Abstract
Embryonic stem cells (ESCs) are pluripotent with multilineage potential to differentiate into virtually all cell types in the organism and thus hold a great promise for cell therapy and regenerative medicine. In vitro differentiation of ESCs starts with a phase known as embryoid body (EB) formation. EB mimics the early stages of embryogenesis and plays an essential role in ESC differentiation in vitro. EB uniformity and size are critical parameters that directly influence the phenotype expression of ESCs. Various methods have been developed to form EBs, which involve natural aggregation of cells. However, challenges persist to form EBs with controlled size, shape, and uniformity in a reproducible manner. The current hanging-drop methods are labor intensive and time consuming. In this study, we report an approach to form controllable, uniform-sized EBs by integrating bioprinting technologies with the existing hanging-drop method. The approach presented here is simple, robust, and rapid. We present significantly enhanced EB size uniformity compared to the conventional manual hanging-drop method.
INTRODUCTION
Embryonic stem cells (ESCs) display indefinite self-renewal and they are a pluripotent cell source with multilineage differentiation potential.1, 2 The unique features of pluripotency make ESCs an ideal source for tissue replacement and regenerative medicine for diseases and injuries.3, 4In vitro differentiation of ESCs into other phenotypes is preceded by the formation of embryoid bodies (EBs). EBs are three dimensional (3D) aggregates of ESCs with characteristics of the early stages of embryogenesis and play a critical role during in vitro differentiation of ESCs. The lack of uniformity in EB size may result in nonhomogeneous and asynchronous differentiation of the residing cells.5, 6 Therefore, formation of EBs with uniform sizes is needed to effectively employ ESCs in regenerative medicine.
Three germ layers form in the early stages of embryogenesis in vivo, which are also observed in EB culture in vitro.7 Therefore, EB provides a suitable microenvironment for ESCs in vitro, which facilitates lineage-specific differentiation.8, 9 It was previously shown that EB-mediated differentiation efficiency is dependent on the EB size. Larger EB sizes tend to differentiate toward mesoderm and endoderm, while smaller EB sizes direct their differentiation toward ectoderm.5, 6 It is also reported that smaller sized EBs (100–500 μm in the lateral dimensions and 120 μm in depth) are more likely to allow cardiomyocyte differentiation.10 On the other hand, EB formation from individual ESCs via spontaneous aggregation is inefficient,11, 12 which results in heterogeneous size distribution and noncontrolled differentiation lineage.13, 14
Although various methods have been developed to promote EB formation through natural aggregation or artificial cell-cell interactions, it is still challenging to obtain controllable, uniform-sized EBs. For instance, enzymatic digestion of the ESC colonies and rotary mass suspension resulted in heterogeneous size distribution of EBs,13, 15, 16 while methods based on surface patterning can only control the initial EB size.6, 17, 18, 19 The hanging-drop method (based on manual pipetting) is commonly used in ESC cultures to form EBs. However, the EB size through this method is a variable due to variation during pipetting, such as droplet volume and number of cells per droplet. Additionally, these manual methods are labor intensive and time consuming, and the reproducibility of the results varies between operators. Nonadhesive microwell arrays of various aspect ratios, sizes, and shapes have been developed to control the uniformity of EB size and shape through physically controlling the size of growing EBs.14, 20, 21, 22 However, ESCs formed disk shaped EBs on microwell arrays, while they aggregated in spherical form in suspension cultures,20 suggestive of different phenotypes.23 Furthermore, the mechanical stress induced on ES cells during the forced aggregation process (e.g., rotary mass suspension24, 25, 26 and centrifugation12) may disrupt the cell-cell signaling10 and damage the fragile cellular components affecting subsequent cell differentiation.27 Methods for sorting EBs of heterogeneous size into uniform size groups have also been developed.27 However, the separation methods generally involve external force fields such as microfluidics27 that may damage ESCs and affect subsequent cell differentiation.
We hypothesized that the recent advances in bioprinting technologies would facilitate the formation of uniform-sized EBs in a reproducible manner, addressing the challenges associated with the current methods. To validate this hypothesis, we integrated a cell printing technique28, 29, 30 with the existing hanging-drop culture method. Although a variety of cell bioprinting methods, such as acoustic printing,31, 32, 33 valve based printing,34, 35, 36 and ink-jet printing,37, 38 have been used to encapsulate cells in microdroplets,39, 40 the combination of these cell printing techniques with the hanging-drop method for EB formation has not yet been evaluated. Here, we present a new method based on bioprinting and hanging-drop methods that results in controllable uniform-sized EBs. The developed method provides a reproducible, efficient, and scalable alternative to the currently available methods. These uniform-sized EBs would be highly applicable towards regenerative medicine and tissue replacement.
