 generations and, by placing them in free-ranging enclosures for 11 weeks, forced them to compete for access to resources and mates. Parentage analyses revealed that female reproductive success was not influenced by selection history, but there was a significant paternity bias towards males from the polygamous selection lines. Therefore, I show that female house mice benefit from polyandry by producing sons that achieve increased fitness in a semi-natural environment.
Keywords: sperm competition, house mice, genetic benefits, experimental evolution
1. Introduction
Mating with multiple partners (polygamy) is a common reproductive strategy observed across a wide range of species. Soliciting and copulating with multiple mates may be costly for individuals [1], as it requires high levels of energy and may increase the risk of disease [2,3] or predation [4,5]. Despite these costs, polygamy allows males to maximize their fitness as male reproductive success depends largely on the number of mating partners [6]. Conversely, female fitness is more constrained, for example, owing to a restriction in the number of ova that can be produced and/or the number of altricial young that can be nurtured [7]. Certainly, a single fertile mating should be sufficient to maximize female fitness. Still, female multiple mating (polyandry) is prolific among many taxa [8,9]. A number of hypotheses have been proposed to account for the evolution and maintenance of this reproductive strategy [10,11].
In some species females have been shown to benefit directly from multiple mating, for example, through receiving nutrients at the time of copulation [12], or as assurance against infertility [13]. However, in many cases the benefit/s of polyandry are not clear [10]. When females mate multiply and ejaculates overlap in the reproductive tract, sperm from different males are forced to compete for fertilizations [14]. Thus, polyandry ensures that post-copulatory sexual selection occurs in the form of sperm competition. It has been suggested that females may use polyandry as a means of acquiring genetic benefits for their offspring. If females are able to make a pre-copulatory assessment of male genetic quality, polyandry may occur due to females ‘trading up’ from a lower quality mate to one of higher genetic quality [15]. Alternatively, females may solicit multiple partners to incite sperm competition and accrue good genes for their offspring. The intrinsic male quality hypothesis suggests that polyandry increases the probability that a male of high genetic quality will be successful in sperm competition and sire the offspring [16,17]. Support for this hypothesis is limited, as only a few studies have reported a link between sperm competitiveness and offspring viability and/or performance [18–20]. If competitive fertilization success is heritable, females may benefit from polyandry by producing sons that are superior sperm competitors [21]. Alternatively, polyandry has been suggested to allow females to avoid reproductive failure resulting from incompatibilities between parental genotypes [22]. Indeed, in some species polyandry may operate as a post-copulatory mechanism of inbreeding avoidance [23].
Polyandry also has significant evolutionary implications for males. Theory suggests that sperm competition should be a pervasive force in the evolution of testes size and sperm number [24], and selection experiments of invertebrates maintained under polygamous and monogamous mating regimes have provided conclusive evidence that sperm competition selects for increased testes mass [25–27]. Additionally, in the yellow dung fly (Scathophaga stercoraria) and the dung beetle (Onthophagus tarus) males from polygamous lines were also shown to have a competitive fertilization advantage when engaged in sperm competition with males from the monogamous lines [25,27]. Although these studies are taxonomically restricted to invertebrates, they do provide empirical support for sperm competition theory and for the observed macro-evolutionary patterns of testes size evolution.
Only a single study of this kind has been performed on a vertebrate, the house mouse (Mus domesticus). Following eight generations of selection, despite there being no divergence in testis size, males evolving under a polygamous regime produced ejaculates with greater numbers of sperm and better swimming performance when compared with males evolving under a monogamous regime [28]. Competitive mating trials at generation 12 revealed that polygamous line males obtained a significant paternity bias over monogamous line males, and thus had evolved to be superior sperm competitors [29]. These results, taken with those of the invertebrate studies, suggest that changes in testes efficiency and sperm quality occur rapidly in response to sexual selection, while shifts in testes mass require longer periods of sustained selection. In the study of house mice, there was a correlative response in litter size; the average number of pups born to females from the polygamous lines steadily increased across generations, while average litter sizes of the monogamous lines remained comparable to the source population [28]. Larger litters in the polygamous lines could be explained by increased male fertility and/or increased embryo viability due to a genetic benefit associated with polyandry.
Thus, laboratory investigations across different taxa have provided evidence for the important evolutionary implications of polygamy. However, it is difficult to simultaneously address all the mechanisms of sexual selection under controlled laboratory conditions [30]. Experiments under semi-natural conditions may offer a more rigorous test of whether polygamy selects for increased fitness, by ensuring competition between individuals for resources and mates, and thus exerting selection among individuals and their offspring.
Here, I compared the reproductive performance of mice that had been evolving with (polygamous) and without (monogamous) sperm competition for 16 generations. I removed animals from laboratory-reared monogamous and polygamous selection lines and placed them in large enclosures, allowing them to roam and interact freely. In doing so I ensured that male mice had to defend nest boxes and compete for territories [19], and all individuals competed for mates. After 11 weeks, the founder individuals and their offspring were removed from the enclosures and genotyped for parentage analyses. Although there was no difference in the reproductive performance of females, there was a significant paternity bias towards males from the polygamous selection lines. Thus, I found that the sons of females that had evolved under a polygamous mating regime achieved greater reproductive success than those under a monogamous mating regime. Consequently, I show that female house mice benefit from polyandry by producing sons that achieve greater fitness in a competitive situation.
