Abstract
As advances in microfluidics continue to make contributions to diagnostics and life sciences, broader awareness of this expanding field becomes necessary. By leveraging low-cost microfabrication techniques that require no capital equipment or infrastructure, simple, accessible, and effective educational modules can be made available for a broad range of educational needs from middle school demonstrations to college laboratory classes. These modules demonstrate key microfluidic concepts such as diffusion and separation as well as “laboratory on-chip” applications including chemical reactions and biological assays. These modules are intended to provide an interdisciplinary hands-on experience, including chip design, fabrication of functional devices, and experiments at the microscale. Consequently, students will be able to conceptualize physics at small scales, gain experience in computer-aided design and microfabrication, and perform experiments—all in the context of addressing real-world challenges by making their own lab-on-chip devices.
INTRODUCTION
With numerous advantages over traditional processes, including minimal reagent consumption, faster reaction times, higher signal to noise ratios, manufacturability, and portability, lab-on-a-chip (LOC) technology has experienced a rapid growth within the past decade.1, 2, 3, 4, 5, 6 By leveraging and manipulating transport processes at small length scales, microfluidics enables processes such as purification, chemical reactions, and biological assays to be integrated into LOC technology.7, 8 Through the integration of multiple processes onto a single platform, complete cellular and molecular assays can be performed on-chip, thus negating the need for dedicated laboratories and lab-based infrastructure; such micrototal analysis systems can reduce the complexity as well as improve the efficacy of analytical assays.9, 10
By leveraging traditional fabrication methods on glass and silicon substrates including standard photolithography, direct etching, and thin film deposition, complex LOC devices have been fabricated to perform such processes as immunoassays, polymerase chain reaction, and single cell analysis.3, 11, 12, 13, 14, 15, 16, 17, 18 Thus, the contributions of traditional fabrication methods have enabled the high resolution patterning of biological substrates and structural features at the micro- and nanoscale. This, in turn, has given us unprecedented control over cellular microenvironment and the ability to carry out molecular assays and single cell interrogation.12, 19, 20, 21 However, due to their adaptation from the semiconductor industry, these fabrication methods are inherently expensive and limited in resolution by their tooling devices.
With the introduction of poly(dimethylsiloxane) (PDMS) in the 1998 seminal paper by Duffy et al., progress in the field of microfluidics has greatly accelerated, attributing primarily to the rapid prototyping ability of PDMS.22 By utilizing the soft lithographic method of molding and patterning structural features with PDMS, microfluidic devices can be easily fabricated in academic institutions, thus facilitating the growth of academic research in LOC technology.8 Specifically, the ability to mold PDMS from glass and silicon substrates and bond PDMS to glass slides to form complete microfluidic channels reduced the need for expensive tooling costs associated with previous LOC devices. Although this process alleviated some of the expenses of microfabrication, glass and silicon masters still require photolithography. The need for costly capital equipment, supplies, and engineering expertise has limited accessibility to such technologies and has spurred the search for low-cost fabrication alternatives. In lieu of the traditional silicon-based fabrication method, recent advances have provided low-cost alternative fabrication approaches including the use of paper,23, 24, 25, 26 cotton,27, 28 thermoplastics,29, 30 wax,31, 32, 33, 34 and photocurable polymers.35 By adapting the substrates to their most suitable application, these methods have provided niche-specific low-cost fabrication alternatives. For example, by leveraging the accessibility and hydrophilic properties of paper and cotton, these substrates have been fashioned for enzyme linked immunosorbent assay25 and colorimetric assays.27, 28 Thermoplastics and photocurable polymers are particularly well suited for device fabrication due to their robustness and ease of fabrication. In addition, processes which utilize the printing of ink and wax onto transparencies and polyimide films have yielded ultrarapid, on-demand techniques for fabricating microfluidic devices.36, 37 These processes, however, are not without their limitations. For example, the printed patterns with these methods are limited by the resolution of the printer as well as the amount of deposited ink.37 In the wax printing technique, for example, the need for a specialized solid ink printers as well as the use of harsh chemical etchants has stymied its accessibility.31, 34, 36, 37
Microfluidics in the classroom
With increasingly broad adoption and application of microfluidics, the need for general awareness as well as in depth education becomes necessary. However, most microfluidic education is not introduced until the graduate level. While some undergraduate courses exist, science and engineering curriculum in general suffers from a dearth of hands-on application driven courses. In response, these modules outlined here seek to combine compelling, real-world issues by designing and developing cutting edge technologies in a rapid, simple, and inexpensive platform to facilitate interdisciplinary problem based learning that can be introduced at earlier levels of education. Integration of microfluidics in undergraduate studies as well as high school and middle school would ensure the exposure of this technology to the next work-force generation. Further, this exposure can provide the opportunity to demonstrate the increasingly interdisciplinary approach to problem solving in science and engineering. To facilitate this educational program, microfluidic courses should be fun and engaging as to promote creativity and imagination as well as be safe and affordable to ensure accessibility.