MATERIALS AND METHODS
ESC culture
In this study, we used genetically engineered mouse ESCs (mESCs, line E14). These cells expressed green fluorescent protein (GFP) upon initiation of gene transfection at the Oct4 promoter at goosecoid (Gsc) gene locus. The mESCs were cultured in high glucose-Dulbecco’s modified eagles medium (Gibco) supplemented with 10% (vol∕vol) ES qualified phosphate buffered saline (PBS) (Gibco), 1 mM L-glutamine (Gibco), 0.1 mM β-mercaptoethanol (Sigma, St. Louis, MO), 100 U∕ml penicillin, 100 μg∕ml streptomycin (Gibco), and 1000 U∕ml of leukemia inhibitory factor (Chemicon).
Embryoid body formation
Bioprinted hanging-drop method
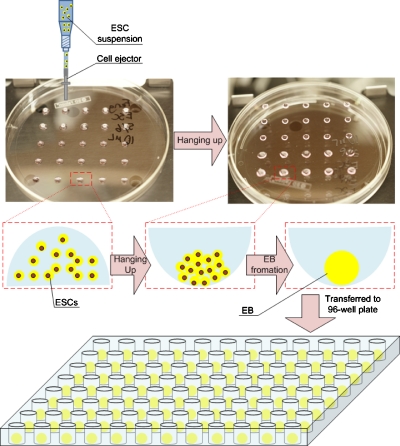
The mESC suspensions were prepared at three concentrations (0.1×106, 0.5×106 and 1.0×106 cells∕ml) in basic EB medium containing alpha minimal essential medium (α-MEM; Gibco) supplemented with 15% (v∕v) heat-inactivated FBS (Invitrogen, Carlsbad, CA) and 1% (v∕v) penicillin∕streptomycin. Droplets of mESC suspension at controlled volumes (1, 4, 10, and 20 μl) were bioprinted onto inside surface of a Petri dish lid in an array format by a cell printer (Fig. 1) mimicking the existing hanging-drop approach. In hanging-drop method, droplets of ESC suspension were generated by manual pipetting.9 The cells were allowed to aggregate with the help of gravity when the Petri dish was reversed. The bioprinted droplets were hung in the Petri dish for 24 h (Fig. 1). The dish bottom was then filled with PBS to prevent drying of bioprinted droplets. The aggregated mESCs within the bioprinted droplets were then transferred to low-adherence 96-well plates and cultured for 96 h to generate EBs (Fig. 1).
Figure 1.
Schematic of the EB formation process using bioprinting approach. Droplets of cell-medium suspension were bioprinted onto the lid of a Petri dish and were hung up for 24 h to allow for EB aggregation. The formed EBs were transferred to a 96-well plate for additional culture up to 96 h.
Standard hanging-drop method using manual pipetting
For control groups, the same droplet size and cell concentrations were used as the bioprinted hanging-drop method. EBs formed by the standard hanging-drop approach using manual pipetting were used as a control group following the commonly used protocols.9 ESCs were seeded onto the Petri dish lids under similar conditions as the bioprinting method. ESCs aggregated within the droplet for 24 h, and then these aggregates were transferred into 96-well plates. EBs formed using this method were cultured for 96 h.
EB morphological observation and EB cell death
The EB sizes formed using different initial cell seeding density (0.1×106, 0.5×106, and 1.0×106 cells∕ml), droplet size (1, 4, 10, and 20 μl), and culture time (t=24, 48, 72, and 96 h) were analyzed for control and bioprinting groups. The morphology of EBs and Oct4-GFP expression of ESCs within EBs were observed at different time points after bioprinting (t=24, 48, 72, and 96 h) using an inverted fluorescent microscope (Nikon Eclipse T2000). The EB sizes were measured from the collected micrographs using the NIH IMAGEJ program (developed at the U.S. National Institutes of Health and is available at http:∕/rsb.info.nih.gov∕nih-image∕). In this study, we used GFP expressing ES cells. These cells show green fluorescence, when they are undifferentiated. EB cell death was analyzed at t=96 h by incubating EBs only in 4 μM ethidium homodimer (Molecular Probes Inc.) in PBS for 10 min at 37 °C to indicate the dead cells. The size uniformity and the resulting diameters at the end of the culture period were assessed and compared to the control and bioprinted groups.