2. Material and methods
(a). Selection lines and experimental animals
A detailed description of the source population, and the mating design of the selection regimes is provided elsewhere [28]. Briefly, the selection lines were established from a colony of wild-derived mice held at the Animal Resources Centre (Murdoch, Western Australia). Monogamous and polygamous selection lines were established with 18 males and 18 females in each. Subsequently, 18 males and 18 females contributed to each generation. In the monogamous lines male and female pairs were inspected daily and separated once a mating plug was detected. In the polygamous lines, males and females were paired, and female oestrous condition was monitored [31]. Once oestrus was detected, females were inspected every half hour for a mating plug. Following their first mating, females mated consecutively with two other males. Microsatellite data generated after 12 generations of selection revealed that individuals from the two selection treatments did not differ in the level of heterozygosity, or average inbreeding coefficient [29].
Here, following 16 generations of selection, I used animals from the lines to assess whether polygamy selects for increased male and female fitness in a competitive situation. A divergence in ejaculate quality and a trend for polygamous females to produce larger litters than monogamous females was observed at this generation [32]. I randomly selected one male and one female from three of the replicate monogamous lines and three of the replicate polygamous lines, and released them into free-ranging enclosures. Thus, there were six founder males and six founder females within each enclosure. The density of natural mouse populations varies considerably (e.g. [33]). However, to ensure intense competition and strong selection, it was important that the enclosures had a high founding density (0.75 mice m−2).
(b). The enclosures
The experiment was conducted for 11 weeks within an animal care unit at the University of Western Australia (UWA). Each of the four enclosures was constructed by adhering aluminium barriers to the walls of a room (electronic supplementary material, plate S1). Each enclosure covered the same area of 16 m2. To ensure that each enclosure was completely sealed, the gap between the barrier and the floor was filled with silicon. The enclosure floors were covered with a 5 cm layer of sand. Standard mouse pellets and water were provided ad libitum to animals, and were replenished at approximately the same time each day. Wooden panelling and plastic piping created subsections within the enclosures (electronic supplementary material, plate S1). Each enclosure contained six wooden nest boxes (22 × 13 × 13 cm). Thus, there were half as many nests boxes as there were founder mice. Empty egg cartons were provided for additional shelter (four per enclosure). Hay was provided as nesting material. The wooden panelling, plastic piping, nest boxes and feed/water bowls were placed in the same position within each enclosure, so that all four enclosures had the same design (electronic supplementary material, figure S1 and plate S1). Each room was maintained under natural lighting. Daily minimum/maximum temperatures of each room were monitored via a digital thermometer affixed to the wall. Prior to releasing the experimental animals, a group of seven females was housed in each enclosure for 5 days to ensure that the animals could not escape.
(c). The founders
The founders of each enclosure population were released on the same day (day 1), and left to roam and interact freely. The founders were eight weeks old at the time of release, and thus sexually mature with the potential to breed immediately but had no prior sexual experience. Across the populations, average body size did not differ between founding males from the monogamous (24.54 ± 0.72 g) and polygamous (23.04 ± 0.77 g) selection lines (F1,22 = 2.016, p = 0.170; electronic supplementary material, table S1). There was also no difference in the average size of founding monogamous (19.27 ± 0.40 g) and polygamous (19.98 ± 0.51 g) females (F1,22 = 1.226, p = 0.280). On day 25 and day 53 of the experiment, a small amount of the top layer of the sand within each enclosure was removed and replaced with clean sand, and the hay piles were replenished. Upon discovery, deceased animals were removed from the enclosures, sexed and a small amount of ear tissue was extracted and stored in 100 per cent ethanol for genotyping.
House mice have an average gestation period of 18–21 days, and exhibit postpartum oestrous [34]. Thus, the founder females had the potential to produce young within three weeks of release, and produce three litters throughout the duration of the experiment. The oldest F1 males (i.e. those produced in the first three weeks of the experiment) were sexually mature by the end of the experiment, and thus had the potential to sire the youngest, in utero offspring. Similarly, the oldest F1 females had the potential to be pregnant at the end of the 11 week experiment.
(d). Genotyping and parentage analysis
Prior to release, the founding mice were ear clipped to allow for individual identification and to obtain a tissue sample for parentage assignment. Upon the completion of the experiment (day 77), the surviving founding animals and their progeny were collected from the enclosures and sacrificed via lethal injection. Each individual was sexed, weighed and assigned to an age cohort based on body weight and stage of development: (i) pup (less than 3 g, dependent on mother); (ii) juvenile (3–10 g, independent from mother); (iii) sub-adult (10–15 g, not sexually mature); (iv) adult (greater than 15 g, sexually mature); or (v) founder (identified by ear tags). Gravid females were dissected and the embryos were preserved in 100 per cent ethanol. Founder and sexually mature F1 females, which were not obviously pregnant at the time of collection, were housed in boxes with nesting material for two weeks, following which they were sacrificed and dissected. The embryos of pregnant females were removed from the reproductive tract and preserved for paternity analysis.
For parentage and paternity assignment DNA was extracted from preserved tissue samples using the EDNA HISPEX extraction kit (Fisher Biotec, Subiaco, Western Australia). I genotyped the founders (48) and their offspring (304) at 10 microsatellite loci: D4Mit1, D10Mit14, D13Mit1, D6Mit138, D14Mit132, D2Mit277, D15Mit13, D11Mit29, D15Mit174 and D17Mit51 [23,35]. Labelled primers were obtained from GeneWorks (Hindmarsh, South Australia; FAM) and Applied Biosystems (Foster City, CA, USA; NED, PET, VIC) and unlabelled primers from GeneWorks. Primers were multiplexed in 10 µl reactions in a PTC-0200 DNA engine (GeneWorks). Reactions contained 5 or 6 µl of a multiplex kit (Qiagen, Doncaster, Victoria), 0.25 µM of forward labelled primer 0.25 µM of reverse primer, and approximately 200 ng of template DNA. The thermocycling profile for all loci was: 5 min denature at 95°C, 50 cycles of 90°C for 20 s, 55°C for 20 s and 72°C for 30 s, followed by 72°C for 3 min. Polymerase chain reaction products (1.5 µl) were run on a ABI3730 Sequencer, sized using Genescan-500 LIZ size standard and genotyped using Genemapper software (v. 3.0; Applied Biosystems).