Recently, several papers have invoked much creative thinking due to their simple yet elegant approach to scientific education.38, 39 In a paper by Yang et al., the dessert Jell-O was used in the process of soft lithography to demonstrate microfluidic technology.38 By molding coffee stirrers of various patterns overnight, Jell-O channels mimicking microfluidic channels were fabricated. These molds were then placed onto aluminum plates to demonstrate flow patterns. Importantly, this method allowed for an affordable, fun way to deliver microfluidics to the classroom by demonstrating key concepts such as flow, diffusion, and even chemical reaction in the form of pH sensing. Although this creative approach to education introduces microfluidics to the classroom, the inherently large size of these devices does not illustrate the physics that occurs at the microscale and does not take advantage of small reagent volume size typical of microfluidics.
Educational kits emphasizing microfabrication for microchip technologies have been developed. For example, Berkowski et al. utilize photosensitive polymers to demonstrate photolithography to junior high and high school students.40 Polymer thin film objects can be created within minutes by UV-initiated photoresist cross-linking. Although the experiment teaches concepts of photolithography, polymerization, chemical cross-linking, and pattern formation, no concepts in or applications of microfluidics are performed.
For microfluidics demonstration, an approach similar to LEGO® blocks using “plug-n-play” pieces has been demonstrated and integrated into educational modules.41 Similar to circuits, the pieces can be put together on a “motherboard” that houses a variety of devices. With these devices, students can learn about small working volumes, microfluidic flow regimes, and real-world applications of microfluidic devices. One major drawback, however, is that the expense and accessibility of the kits limit adoption.
These academic modules have laid excellent groundwork in promoting awareness of microfluidics in the classroom, but there are several key factors that can further be improved upon. A combination of accessibility and concepts such as computer-aided design (CAD) of microchannels, standard soft lithography, and the understanding of working volumes of microfluidics is overlooked in some or all of these experiments.
Shrink-induced fabrication of microfluidics
The use of low-cost thermoplastics for microfluidic applications has been demonstrated and developed in recent years.29, 30 Processes such as direct patterning of microstructures into plastics and printing onto plastics to generate high-aspect micromolds are attractive venues for the production of low-cost, disposable point-of-care (POC) diagnostic devices.30, 42 In our previous efforts, we have leveraged printing and etching prestressed polystyrene and polyolefin (PO), which retracted in-plane by approximately 60% and 95%, respectively.29, 30, 42, 43, 44 This strategy allows for rapid fabrication of high-aspect micromolds, which can be used for standard soft lithography, negating the use of expensive patterning tools and microfabrication infrastructure. Importantly, this technique improves upon the printed resolution as the features shrink with the retracting substrate.
Here, we demonstrate the ease of integration of microfluidics into the teaching environment through the use of thermoplastic patterning. Notably, by utilizing commercially available shrink-film (PO) which yields approximately 95% reduction in surface area, high-aspect masters can be generated by using a standard laser-jet printer with a single print. This facilitates the formation of tunable and rounded microchannels, which can be molded with PDMS to create microfluidic channels. To demonstrate the feasibility of teaching key concepts of microfluidics, we took inspiration from previously reported educational microfluidic kits, which have been implemented within the classroom.38, 45 Key concepts such as separations, flowrate measurements, chemical gradients, and chemical reactions can be easily performed on our simple platform, yielding reproducible results. These concepts can be integrated into teaching modules and tailored to meet the criteria and academic level of the institution.