Statistical analysis
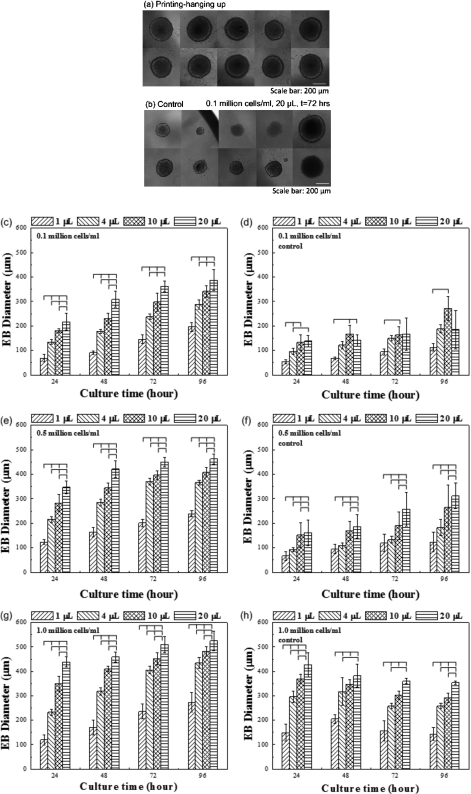
The experimental results for both control and bioprinted groups were initially tested for normal distribution using Anderson–Darling test. The sample size used for each experimental group was between 10 to 25 droplets. The effects of droplet size, initial cell seeding density, and culture time on the EB sizes were analyzed with one way analysis of variance with Tukey post hoc comparisons. The EB size uniformity was statistically assessed with Levene’s test for equality of variances at the end of the 96 h culture period for all initial cell seeding densities (0.1×106, 0.5×106, and 1.0×106 cells∕ml) and droplet sizes (1, 4, 10, and 20 μl). The uniformity of the EB sizes was assessed based on the variance in the data sets, where a smaller variance indicated higher uniformity in the resulting EB sizes. The EB diameters at the end of the culture period were compared statistically between control group and bioprinted groups with Mann–Whitney U test for pairwise comparison. The statistical significance threshold was set at 0.05 for all tests (with p<0.05). Error bars in the figures represented standard deviation (Figs. 34).
Figure 3.
The effect of initial cell density, droplet volume, and culture time on the EB size for bioprinting and control with manual pipetting. EBs retrieved from bioprinted droplets after 24 h culture in vitro (a) displayed more uniform size distribution compared to control method (i.e., pipetting based manual hanging-droplet) (b). Statistical analysis of EB size with different initial cell concentrations (0.1×106, 0.5×106, and 1.0×106 cells∕ml) formed by the bioprinting method [(c), (e), and (g)] and by the control method [(d), (f), and (h)]. The EB sizes formed by bioprinting were well controlled by varying the droplet size (1, 4, 10, and 20 μl) and the culture time (24, 48, 72, and 96 h).
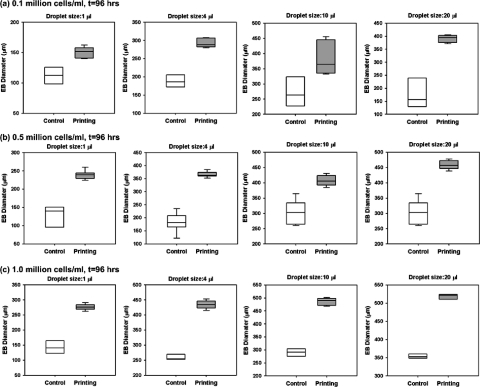
Figure 4.