Parentage was assigned via a maximum-likelihood approach using Cervus (v. 3.0) [36]. The F1 individuals were analysed using the six female founders and six male founders of each population as candidate parents. Paternity was assigned to unborn embryos using a known mother's genotype and candidate sires, which included the founder males and adult F1 males.
(e). Statistical analyses
To assess whether selection history influenced the reproductive success of males and females, I performed Pearson's χ2-tests based on the expected frequency of equal reproductive success (i.e. 0.5) for each of the four enclosure populations. I applied the weighted Z-method for combining probability from independent tests to determine whether collectively there was an effect of selection history on female and/or male reproductive success [37]. Unless otherwise stated, ANOVAs were applied in all other tests.
3. Results
(a). Population characteristics and survival
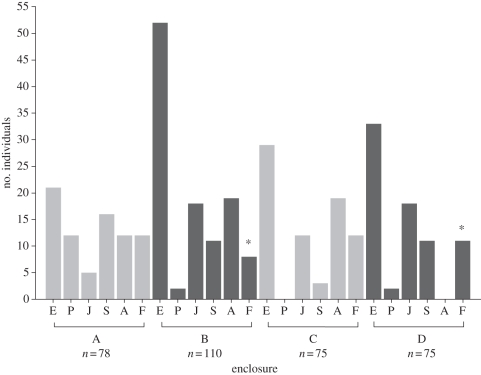
At the end of the experiment the characteristics of the replicate enclosure populations were relatively similar, with most age cohorts being represented in each population (figure 1 and electronic supplementary material, table S2). Enclosure B population had the most individuals, attributable to the large number of pregnant females and in utero embryos (figure 1). This could be explained by the death of four founders during the fifth week of the experiment, which may have resulted in less competition among individuals and thus high fecundity of adult females. Given that these four deceased founders consisted of a monogamous male and female and a polygamous male and female, it is unlikely that their deaths affected the parentage results. There were no adult individuals in enclosure D, but a substantial number of young offspring, suggesting that the founders of this population did not reproduce as quickly as those in the other three replicate populations. Alternatively, the population in enclosure C had no altricial pups but a large number of pregnant females bearing F2 progeny (figure 1).
Figure 1.
The number of individuals belonging to each age cohort for each of the four enclosure populations (E, embryo; P, pup; J, juvenile; S, sub-adult; A, adult; F, founder). Five founders (asterisks) died before the end of the 11 weeks (B, four; D, one). Deceased offspring were found in three enclosures (A, four; B, two; C, three).
The number of litters produced by the founder females did not differ among the enclosure populations (table 1). Additionally, the average number of litters produced by females was not influenced by selection history or replicate selection line (table 1). The average size of litters did not differ among the enclosure populations, or between females with different selection histories (table 1). Parity of female mice is known to effect litter size [38], however, there was no effect of litter order on litter size (table 1). The average number of sires per litter did differ among the enclosure populations (table 1), with enclosure C having on average more sires per litter than the other three populations (table 2). The frequency of multiple paternity also differed among the replicate enclosure populations, ranging from 0.45 to 1.00 (table 2). Given the high stocking densities of the enclosure populations it was not surprising to find that the occurrence of multiply sired litters (0.66 ± 0.13) was more frequent than was reported for seven natural populations with lower densities (mean = 0.26, range = 0.06–0.43; [33]).
Table 1.
ANOVAs of litter characteristics for females from the enclosure populations. Females had either monogamous or polygamous selection history. To account for the fact that females from the same replicate line were not statistically independent, replicate selection line was nested within selection history. Significant values (p < 0.05) are given in bold. SS, sum of squares; MS, mean square.
| effect | SS | d.f. | MS | F | p |
|---|---|---|---|---|---|
| number of litters | |||||
| enclosure | 0.231 | 3 | 0.077 | 0.060 | 0.980 |
| selection history | 0.001 | 1 | 0.001 | 0.001 | 0.978 |
| line [selection history] | 4.359 | 4 | 1.090 | 0.848 | 0.516 |
| error | 19.269 | 15 | |||
| litter size | |||||
| enclosure | 5.966 | 3 | 1.989 | 0.341 | 0.796 |
| selection history | 0.004 | 1 | 0.004 | 0.001 | 0.972 |
| line [selection history] | 12.379 | 4 | 3.095 | 0.531 | 0.714 |
| litter order | 22.482 | 3 | 7.494 | 1.286 | 0.299 |
| error | 163.160 | 28 | |||
| number of sires per litter | |||||
| enclosure | 11.473 | 3 | 3.824 | 5.163 | 0.005 |
| selection history | 0.033 | 1 | 0.033 | 0.112 | 0.747 |
| line [selection history] | 0.972 | 4 | 0.243 | 0.328 | 0.857 |
| error | 22.960 | 31 | |||
Table 2.