TEACHING OBJECTIVES
The objectives of these teaching modules are to demonstrate key concepts in and applications of microfluidics. Fundamental concepts in microfluidics include principles of separation, diffusion, and flow regimes while LOC applications can be demonstrated through simple separation processes analogous to real-world POC diagnostics and immunoassays. Each laboratory exercise was further designed to enable data acquisition with minimal laboratory equipment.
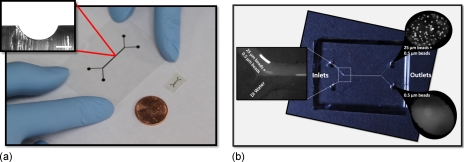
As outlined in Table 1, module 1 is designed for middle school students and will teach CAD of microchannels, soft lithography, pressure driven flow, and most importantly, the working volumes of microfluidics. Concepts such as separation and diffusion, which play an integral part in LOC technology and POC diagnostics, will be covered in this same module. As an example of such processes, we have chosen the well-known H-filter,46, 47 a classic microfluidic device, for the separation of analyte and whole blood in LOC technology. To demonstrate the sample preparations of whole blood and saliva in POC technology, fluorescent microbeads on the same size scale as cells are used in the H-filter. Using fluorescent beads mimics cells as to simulate biological applications with microfluidics.
Table 1.
Table of teaching objectives for each module. Each module is inclusive of previous modules so that an accumulative understanding of the material is achieved. Module 1 teaches fabrication and design of microfluidics as well as basic concepts of microfluidics and focuses on separation and diffusion using an H-filter. Modules 2 and 3 use a gradient generator to teach calculations and perform an immunoassay.
| Module 1 | Module 2 | Module 3 | |
|---|---|---|---|
| Name | Separation and diffusion | Gradient generation | Immunoassay |
| Device | H-filter | Gradient generator | Gradient generator |
| Target audience | Middle school | High school | College undergraduate |
| Learning objectives | 1. Engineering drafting software | 1. Inclusive of module 1 | 1. Inclusive of modules 1 and 2 |
| 2. Design of microfluidic patterns | 2. Gradient generator | 2. Dimensionless groups (Reynolds and Stokes numbers) | |
| 3. Soft lithography | 3. Concentration profiles | 3. Biological assay with antibodies | |
| 4. Pressure driven flow | 4. Image processing software | 4. Fluorescence microscopy | |
| 5. Length scale and working volume | 5. Flowrate calculations | ||
| 6. Channel dimensions | |||
| 7. Laminar flow | |||
| 8. Separation and mixing | |||
| 9. Empirical data acquisition | |||
| Focus questions | 1. How are channels formed in microfluidic devices? Describe this process. | 1. Inclusive of module 1 | 1. Inclusive of modules 1 and 2 |
| 2. Can fluid flow through a device if there is only one inlet and no outlet? Why or why not? | 2. How is a gradient formed in the device and why does the fluid not become homogeneous throughout? | 2. Calculate the Reynolds number. Is the flow laminar? | |
| 3. If pressure increases, does flowrate increase or decrease? | 3. Calculate the theoretical flowrate within the device using the channel dimensions. | 3. What characteristic lengthL should be used? Why? | |
| 4. What are the two reasons why microfluidic devices are advantageous over other techniques? | 4. Generate a gradient profile using image processing software and explain why this happens. | 4. Calculate the Stokes number. What is the importance of the Stokes number? | |
| 5. Calculate the area of the channels. What is unique about the dimensions? | 5. How would you determine the concentration at a specific location based on the intensity profile? | 5. Why is a gradient formed using antibodies? | |
| 6. What is high-aspect ratio and why is it important for microfluidics? | 6. What important parameters are necessary to sustain the gradient? | ||
| 7. What is laminar flow and why does it occur at the microlevel? | |||
| 8. For separation, is a faster or slower flowrate desirable? What about for mixing? |
To accommodate the more advanced high school level, module 2 is inclusive of module 1 and will elaborate on the concept of laminar flow as well as demonstrate a chemical gradient using the gradient generator, which plays a crucial role in both biomolecular assays and live cell imaging.48, 49, 50, 51 A gradient can easily be generated with the use of the fluorophore fluorescein or food coloring. Once the students generate a gradient using fluorescein (or food coloring) and de-ionized (DI) water, they will learn image processing software such as with the openware IMAGEJ52 to process their data and produce an intensity graph to show the gradient from the gradient generator device. High school students will also learn to calculate channel dimensions and fluid flowrates relative to microfluidics.