Comparison of the uniformity and the size of the EBs formed by control (manual hanging-drop method) and bioprinted hanging-drop methods. The statistical comparison of the EB size uniformity was performed with Levene’s test for equality of variances (p<0.05) at the end of the 96 h culture period for different initial cell seeding densities and droplet sizes. The uniformity of the sizes was assessed based on the variance in the data sets. Less variance in the data indicated higher uniformity in the resulting EB sizes. The diameters of the resulting EBs were compared statistically for control and bioprinting methods with Mann–Whitney U test for pairwise comparisons (p<0.05). (a) For 0.1×106∕ml initial cell seeding density, bioprinting method resulted in more uniform EB sizes at the end of the 96 h culture period for 1, 4, and 20 μl droplet sizes. (b) For 0.5×106∕ml initial seeding density, bioprinting resulted in significantly higher uniformity in the EB sizes for all droplet sizes compared to control method. (c) For 1×106∕ml initial cell seeding density, higher uniformity in EB sizes was observed for the droplet size of 1 μl. Overall, bioprinting method resulted in statistically greater EB sizes compared to control method at the end of the 96 h culture period in all groups.
RESULTS AND DISCUSSION
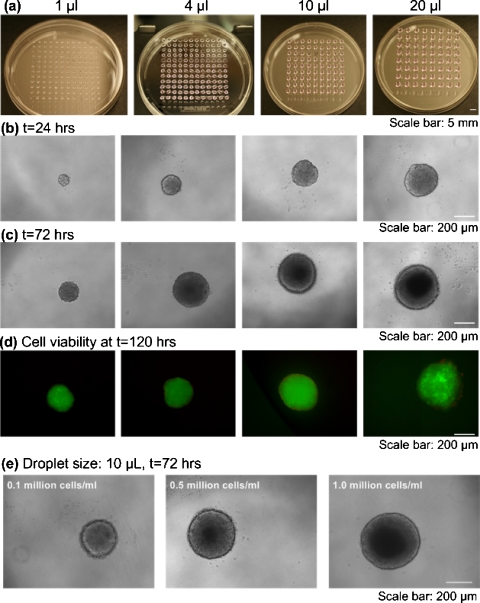
In this study, we assessed the feasibility of using a cell bioprinting based hanging-drop method to form EBs with controllable and uniform sizes. The effects of cell seeding concentration (0.1×106, 0.5×106, and 1.0×106 cells∕ml), droplet volume (1, 4, 10, and 20 μl), and culture time (24, 48, 72, and 96 h) on the EB size were analyzed by both methods. Morphological assessment of the EBs showed that EB sizes increased with both increasing bioprinted droplet size [Figs. 2a, 2b] and culture time [Fig. 2c]. We evaluated the cell death in the EBs formed using the bioprinting method. The results showed that cells were viable throughout the culture period independent of the droplet size [Fig. 2d]. However, a small number of dead cells were observed in EBs generated by larger droplet volumes (i.e., 20 μl), which amounted to less than 1% of the total number of cells. Larger EB sizes were obtained at the end of the culture period (t=96 h) with higher initial cell seeding densities [Fig. 2e].
Figure 2.
EB formation using bioprinting method. (a)–(c) Images of formed EBs with droplet sizes of 1, 4, 10, and 20 μL at a cell density of 105 cells∕ml. (a) Uniform-sized droplets encapsulating ESCs were generated by bioprinting. (b) Phase contrast images of EBs formed after hanging for 24 h and (c) after culture for 72 h in a 96 multiwell plate. (d) Fluorescent images of GFP positive EBs at t=96 h stained with ethidium homodimer. (e) Images of EBs formed with printed droplet size of 10 μl at t=72 h at different cell concentrations.
Next, we quantitatively evaluated the EB size change over 96 h of culture period, which indicated that EB sizes continuously increased, when bioprinting method was used (Fig. 3). In addition, increasing volume of bioprinted droplets resulted in larger EB sizes for all groups at all time points [Figs. 3c, 3e, 3g]. A similar consistent trend was not observed in dependence of the EB sizes on culture time and droplet volume in controls [Figs. 3d, 3f, 3h], which indicated that it is difficult to achieve a controlled EB size with the control method. Overall, it was observed that EB sizes formed using the bioprinting method presented a significant increase in response to increasing initial droplet size, cell concentration, and culture time, which was not observed for the EBs formed with the control method. Therefore, these results indicated that controllable EB sizes can be achieved by varying droplet volume, ESC seeding density, and culture time using the bioprinting method.