Mean (±s.e.) size and number of sires of litters of females with either a monogamous (M) or polygamous (P) selection history from each of the four enclosure populations. The frequency of multiple paternity (MP) is also given.
| n | litter size |
number of sires |
frequency of MP |
||||
|---|---|---|---|---|---|---|---|
| M | P | M | P | M | P | ||
| A | 11 | 6.3 ± 0.9 | 5.9 ± 0.8 | 1.8 ± 0.3 | 2.0 ± 0.4 | 0.75 | 0.57 |
| B | 16 | 7.1 ± 0.6 | 5.4 ± 0.4 | 1.8 ± 0.3 | 1.5 ± 0.2 | 0.50 | 0.50 |
| C | 9 | 7.2 ± 1.6 | 6.3 ± 1.6 | 2.8 ± 0.6 | 3.0 ± 0.4 | 1.00 | 1.00 |
| D | 10 | 4.3 ± 0.7 | 7.0 ± 0.9 | 1.3 ± 0.3 | 1.7 ± 0.3 | 0.33 | 0.57 |
| total | 46 | 6.6 ± 0.5 | 6.1 ± 0.4 | 2.0 ± 0.2 | 1.9 ± 0.2 | 0.65 ± 0.15 | 0.66 ± 0.11 |
Of the 48 population founders, three monogamous line founders and two polygamous line founders did not survive to the end of the experiment. Nine offspring were found deceased during the experiment, of which four had complete polygamous parentage, two had complete monogamous parentage and three had mixed polygamous/monogamous parentage.
I did not observe which individuals inhabited the nest boxes. However, in a similar study it has been shown that male house mice defend areas around nest boxes (‘territories’) by attacking and driving away intruding males [39]. Conversely, female house mice were reported to inhabit male territories but not defend their own [39]. General observations made throughout the course of the current study did reveal that feed and water bowls were not defended by individual mice.
Temperatures did not differ greatly across the enclosure rooms (electronic supplementary material, table S3). However, there was a significant difference in the average maximum temperature, whereby some rooms differed from others (see electronic supplementary material, table S3 for analysis).
(b). Parentage and paternity analysis
Of the total 169 free-living offspring, Cervus assigned parentage with more than 95% confidence to 135 individuals, and more than 80% confidence to another 17 individuals. For the remaining 17 individuals Cervus returned candidate parents with less than 80% confidence. Thus, for these individuals parentage was scored manually via the exclusion method. Paternity was assigned with more than 95% confidence to 128 of the in utero embryos, and more than 80 per cent to four embryos. Owing to repeated failed genotyping, paternity could not be assigned to two embryos. Based on shared maternal origin and stage of development I grouped siblings together, and thus segregated the offspring into litters. Across the four populations, the adult F1 males sired 14 embryos. Of these seven F1 sires, six were the sons of polygamous line males.
(c). Reproductive success
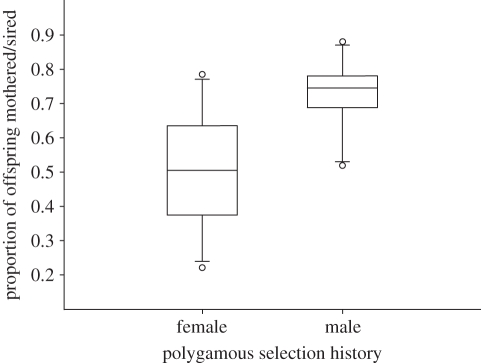
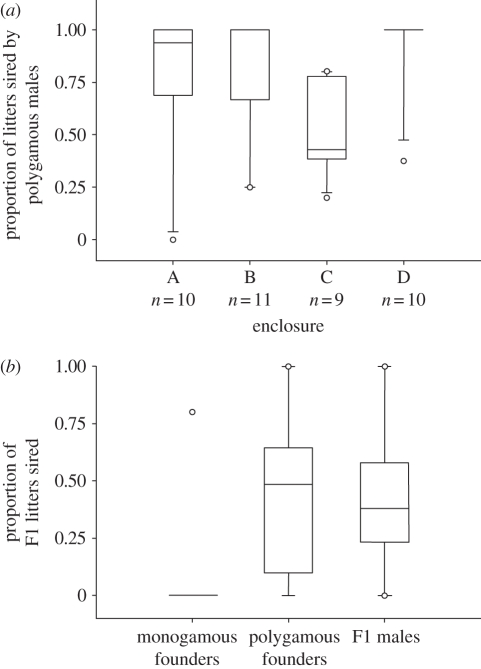
I performed χ2 analyses with the expectation of random mating and equal reproductive success of founder individuals to determine whether selection history affected female and/or male fitness within each enclosure population. In two of the enclosure populations, the monogamous and polygamous females had equivalent reproductive success. However, in enclosure B the monogamous females had greater reproductive success than the polygamous females, and vice versa in enclosure D (table 3). Therefore, although the combined p-value reflected divergence from the expectation of equal reproductive success (Zw = 5.107, p < 0.001), there was no consistent bias in reproductive success towards females with either selection history (table 3). Indeed, the median value for the success of the polygamous females was 0.52 (figure 2). Conversely, selection history did influence male reproductive success (Zw = 6.972, p < 0.001; figure 2); in three of the four replicate populations males with a polygamous selection history sired more offspring than was expected under the assumption of random mating and equal reproductive success (table 3 and figure 3a). The median value for the success of the polygamous males was 0.77 (figure 2). The paternity bias towards the polygamous males was evident in separate analyses of the in utero embryos (Zw = 4.620, p < 0.001) and the free-living offspring (Zw = 6.676, p < 0.001; electronic supplementary material, table S4). The males from one of the replicate polygamous lines were not as successful those from the other two polygamous lines, but on average males from the polygamous lines had greater success than males from the monogamous lines (electronic supplementary material, figure S2). The monogamous males gained representation in 46 per cent of the litters across all four replicate populations, but were represented in 100 per cent of the litters in enclosure population C.