Module 3 challenges the more advanced college students to develop a gradient using the same gradient generator as module 2 and perform a biomolecular assay on-chip. Performing a secondary antibody assay on-chip would mimic the biomolecular assays similar to that of analyte detection and antigen binding to cell receptors. In addition, the concept of dimensionless numbers such as Reynolds number and Stokes number, which plays an important role in the characterization of fluid flow, can be incorporated into the teaching module through simple flow velocity measurements with known device dimensions.45, 51
Together, these modules will teach students fabrication techniques, flow patterns in microfluidics, relevant calculations for microfluidics, and data processing and analysis. Most importantly, these educational modules offer a hands-on experience and cover the breadth of important foundations in microfluidics and applications in real-world POC diagnostics.
MATERIALS AND METHODS
An array of microfluidic designs was drafted in AUTOCAD® (AutoDesk, San Rafael, CA, USA) drafting software, although simpler software may be used, and printed using a standard laser-jet printer (Hewlett Packard CP 2025, USA) [Figs. 1a, 1b].29, 30 For ease of handling, 1 mil thick prestressed polyolefin shrink-film (Sealed Air Nexcel multilayer shrink-film 955D, USA) was laminated onto a 3 mil thick nonshrinkable polyester backing.30, 44 During the printing process, ink was deposited onto this prestressed polyolefin film on the polyester backing. For structural support during the printing process, the polyolefin film and polyester backing should be taped onto a flat sheet of paper [Fig. 1b]. Once the devices are printed onto the polyolefin shrink-film and cut into individual devices [Fig. 1c], using a toaster oven they are heated to 160 °C by a ramping method as previously reported or by a heat gun to shrink the devices [Fig. 1d].30 For a slower, more constant heating method and to produce flatter devices, using a toaster oven is preferred, but using a heat gun to shrink is a rapid alternative that can be performed within seconds. During either of these shrinking techniques, the prestressed polyolefin shrinks while the polyester backing does not. After shrinking, the polyolefin sample can therefore be removed from the polyester backing, and the polyester backing can be discarded. Due to the significant size reduction and increase in aspect ratio of the postshrunk polyolefin device [Fig. 1e], well defined channels can be fabricated with only single prints. PDMS was then molded onto the high-aspect masters to create the devices [Figs. 1f, 1g].
Figure 1.
Fabrication process of printed microfluidics. In this process, an array is drafted in AUTOCAD® drafting software. (a) The design is then printed onto polyolefin polymer shrink-film laminated to polyester backing and taped onto printer paper. (b) The printed design is then cut (c) and shrunken using a heat gun. (d) The shrunken polyolefin with high-aspect microfeatures (e) is then molded with PDMS. (f) The PDMS device is then removed (g) and bonded to a glass slide using an O2 plasma to create a functional device (h).
Prior to assembly, the PDMS devices were punctured at channel ends with hollow metal punches with 0.83 mm inner diameter (ID) and 1.1 mm outer diameter (OD) (Technical Innovations, Arlington, TX, USA) to form inlets and outlets for the fluidics. The PDMS devices can be bonded to a glass slide using Scotch double-sided packaging tape (3M, St. Paul, MN, USA). By placing a smooth, flat layer of tape on a glass slide, the PDMS can be adhered well enough to demonstrate pressure driven flow in a microfluidic device.24, 53 For the purpose of demonstrating particle separations to more advanced high school and college classes, however, it is recommended that the device be bonded to glass using the standard oxygen plasma bonding method (SPI Supplies, West Chester, Pennsylvania, USA) or a more cost effective, hand-held corona discharger (Electro-Technic Products, Inc., Chicago, IL, USA).54 Once the surfaces of the PDMS and glass are exposed to oxygen plasma, or corona discharge, for 20 s, the surfaces become hydrophilic and a bond can form when the surfaces come into contact.