To assess the EB size uniformity, we analyzed the EB size obtained by the bioprinting and manual pipetting methods at the end of the 96 h culture period [Fig. 3a]. Variation in the measured data was used to quantify uniformity and compare the two methods used in this study by the Levene’s statistical test for the equality of variances. Overall, bioprinting resulted in significantly lower variation (i.e., 57%–94% less variance), and hence, an improved uniformity in EB sizes at the end of the culture period compared to manual pipetting (Fig. 4). Specifically, with bioprinting method, enhanced uniformity in EB sizes was achieved for (i) 0.1×106∕ml initial cell seeding density with 1, 4, and 20 μl droplet sizes [Fig. 4a], (ii) 0.5×106∕ml initial cell seeding density with all droplet sizes [Fig. 4b], and (iii) 1×106∕ml initial cell seeding density with 1 and 10 μl droplet sizes [Fig. 4c]. Furthermore, bioprinting method resulted in significantly larger EB sizes (i.e., 25%–55% larger EBs with bioprinting) compared to controls at the end of the 96 h culture period in all groups (Fig. 4).
The results presented here suggest that more uniform EB size distribution can be achieved with bioprinting compared to the control method. Furthermore, significantly greater EB sizes can be obtained for the same cell seeding density and droplet volume when bioprinting method is utilized compared to the manual pipetting. The enhanced uniformity and larger size of the EBs formed utilizing the bioprinting method over the manual methods can be explained by the inhomogeneous droplet spread during manual pipetting and cell settling during the time consuming, labor intensive processes, which led to nonuniform cell seeding densities with manual methods. These technical and practical limitations may be responsible for nonuniform droplet geometry and increased mechanical stress on the ESCs within the manually pipetted droplet, thus affecting the EB formation process. Furthermore, the bioprinting system presented here can generate up to 160 droplets∕second. It would take up to ∼10 min to generate that many droplets with the most broadly used manual pipetting methods. This corresponds to at least two orders of magnitude decrease in the speed time that it takes to pattern cells for EB formation. Therefore, bioprinting method presents additional advantages for EB formation over manual pipetting in terms of throughput and user-friendly operation. In addition, our system creates EBs of controllable sizes similar to the hanging-drop method, which can then be collected and cultured together for further studies.
CONCLUSIONS
The EB size and uniformity are critical parameters that play an important role in differentiation efficiency of residing ESCs. In this study, we presented a cell printing based high throughput method to produce EBs with uniform sizes by controlling the cell seeding density, bioprinting volume, and culture time compared to the existing hanging-drop methods. The results showed that the bioprinting approach presented here formed EBs with a high degree of uniformity in size compared to EBs that were generated by using the manual pipetting approach. Furthermore, bioprinting method resulted in significantly larger size EBs at the end of the culture period compared to the manual controls. Therefore, the combination of bioprinting technique with the hanging-drop method provides an effective tool to generate controllable uniform-sized EBs. The EBs formed with this method would be essential for applications in regenerative medicine, investigating stem cell differentiation, screening drug candidates, and evaluating embryonic toxicity.
ACKNOWLEDGMENTS
This work was performed at the Demirci Bio-Acoustic MEMS in Medicine (BAMM) Laboratories at the HST-BWH Center for Bioengineering, Harvard Medical School. This work was also supported by NIHR21 (Grant No. AI087107), the W.H. Coulter Foundation Young Investigator Award, and the Center for Integration of Medicine and Innovative Technology under U.S. Army Medical Research Acquisition Activity Cooperative Agreement (Nos. DAMD17-02-2-0006, W81XWH-07-2-0011, and W81XWH-09-2-0001). Also, this research was made possible by a research grant that was awarded and administered by the U.S. Army Medical Research and Materiel Command (USAMRMC) and the Telemedicine and Advanced Technology Research Center (TATRC) at Fort Detrick, MD. The information contained herein does not necessarily reflect the position or policy of the Government, and no official endorsement should be inferred.
U.D. proposed the idea; F.X. and U.D designed the research; F.X., B.S., and B.S. performed research; F.X., S.Q.W. B.S., and U.A.G. analyzed the data; and F.X., S.Q.W., U.A.G., and U.D. wrote the paper.