Table 3.
Pearson's χ2 analyses with the expectation of random mating and equal reproductive success to determine whether selection history affected female and/or male fitness within each enclosure population. The weighted Z-method for combining probabilities was applied to test whether there was an overall effect of selection history on female and/or male reproductive success (see text). The proportion of offspring mothered/sired by the monogamous (PM) and polygamous (PP) individuals is given. Significant values (p < 0.05) are given in bold.
| enclosure | d.f. | n | PM | PP | χ2 | p | z | |
|---|---|---|---|---|---|---|---|---|
| females | A | 1 | 63 | 0.41 | 0.59 | 1.921 | 0.166 | 0.971 |
| B | 1 | 78 | 0.76 | 0.24 | 20.513 | <0.001 | 4.380 | |
| C | 1 | 66 | 0.58 | 0.42 | 0.970 | 0.325 | 0.455 | |
| D | 1 | 64 | 0.23 | 0.77 | 18.063 | <0.001 | 4.092 | |
| males | A | 1 | 63 | 0.25 | 0.75 | 15.254 | <0.001 | 3.735 |
| B | 1 | 78 | 0.26 | 0.74 | 18.513 | <0.001 | 4.147 | |
| C | 1 | 66 | 0.47 | 0.53 | 0.242 | 0.623 | 0.313 | |
| D | 1 | 63 | 0.13 | 0.87 | 35.063 | <0.001 | 5.806 |
Figure 2.
The total proportion of offspring sired by female and male founders with a polygamous selection history for the four enclosure populations. The line of each box represents the median value (females, 0.52; males, 0.77), the box represents 90% of the variation, and the whiskers represent 95% of the variation (outliers are represented by circles).
Figure 3.
Male reproductive success. (a) The proportion of litters produced by the founder females and sired by males with a polygamous selection history for each of the replicate enclosure populations. (b) The proportion of litters produced by the F1 females and sired by the monogamous founder males, the polygamous founder males, and the F1 males. The line of each box represents the median value, the box represents 90% of the variation, and the whiskers represent 95% of the variation (outliers are represented by circles).
The F1 males did not sire any of the offspring produced by the founder females. However, they were successful in gaining paternity within litters produced by the F1 females (enclosures A and B). A χ2 analysis revealed that both the F1 males and polygamous founder males sired equivalent proportions of the F2 offspring (0.44), and more than the monogamous founder males (0.12) (χ2 = 6.248, p = 0.012; figure 3b).
In further analyses, I assessed whether paternity (based on male selection history) was repeatable across multiple litters produced by the same females (n = 17). Although the repeatability estimate was low (r = 0.104; [40]), ANOVA revealed that there was significantly more variation between females than within females in the proportion of litters sired by polygamous males (F16,24 = 2.168, p = 0.042). Thus, paternity was repeatable across litters. I was then able to test whether there was a paternity bias towards males from the same replicate selection line as the females. To do so I used a one-sample t-test and compared the mean proportion of females' litters sired by males from the same replicate selection line versus the mean proportion of litters sired by males from different replicate selection lines. The analysis revealed that there was no paternity bias towards males from the same replicate line as the female (d.f. = 21, t-value = 1.395, p = 0.178).
4. Discussion
Polygamy is a common mating strategy observed across many different taxa. Laboratory studies of experimental evolution have become a powerful tool for investigating the evolutionary implications of polygamy. Here, I took house mice from selection lines that had been evolving with (polygamous) and without (monogamous) sperm competition for 16 generations and allowed them to compete under semi-natural conditions to assess the benefits of polygamy for each of the sexes in a competitive situation. In natural populations, male–male competition and pre-copulatory female mate choice rarely act independently [41]. Thus, by placing the animals in a semi-natural situation I allowed for both of these processes to operate simultaneously. The frequency of multiple paternity, which is used as a proxy for the frequency of sperm competition, varies considerably among natural populations of house mice. For example, among seven wild mouse populations the percentage of multiply sired litters was reported to range from 6 to 43 per cent [33]. Across the experimental enclosure populations the frequency of multiple paternity was high, most likely due to high stocking densities. I can conclude that there were high levels of sperm competition and post-copulatory sexual selection operating within each of the enclosure populations.
Parentage analyses revealed that female reproductive success was not dependent on selection history. However, selection history did account for male reproductive success; in three of the four replicate enclosure populations the polygamous males sired more offspring than the monogamous males. Despite a collective paternity bias towards the polygamous males, there was close to equal reproductive success of monogamous and polygamous males in one of the replicate populations. It is interesting to note that all of the litters in this population were multiply sired, indicative of extremely high levels of sperm competition. Indeed, on average paternity was distributed across more males in population C compared with the other three replicate populations. The size of the monogamous and polygamous founder males of enclosure population C did differ the most when compared with the other three populations, with the monogamous males being on average heavier than the polygamous males. Consequently, the monogamous males in this population may have gained greater access to territories and/or females compared with the monogamous males in the other populations. Importantly, the weighted Z-method for combining probability from independent tests revealed that globally there was a significant paternity bias towards males from the polygamous selection lines [37]. It is also interesting to note that all but one of the F1 males that gained paternity of the F2 progeny were the sons of males from the polygamous selection lines. Collectively, these results show that polygamy selects for increased male fitness in a semi-natural context.