To connect the device to microfluidic tubing, the inlets and outlets were fitted with hollow 15 mm long stainless steel catheters with an ID of 0.49 mm and OD of 0.90 mm (Instech Solomon, Plymouth Meeting, PA, USA). The catheters were then attached to 20 mm sections of adapter tubing with an ID of 0.76 mm and OD of 2.29 mm (Tygon Tubing, Akron, OH, USA). For extension and flexibility, the adapter tubing was fitted with smaller tubing, 0.30 mm ID and 0.76 mm OD (Cole-Parmer, Vermont Hills, IL, USA). The ends of the extension tubing were again inserted into adapter tubing, which was then fitted to Luer stubs (Instech Solomon, Plymouth Meeting, PA, USA). To control the flow of the fluid, four-way stopcocks (Cole-Parmer, Vermont Hills, IL, USA) were attached to the Luer stubs at one end and to 3 mL syringes (Becton Dickinson and Co., Franklin Lakes, NJ, USA) at the other end. Finally, to complete the setup, the 3 mL syringes were mounted onto a microdialysis infusion pump (KD Scientific, Holliston, MA, USA). This microdialysis infusion pump was used to control the pressure and flowrate of fluid in the microfluidic devices. To visualize the pressure driven flow, food coloring can be flowed through the channels of the microfluidic device, as shown in Fig. 1h. Both the H-filter and the gradient generator devices were fabricated using this method.
To perform the diffusion and separation experiments, the H-filter device was drafted using AUTOCAD® with an initial width of 1 mm for the primary channel and 0.5 mm for the inlet and outlet channels. Upon shrinking, the channel size was reduced to 95% in-plane of the device with a final width of 250±26 μm for the primary channel and 200±20 μm for the inlet and outlet channels. During soft lithography, the H-filter channels yielded a height of 100±10 μm in the primary channel and 80±3 μm for the inlet and outlet channels. Two different size fluorescent beads, 0.5 and 25 μm, were dissolved in DI water to perform the H-filter experiment. One inlet was loaded with a mixture of the different size fluorescent beads and the other inlet was filled with DI water. Solutions were flowed at a constant flowrate of 30 μL∕h through both inlets. The beads separated during the flow process, and the exiting streams were collected to determine the size separation principle. Undergraduate students can image the beads using a fluorescence microscope to observe the separation layer and visualize the separation of smaller from larger particles. The flowrate can also be calculated from the channel dimensions.
For the gradient generator experiments, a gradient device design must be produced in AUTOCAD® and fabricated via the above process. All CAD designs are included as downloadable files in the Appendix for students to print out directly, but it is recommended that students familiarize themselves with the design and software using a CAD software program. In the device, a gradient was first developed by flowing fluorescein in one inlet and DI water in the other inlet at 5 μl∕h through the gradient generator device to demonstrate the effects of laminar flow and viscous forces. For the visualization of fluorescein within each distinct channel, a low flowrate of 5 μl∕h is recommended as to prevent the generation of discrete concentration bands within each of the five distinct channels as a result of inefficient mixing. A gradient is therefore formed only in between the five outlet channels (instead of within each of the channels as well). (Food coloring can be used as a substitute for fluorescein.) To perform an immunoassay, 100 μg∕ml of primary rabbit IgG (Invitrogen, Carlsbad, California, USA) dissolved in 1x phosphate buffered saline (PBS) was introduced into both inlets at a flowrate of 30 μL∕h. Once all the channels were filled, the flow was stopped and the device was incubated at room temperature for 2 h to allow binding. Next, channels were then washed with 1xPBS and filled with 1 mg∕ml of bovine serum albumin blocking solution for 1 h to prevent nonspecific binding of secondary antibodies.44 It should be noted that for the loading of primary antibody and rinsing steps, generation of a gradient was not necessary. For the loading of secondary antibody, channels were then filled as a gradient with one inlet containing 50 μg∕ml secondary antirabbit IgG conjugated fluorophore and the other inlet containing 1xPBS at a steady flowrate of 30 μL∕h. In this procedure, the high flowrate of 30 μL∕h minimizes the chances of backflow and upstream mixing that might induce gradients in the five channels. Once the gradient was formed in the outlet channels, the flow was stopped by plugging the inlets and outlets. Note that no mixing occurred during the plugging process between distinct channels. The gradient was then incubated for 2 h at room temperature. Once the incubation process was finished, the channels were rinsed with 1xPBS.