References
- Murry C. E. and Keller G., Cell 132, 661 (2008). 10.1016/j.cell.2008.02.008 [DOI] [PubMed] [Google Scholar]
- Pouton C. W. and Haynes J. M., Nat. Rev. Drug Discovery 6, 605 (2007). 10.1038/nrd2194 [DOI] [PubMed] [Google Scholar]
- Heit J. J. and Kim S. K., Pediatr. Diabetes 5, 5 (2004). 10.1111/j.1399-543X.2004.00074.x [DOI] [PubMed] [Google Scholar]
- Xu Y., Shi Y., and Ding S., Nature (London) 453, 338 (2008). 10.1038/nature07042 [DOI] [PubMed] [Google Scholar]
- Peerani R., Rao B. M., Bauwens C., Yin T., Wood G. A., Nagy A., Kumacheva E., and Zandstra P. W., EMBO J. 26, 4744 (2007). 10.1038/sj.emboj.7601896 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Park J., Cho C. H., Parashurama N., Li Y., Berthiaume F., Toner M., Tilles A. W., and Yarmush M. L., Lab Chip 7, 1018 (2007). 10.1039/b704739h [DOI] [PubMed] [Google Scholar]
- Itskovitz-Eldor J., Schuldiner M., Karsenti D., Eden A., Yanuka O., Amit M., Soreq H., Benvenisty N., Mol. Med. 6, 88 (2000). [PMC free article] [PubMed] [Google Scholar]
- Koike M., Sakaki S., Amano Y., and Kurosawa H., J. Biosci. Bioeng. 104, 294 (2007). 10.1263/jbb.104.294 [DOI] [PubMed] [Google Scholar]
- Kurosawa H., J. Biosci. Bioeng. 103, 389 (2007). 10.1263/jbb.103.389 [DOI] [PubMed] [Google Scholar]
- Mohr J. C., Zhang J., Azarin S. M., Soerens A. G., de Pablo J. J., Thomson J. A., Lyons G. E., Palecek S. P., and Kamp T. J., Biomaterials 31, 1885 (2010). 10.1016/j.biomaterials.2009.11.033 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Reubinoff B. E., Pera M. F., Fong C. Y., Trounson A., and Bongso A., Nat. Biotechnol. 18, 399 (2000). 10.1038/74447 [DOI] [PubMed] [Google Scholar]
- Burridge P. W., Anderson D., Priddle H., Barbadillo Muñoz M. D., Chamberlain S., Allegrucci C., Young L. E., and Denning C., Stem Cells 25, 929 (2007). 10.1634/stemcells.2006-0598 [DOI] [PubMed] [Google Scholar]
- Singla D. K., Jayaraman S., Zhang J., and Kamp T. J., in Human Cell Culture 6: Embryonic Stem Cells, edited by Master J. R., Palsson B. O., and Thomson J. A. (Springer-Verlag, New York, 2007), pp. 211–234. [Google Scholar]
- Choi Y. Y., Chung B. G., Lee D. H., Khademhosseini A., Kim J. -H., and Lee S. -H., Biomaterials 31, 4296 (2010). 10.1016/j.biomaterials.2010.01.115 [DOI] [PubMed] [Google Scholar]
- Rohani L., Karbalaie K., Vahdati A., Hatami M., Nasr-Esfahani M. H., and Baharvand H., Int. J. Artif. Organs 31, 258 (2008). [DOI] [PubMed] [Google Scholar]
- Youn B. S., Sen A., Behie L. A., Girgis-Gabardo A., and Hassell J. A., Biotechnol. Prog. 22, 801 (2006). 10.1021/bp050430z [DOI] [PubMed] [Google Scholar]
- Bauwens C. L., Peerani R., Niebruegge S., Woodhouse K. A., Kumacheva E., Husain M., and Zandstra P. W., Stem Cells 26, 2300 (2008). 10.1634/stemcells.2008-0183 [DOI] [PubMed] [Google Scholar]
- Ungrin M. D., Joshi C., Nica A., Bauwens C., and Zandstra P. W., PLoS ONE 3, e1565 (2008). 10.1371/journal.pone.0001565 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gothard D., Roberts S. J., Shakesheff K. M., and Buttery L. D., Cytotechnology 61, 135 (2009). 10.1007/s10616-010-9255-3 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Karp J. M., Yeh J., Eng G., Fukuda J., Blumling J., Suh K. -Y., Cheng J., Mahdavi A., Borenstein J., Langer R., and Khademhosseini A., Lab Chip 7, 786 (2007). 10.1039/b705085m [DOI] [PubMed] [Google Scholar]