The increased fitness of males from lineages evolving with sperm competition may be explained by a genetic benefit associated with polyandry. A long evolutionary history of multiple mating in the polygamous lines may have allowed for the selection of good genes, such that those males that were successful in sperm competition had an intrinsic quality that resulted in offspring of greater fitness [17]. Across the first 10 generations of selection it was shown that litters born to females of the polygamous lines steadily increased while the litters born to monogamous line females remained comparable to the source population [28]. Indeed, this increase in female fecundity may be attributable the post-copulatory selection of good genes and the production of higher quality offspring [28,32]. Further evidence comes from this study where there was a significant paternity bias to males from the polygamous lines but no effect of female selection history on average litter size. Thus, founding males from the polygamous selection lines may have been of higher genetic quality than founding males from the monogamous lines. Consequently, females may have preferentially mated with the polygamous line males to obtain genetic benefits for their offspring. Alternatively, the polygamous males may have attained superiority in dominance over the monogamous males, which allowed them to better defend territories and ensued greater access to females. However, males from the monogamous selection lines gained paternity representation in approximately half of the litters, suggesting that they had adequate access to females. Indeed, it has been shown that in a single oestrous period female house mice will copulate with both dominant and subordinate males, but more frequently with dominant males that may be deemed to be of higher genetic quality [42].
It is possible that increased reproductive success of the polygamous males was attributable to, or magnified by, preferential embryo survival. A comparative analysis across mammals suggested that polyandry may allow paternity to be biased towards compatible genotypes and, therefore, enhance embryo survival during development [43], and offspring born to female house mice mated polyandrously were shown to have greater survival rates when compared with offspring born to females mated monandrously [44]. Consequently, maternal effects such as differential female investment in developing embryos or altricial pups may have also contributed to a paternity bias towards males with a polygamous selection history.
Covert male–male battles in the form of sperm competition could also explain the increased reproductive success of the polygamous males. It has been shown that males in the polygamous selection lines produce ejaculates with more sperm of higher quality compared with males from the monogamous lines [28,32]. Thus, alternatively and/or additionally to polygamous males being of greater genetic quality, a paternity bias towards these males may be due to their superior competitive fertilization ability [29]. While the average number of sires per litter among the enclosure populations was comparable to natural populations, it is known that not all males engaging in sperm competition will obtain paternity representation [33]. Thus, success of monogamous males in achieving copulations may be overshadowed by the superior competitive fertilization success of the polygamous males. Further experimentation is required to elucidate whether males from the polygamous selection lines are of greater genetic quality than males from the monogamous lines, and differentiate between the pre-copulatory and post-copulatory success of these males. Nevertheless, given that the sons of females from polygamous lineages had increased fitness over the sons of females from monogamous lineages, I have shown that female house mice benefit from polyandry.
Previous studies of mammals have shown that polyandry can improve offspring fitness and performance in natural, competitive environments. In the Australian marsupial Antechinus stuartii, it was shown that females that mated with three males produced more offspring than females mated three times to the same male, and after being released into the wild that offspring survival to weaning was higher for polyandrous females [18]. Indeed, this experiment provided support for the ‘good sperm’ hypothesis and showed that males that were successful in sperm competition sired more viable offspring [18]. In the current study, the animals were provided with plentiful resources and shelter. Thus, it is not surprising that there was very little animal mortality. However, the results of my experiment are strikingly similar to a study of voles, which showed that the sons of polyandrously mated females had greater reproductive success than the sons of monandrously mated females in a semi-natural environment [45]. Additionally, there was no effect of treatment on offspring survival despite the animals being housed in outdoor enclosures over winter [45]. Collectively, these results show that female mammals gain both immediate and long-term benefits from polyandry.
In conclusion, I have shown that male house mice with a polygamous selection history have greater reproductive success than males with a monogamous selection history when they are forced to compete for mates in a semi-natural environment. This result is most parsimoniously explained by selection for genetically superior males, who have greater competitive ability, which supports the ‘good genes’ model for the evolution of polyandry. Thus, I provide evidence that female house mice benefit from polyandry by producing sons that achieve increased reproductive success in a competitive environment.
Acknowledgements
This research was funded by a UWA Research Development Award and approved by the UWA Animal Ethics Committee (approval no. 3/100/299).
I thank Maria Almbro for technical assistance; John Fitzpatrick for statistical advice; and Paco García-González, Ines Klemme and an anonymous reviewer for comments on the manuscript.