If time is constrained for the classroom laboratory session, a separate set of devices can be prepared by incubating overnight with primary antibody. This would allow the students to load secondary antibody on a separate set of chips on the same day as loading the primary antibody by eliminating the incubation time. Thus, by staggering the experimental procedure, students can learn to load samples at different stages, and observe key concepts of each step such as the loading of primary antibody. For image analysis, images of both the fluorescein and the antibody gradients were captured in gray scale and analyzed using the imaging software QCAPTURE PRO® (QImaging, Surrey, BC, Canada). A gradient profile was generated as an intensity curve using IMAGEJ software. A detailed materials list as well as the microfluidic designs is available in the supplemental information section.
MODULE 1: SEPARATION AND DIFFUSION
While LOC technology offers many advantages to POC diagnostics, it also presents several challenges due to the physics at the microscale. To address such issues, designs that leverage the properties of laminar flow and diffusive properties of particles have been developed to mimic standard procedures. Considered to be an alternative to centrifugation, the H-filter is a nonmembrane based microfabricated device which utilizes the unique flow property at low Reynolds number to separate particles based on size. In processes such as the purification of plasma from blood or the separation of analytes in solution, the H-filter offers a simple on-chip platform which can perform these preconditioning steps.49 The design was pioneered by Brody and Yager and was based off the original split-flow-thin fractionation device reported by Giddings et al.55 The H-filter utilizes the diffusion dominant mixing within microfluidic devices to sort particulates based on size corresponding to their diffusive properties.47, 55, 56, 57
In fluid mechanics, the Reynolds number is a dimensionless number which essentially defines the ratio of inertial force to viscous force for a given flow condition. Thus, Reynolds number is an important parameter in assessing the relative importance of the two forces and is defined as
| (1) |
where ρf is the fluid density, L is the characteristic length scale, νav is the average velocity of the fluid, and μ is the viscosity. The Reynolds number in microfluidic devices is typically <1 and therefore laminar.
Stokes number describes the interaction of the particle to its surrounding flow field and is given by the ratio of the particle relaxation time to the characteristic time of the fluid. A Stokes number⪡1 indicates that the motion of the particle is coupled to that of the flow field and will follow the fluid motion, while a Stokes number⪢1 indicates that the motion of the particle is decoupled from the flow field, and thus its movement is less responsive to the flow regime. Stokes number is defined as
| (2) |
where τr is the particle relaxation time and τf is the characteristic time, and τr and τf can be respectively defined as
| (3) |
| (4) |
where ρp is the particle density, dp is the particle diameter, μ the fluid viscosity, L the characteristic length scale, and νav is the average velocity of the fluid.
Due to the small length scale, the Reynolds number is low, and therefore the flow within the microchannels is laminar at these flow conditions. H-filters separate particles based on the principle of the nonmixing property of laminar flow along with the relative diffusivities of different size molecules.47, 48, 51, 55, 56, 57 To demonstrate separation, crucial in the miniaturization and adaptation of microfluidics to LOC technology and POC diagnostics, fluorescent microbeads of 0.5 and 25 μm diameter were mixed with water and introduced to one inlet of the device. Next, pure DI water was introduced into the other inlet, and a constant flowrate of 30 μL∕h was applied to both inlets. By collecting the outlet streams, the composition of the exiting streams can be observed, and the separation of smaller particles from the larger ones can clearly be seen, as shown in Fig. 2b. In the case where fluorescent microscopy is unavailable, this method can be used to demonstrate the size separation at the outlet. In a teaching laboratory setting where a fluorescence microscope is available, a clear separation layer can be observed, and the separation of smaller particles from the larger ones can clearly be seen, as shown in Fig. 2b. Based upon the size-dependent diffusive properties of the particles and the inherent limitation of mixing in laminar flow, the concept of separation was demonstrated. In addition to the questions outlined in Table 1, students are encouraged to “play around” with the modules to provide flexibility. For example, they are encouraged to put the two different sized particles into separate channels and observe and comment on what happens.
Figure 2.
Particle separation using H-filter. (a) An H-filter design was printed onto polyolefin thin film. Scale bar is 100 μm. (b) To demonstrate the separation of particles based on size differences, a mixture of 25 and 0.5 μm beads was flowed in at one inlet, while water is flowed into the other inlet. As a result of diffusion, the 0.5 μm beads are seen in both outlets, while the 25 μm beads are only seen in one outlet.