- Hwang Y. S., Chung B. G., Ortmann D., Hattori N., Moeller H. C., and Khademhosseini A., Proc. Natl. Acad. Sci. U.S.A. 106, 16978 (2009). 10.1073/pnas.0905550106 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lee W. G., Ortmann D., Hancock M. J., Bae H., and Khademhosseini A., Tissue Eng. 16, 249 (2010). [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nelson C. M., Jean R. P., Tan J. L., Liu W. F., Sniadecki N. J., Spector A. A., and Chen C. S., Proc. Natl. Acad. Sci. U.S.A. 102, 11594 (2005). 10.1073/pnas.0502575102 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Abilez O., Benharash P., Mehrotra M., Miyamoto E., Gale A., Picquet J., Xu C., and Zarins C., J. Surg. Res. 132, 170 (2006). 10.1016/j.jss.2006.02.017 [DOI] [PubMed] [Google Scholar]
- Niebruegge S., Nehring A., Baer H., Schroeder M., Zweigerdt R., and Lehmann J., Tissue Eng A 14, 1591 (2008). 10.1089/ten.tea.2007.0247 [DOI] [PubMed] [Google Scholar]
- Ng E. S., Davis R. P., Azzola L., Stanley E. G., and Elefanty A. G., Blood 106, 1601 (2005). 10.1182/blood-2005-03-0987 [DOI] [PubMed] [Google Scholar]
- Lillehoj P. B., Tsutsui H., Valamehr B., Wu H., and Ho C. M., Lab Chip 10, 1678 (2010). 10.1039/c000163e [DOI] [PMC free article] [PubMed] [Google Scholar]
- Moon S., Hasan S. K., Song Y. S., Xu F., Keles H. O., Manzur F., Mikkilineni S., Hong J. W., Nagatomi J., Haeggstrom E., Khademhosseini A., and Demirci U., Tissue Eng Part C Methods 16, 157 (2010). 10.1089/ten.tec.2009.0179 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Xu F., Moon S. J., Emre A. E., Turali E. S., Song Y. S., Hacking S. A., Nagatomi J., and Demirci U., Biofabrication 2, 014105 (2010). 10.1088/1758-5082/2/1/014105 [DOI] [PubMed] [Google Scholar]
- Xu F., Celli J., Rizvi I., Moon S., Hasan T., and Demirci U., Biotechnol. J. 6, 204 (2011). 10.1002/biot.201000340 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Demirci U. and Montesano G., Lab Chip 7, 1139 (2007). 10.1039/b704965j [DOI] [PubMed] [Google Scholar]
- Demirci U., J. Microelectromech. Syst. 15, 11 (2006). [Google Scholar]
- Demirci U., Yaralioglu G. G., Haeggstrom E., Percin G., Ergun S., and Khuri-Yakub B. T., IEEE Trans. Semicond. Manuf. 18, 709 (2005). 10.1109/TSM.2005.858500 [DOI] [Google Scholar]
- Demirci U. and Montesano G., Lab Chip 7, 1428 (2007). 10.1039/b705809h [DOI] [PubMed] [Google Scholar]
- Song Y. S., Adler D., Xu F., Kayaalp E., Nureddin A., Anchan R. M., Maas R. L., and Demirci U., Proc. Natl. Acad. Sci. U.S.A. 107, 4596 (2010). 10.1073/pnas.0914059107 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Moon S., Kim Y. -G., Dong L., Lombardi M., Haeggstrom E., Jensen R. V., Hsiao L. -L., and Demirci U., PLoS ONE 6, e17455 (2011). 10.1371/journal.pone.0017455 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ringeisen B. R., Othon C. M., Barron J. A., Young D., and Spargo B. J., Biotechnol. J. 1, 930 (2006). 10.1002/biot.200600058 [DOI] [PubMed] [Google Scholar]
- Boland T., Xu T., Damon B., and Cui X., Biotechnol. J. 1, 910 (2006). 10.1002/biot.200600081 [DOI] [PubMed] [Google Scholar]
- Geckil H., Xu F., Zhang X., Moon S., and Demirci U., Nanomedicine 5, 469 (2010). 10.2217/nnm.10.12 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Samot J., Moon S., Shao L., Zhang X., Xu F., Song Y., Keles H. O., Matloff L., Markel J., and Demirci U., PLoS ONE 6, e17530 (2011). 10.1371/journal.pone.0017530 [DOI] [PMC free article] [PubMed] [Google Scholar]