References
- 1.Daly M. 1978. The cost of mating. Am. Nat. 112, 771–774 10.1086/283319 (doi:10.1086/283319) [DOI] [Google Scholar]
- 2.Sheldon B. C. 1993. Sexually transmitted disease in birds: occurrence and evolutionary significance. Phil. Trans. R. Soc. Lond. B 339, 491–497 10.1098/rstb.1993.0044 (doi:10.1098/rstb.1993.0044) [DOI] [PubMed] [Google Scholar]
- 3.Thrall P. H., Antonovics J., Bever J. D. 1997. Sexual transmission of disease and host mating systems: within-season reproductive success. Am. Nat. 149, 485–506 10.1086/286001 (doi:10.1086/286001) [DOI] [Google Scholar]
- 4.Arnqvist G. 1989. Multiple mating in the water strider: mutual benefits or intersexual conflict? Anim. Behav. 38, 749–756 10.1016/S0003-3472(89)80107-1 (doi:10.1016/S0003-3472(89)80107-1) [DOI] [Google Scholar]
- 5.Fairbarn D. J. 1993. Costs of loading associated with mate-carrying in the water strider, Aquarius remigis. Behav. Ecol. 4, 224–231 10.1093/beheco/4.3.224 (doi:10.1093/beheco/4.3.224) [DOI] [Google Scholar]
- 6.Bateman A. J. 1948. Intra-sexual selection in Drosophila. Heredity 2, 349–368 10.1038/hdy.1948.21 (doi:10.1038/hdy.1948.21) [DOI] [PubMed] [Google Scholar]
- 7.Trivers R. L. 1972. Parental investment and sexual selection. In Sexual selection and the descent of man (ed. Campbell B.), pp. 136–179 Chicago, IL: Aldine-Atherton [Google Scholar]
- 8.Birkhead T. R., Møller A. P. 1998. Sperm competition and sexual selection. London, UK: Academic Press [Google Scholar]
- 9.Simmons L. W. 2001. Sperm competition and its evolutionary consequences in the insects. Princeton, NJ: Princeton University Press [Google Scholar]
- 10.Jennions M. D., Petrie M. 2000. Why do females mate multiply? A review of the genetic benefits. Biol. Rev. 75, 21–64 10.1017/S0006323199005423 (doi:10.1017/S0006323199005423) [DOI] [PubMed] [Google Scholar]
- 11.Tregenza T., Wedell N. 2000. Genetic compatibility, mate choice and patterns of parentage: invited review. Mol. Ecol. 9, 1013–1027 10.1046/j.1365-294x.2000.00964.x (doi:10.1046/j.1365-294x.2000.00964.x) [DOI] [PubMed] [Google Scholar]
- 12.Arnqvist G., Nilsson T. 2000. The evolution of polyandry: multiple mating and female fitness in insects. Anim. Behav. 60, 145–164 10.1006/anbe.2000.1446 (doi:10.1006/anbe.2000.1446) [DOI] [PubMed] [Google Scholar]
- 13.Sheldon B. C. 1994. Male phenotype, fertility, and the pursuit of extra-pair copulations by female birds. Proc. R. Soc. Lond. B 257, 25–30 10.1098/rspb.1994.0089 (doi:10.1098/rspb.1994.0089) [DOI] [Google Scholar]
- 14.Parker G. A. 1970. Sperm competition and its evolutionary consequences in the insects. Biol. Rev. 45, 525–567 10.1111/j.1469-185X.1970.tb01176.x (doi:10.1111/j.1469-185X.1970.tb01176.x) [DOI] [Google Scholar]
- 15.Halliday T. R. 1983. The study of mate choice. In Mate choice (ed. Bateson P.). Cambridge, UK: Cambridge University Press [Google Scholar]
- 16.García-González F., Simmons L. W. 2005. Sperm viability matters in insect sperm competition. Curr. Biol. 15, 271–275 10.1016/j.cub.2005.01.032 (doi:10.1016/j.cub.2005.01.032) [DOI] [PubMed] [Google Scholar]
- 17.Yasui Y. 1997. A ‘good sperm’ model can explain the evolution of costly multiple mating by females. Am. Nat. 149, 573–584 10.1086/286006 (doi:10.1086/286006) [DOI] [Google Scholar]
- 18.Fisher D. O., Double M. C., Blomberg S. P., Jennions M. D., Cockburn A. 2006. Post-mating sexual selection increase lifetime fitness of polyandrous females in the wild. Nature 444, 89–92 10.1038/nature05206 (doi:10.1038/nature05206) [DOI] [PubMed] [Google Scholar]
- 19.García-González F., Simmons L. W. 2007. Shorter sperm confer higher competitive fertilization success. Evolution 61, 816–824 10.1111/j.1558-5646.2007.00084.x (doi:10.1111/j.1558-5646.2007.00084.x) [DOI] [PubMed] [Google Scholar]
- 20.Hosken D. J., Garner T. W. J., Tregenza T., Wedell N., Ward P. I. 2003. Superior sperm competitors sire higher-quality young. Proc. R. Soc. Lond. B 270, 1933–1938 10.1098/rspb.2003.2443 (doi:10.1098/rspb.2003.2443) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Keller L., Reeve H. K. 1995. Why do females mate with multiple males? The sexually selected sperm hypothesis. Adv. Study Behav. 24, 291–315 10.1016/S0065-3454(08)60397-6 (doi:10.1016/S0065-3454(08)60397-6) [DOI] [Google Scholar]
- 22.Zeh J. A., Zeh D. W. 1997. The evolution of polyandry II: post-copulatory defences against genetic incompatibility. Proc. R. Soc. Lond. B 264, 69–75 10.1098/rspb.1997.0010 (doi:10.1098/rspb.1997.0010) [DOI] [Google Scholar]
- 23.Firman R. C., Simmons L. W. 2008. Polyandry facilitates post-copulatory inbreeding avoidance in house mice. Evolution 62, 601–611 10.1111/j.1558-5646.2007.00307.x (doi:10.1111/j.1558-5646.2007.00307.x) [DOI] [PubMed] [Google Scholar]