MODULE 2: GRADIENT GENERATION
Gradients play an important role in many chemical and biological processes such as chemotaxis, crystal growth, morphogenesis, and nerve growth cone guidance.58, 59 Specifically, in chemotaxis, chemical gradients play an essential role in wound inflammation, wound healing, and cancer metastasis by initiating cell recruitment and migration.50, 60, 61, 62In vivo, biomolecular concentration gradients can be observed through the growth, differentiation, and migration of various cell types. Much effort has been taken to mimic these gradients in vitro for studying the response of cells to chemical cues. Assays such as the Boyden chamber, Zigmond chamber, Dunn chamber, and the micropipette are used to study the phenomenon of chemotaxis.61, 62, 63 While all these assays play an important role in elucidating the mechanisms of chemotaxis, several challenges remain such as the inability to maintain a chemical gradient over long periods of time and consequently observe real-time cell migration.50, 62 In addition, the gradient generator has also been adapted for the immunological assays as a fluorescent-based detection method in biological assays.64, 65
Microfluidics has emerged as one of the powerful technologies that allows one to manipulate the physiological and chemical environment at very small spatial scales without the use of excessive amount of reagents.50, 62 Jeon et al. and Dertinger et al. demonstrated the use of a microfluidic gradient generator from a pyramidal branched array of microchannels that repetitively mix and split solutions of different concentrations.50, 66 One of the advantages of using a microfluidic gradient generator over the conventional methods is the ability to tune and sustain gradients to create different concentration profiles (linear, ramp, parabolic). Furthermore, the ability to maintain a constant linear gradient for a long time-period (or while at steady-state) in real-time has also been of much interest.50, 61
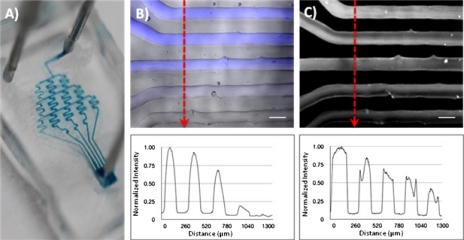
In modules 2 and 3, the gradient generator design was used to demonstrate both the chemical gradient within microfluidic channels using the branching tree design as well as the application of the gradient generator for an immunological assay. In module 2, the gradient generator is tailored for high school students and simple food coloring or the fluorescent dye fluorescein can be used. This design of branching and mixing channels will be suitable for the generation of a chemical gradient and will facilitate the adaptation for the more advanced immunological assay in module 3. With the flow of the blue and yellow food coloring at a steady-state, the visualization of mixing in a gradient can be done with the naked eye or with a microscope. With the availability of a fluorescent microscope in most college level academic settings, stable gradients can be seen through the use of fluorescein [Fig. 3b]. A profile can also be acquired through a gray scale image, as shown in Fig. 3b. This process will allow for students the ability to observe and verify the generation of a stable chemical gradient and to generate quantitative data as well. In addition to the focus questions outlined in Table 1, students are encouraged to try mixing different colored dyes at different flowrates and examining the results.
Figure 3.
Applications of gradient generator. (a) shows food coloring gradient for visual reference. Generation of gradient can be demonstrated with fluorescein (pseudocolored blue) and normalized concentration profile can be obtained through the plot of the gray scale intensity. (b) For demonstration of an immunoassay, antibody detection can be performed on-chip to yield a dose-dependent assay with corresponding gray scale intensity profile. (c) All scale bars are 200 μm.
MODULE 3: IMMUNOASSAY
In a recent report by Chon et al., a similar branching tree gradient generator with separate channels was utilized for on-chip Raman spectroscopy.64 By generating a gradient of hollow gold nanosphere conjugated antibodies, complementary antigen conjugated magnetic beads were flowed in forming a gradient representing a dose-dependent expression of surface-enhanced Raman spectroscopy. To demonstrate the use of gradient generators for immunological assays, the same design from module 2 can be adapted for the use of a fluorescent antibody assay. In module 3, primary rabbit IgG was flowed into the device through both inlets of the device. To generate the antibody fluorescent gradient, secondary goat-antirabbit IgG fluorophore conjugated antibody was flowed into one inlet of the channel, while PBS was flowed into the other. The flow then undergoes splitting and mixing so that a gradient develops where secondary antibody is present. Fluorescent images were taken of the channels near the outlet where the gradient is formed [Fig. 3c]. In addition, by utilizing the gray scale of the fluorescent images, a gradient profile of the assay was generated [Fig. 3c]. Although the immunoassay was performed by utilizing the binding affinity of primary and secondary antibodies, this concept was analogous to the generation of defined chemical gradients in the chemotaxis assay.50, 61, 62, 66 The students are encouraged to think about the gradient profile generated from the stationary immunoassay experiment (module 3) versus the sustained gradient generation (module 2) and explain any discrepancies in gradient profile.