- 24.Parker G. A. 1990. Sperm competition games: raffles and roles. Proc. R. Soc. Lond. B 242, 120–126 10.1098/rspb.1990.0114 (doi:10.1098/rspb.1990.0114) [DOI] [Google Scholar]
- 25.Hosken D. J., Ward P. I. 2001. Experimental evidence for testis size evolution via sperm competition. Ecol. Lett. 4, 10–13 10.1046/j.1461-0248.2001.00198.x (doi:10.1046/j.1461-0248.2001.00198.x) [DOI] [Google Scholar]
- 26.Pitnick S., Miller G. T., Reagan J., Holland B. 2001. Males' evolutionary response to experimental removal of sexual selection. Proc. R. Soc. Lond. B 268, 1071–1080 10.1098/rspb.2001.1621 (doi:10.1098/rspb.2001.1621) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Simmons L. W., García-González F. 2008. Evolutionary reduction in testes size and competitive fertilisation success in response to the experimental removal of sexual selection in dung beetles. Evolution 62, 2580–2591 10.1111/j.1558-5646.2008.00479.x (doi:10.1111/j.1558-5646.2008.00479.x) [DOI] [PubMed] [Google Scholar]
- 28.Firman R. C., Simmons L. W. 2010. Experimental evolution of sperm quality via postcopulatory sexual selection in house mice. Evolution 64, 1245–1256 [DOI] [PubMed] [Google Scholar]
- 29.Firman R. C., Simmons L. W. 2011. Experimental evolution of sperm competitiveness in a mammal. BMC Evol. Biol. 11, 19. 10.1186/1471-2148-11-19 (doi:10.1186/1471-2148-11-19) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Hunt J., Breuker C. J., Sadowski J. A., Moore A. J. 2009. Male–male competition, female mate choice and their interaction: determining total sexual selection. J. Evol. Biol. 22, 13–26 10.1111/j.1420-9101.2008.01633.x (doi:10.1111/j.1420-9101.2008.01633.x) [DOI] [PubMed] [Google Scholar]
- 31.Champlin A. K., Dorr D. L., Gates A. H. 1973. Determining the stage of the estrous cycle in the mouse by the appearance of the vagina. Biol. Reprod. 8, 491–494 [DOI] [PubMed] [Google Scholar]
- 32.Firman R. C., Cheam L. Y., Simmons L. W. In press Sperm competition does not influence sperm hook morphology in selection lines of house mice. J. Evol. Biol. [DOI] [PubMed] [Google Scholar]
- 33.Firman R. C., Simmons L. W. 2008. The frequency of multiple paternity predicts variation in testes size among island populations of house mice. J. Evol. Biol. 21, 1524–1533 10.1111/j.1420-9101.2008.01612.x (doi:10.1111/j.1420-9101.2008.01612.x) [DOI] [PubMed] [Google Scholar]
- 34.Rugh R. 1968. The mouse: its reproduction and development. Minneapolis, MN: Burgess Publishing Company [Google Scholar]
- 35.Dietrich W., Katz H., Lincoln S. E., Shin H. S., Friedman J., Dracopoli N. C., Lander E. S. 1992. A genetic map of the mouse suitable for typing intraspecific crosses. Genetics 131, 423–447 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Kalinowski S. T., Taper M. L., Marshall T. C. 2007. Revising how the computer program Cervus accommodates genotyping error increases success in paternity assignment. Mol. Ecol. 9, 801–808 [DOI] [PubMed] [Google Scholar]
- 37.Whitlock M. C. 2005. Combining probability from independent tests: the weighted Z-method is superior to Fisher's approach. J. Evol. Biol. 18, 1368–1373 10.1111/j.1420-9101.2005.00917.x (doi:10.1111/j.1420-9101.2005.00917.x) [DOI] [PubMed] [Google Scholar]
- 38.König B., Markl H. 1987. Maternal care in house mice I. The weaning strategy as a means for parental manipulation of offspring quality. Behav. Ecol. Sociobiol. 20, 1–9 10.1007/BF00292161 (doi:10.1007/BF00292161) [DOI] [Google Scholar]
- 39.Crowcroft P. 1955. Territoriality in wild house mice, Mus Musculus L. J. Mammal. 36, 299–301 10.2307/1375908 (doi:10.2307/1375908) [DOI] [Google Scholar]
- 40.Becker W. A. 1984. Manual of quantitative genetics. Washington, DC: Pullman [Google Scholar]
- 41.Wong B. B. M., Candolin U. 2005. How is female mate choice affected by male competition? Biol. Rev. 80, 559–571 10.1017/S1464793105006809 (doi:10.1017/S1464793105006809) [DOI] [PubMed] [Google Scholar]
- 42.Rolland C., MacDonald D. W., De Fraipont M., Berdoy M. 2003. Free female mate choice in house mice: leaving the best for last. Behaviour 140, 1371–1388 10.1163/156853903771980639 (doi:10.1163/156853903771980639) [DOI] [Google Scholar]
- 43.Stockley P. 2003. Female multiple mating behaviour, early reproductive failure and litter size variation in mammals. Proc. R. Soc. Lond. B 270, 271–278 10.1098/rspb.2002.2228 (doi:10.1098/rspb.2002.2228) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Firman R. C., Simmons L. W. 2008. Polyandry, sperm competition, and reproductive success in mice. Behav. Ecol. 19, 695–702 10.1093/beheco/arm158 (doi:10.1093/beheco/arm158) [DOI] [Google Scholar]
- 45.Klemme I., Ylönen H., Eccard J. A. 2008. Long-term fitness benefits of polyandry in a small mammal, the bank vole Clethrionomys glareolus. Proc. R. Soc. B 275, 1095–1100 10.1098/rspb.2008.0038 (doi:10.1098/rspb.2008.0038) [DOI] [PMC free article] [PubMed] [Google Scholar]