CONCLUSION
By leveraging our ultrarapid prototyping method on a commercially available shrink-film substrate, we have demonstrated the ease of introducing microfluidic educational modules at the middle school, high school, and college level. With the growing relevance of LOC technology, this prototyping method will allow for the rapid, low-cost production of microfluidic devices that can be utilized in various levels of academic institutions. Since each educational module is inclusive of the previous educational content, this comprehensive teaching package can demonstrate key concepts in and applications of microfluidics in various educational levels. The flexibility of this platform enables students to design and develop their own custom chips on demand as well.
ACKNOWLEDGMENTS
This work was supported in part by the National Institutes of Health through the NIH Director’s New Innovator Award Program (Grant No. 1DP2OD007283), the DARPA MF3 Center, and Shrink Nanotechnologies, Inc. The author (M.K.) is the scientific founder of Shrink but receives no compensation nor does have any financial interest in the company. Terms of this arrangement have been reviewed and approved by UC Irvine in accordance with its conflict of interest policies. We would like to thank Himanshu Sharma for his help.
References
- Arora A., Simone G., Salieb-Beugelaar G. B., Kim J. T., and Manz A., Anal. Chem. 82, 4830 (2010). 10.1021/ac100969k [DOI] [PubMed] [Google Scholar]
- Novak L., Neuzil P., Pipper J., Zhang Y., and Lee S., Lab Chip 7, 27 (2007). 10.1039/b611745g [DOI] [PubMed] [Google Scholar]
- Wood D. K., Weingeist D. M., Bhatia S. N., and Engelward B. P., Proc. Natl. Acad. Sci. U.S.A. 107, 10008 (2010). 10.1073/pnas.1004056107 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Monaghan P. B., McCarney K. M., Ricketts A., Littleford R. E., Docherty F., Smith W. E., Graham D., and Cooper J. M., Anal. Chem. 79, 2844 (2007). 10.1021/ac061769i [DOI] [PubMed] [Google Scholar]
- Chen X., Cui D. F., and Liu C. C., Electrophoresis 29, 1844 (2008). 10.1002/elps.200700551 [DOI] [PubMed] [Google Scholar]
- Sharma H., Nguyen D., Chen A., Lew V., and Khine M., Ann. Biomed. Eng. 39, 1313 (2011). 10.1007/s10439-010-0213-1 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chován T. and Guttman A., Trends Biotechnol. 20, 116 (2002). 10.1016/S0167-7799(02)01905-4 [DOI] [PubMed] [Google Scholar]
- Betancourt T. and Brannon-Peppas L., Int. J. Nanomedicine 1, 483 (2006). 10.2147/nano.2006.1.4.483 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dittrich P. S. and Manz A., Nat. Rev. Drug Discovery 5, 210 (2006). 10.1038/nrd1985 [DOI] [PubMed] [Google Scholar]
- Borenstein J. T., Weinberg E. J., Orrick B. K., Sundback C., Kaazempur-Mofrad M. R., and Vacanti J. P., Tissue Eng. 13, 1837 (2007). 10.1089/ten.2006.0156 [DOI] [PubMed] [Google Scholar]
- Zare R. N. and Kim S., Annu. Rev. Biomed. Eng. 12, 187 (2010). 10.1146/annurev-bioeng-070909-105238 [DOI] [PubMed] [Google Scholar]
- Khine M., Lau A., Ionescu-Zanetti C., Seo J., and Lee L. P., Lab Chip 5, 38 (2005). 10.1039/b408352k [DOI] [PubMed] [Google Scholar]
- Liu Y., Yang D., Yu T., and Jiang X., Electrophoresis 30, 3269 (2009). 10.1002/elps.200900128 [DOI] [PubMed] [Google Scholar]
- Xiang Q., Xu B., and Li D., Biomed. Microdevices 9, 443 (2007). 10.1007/s10544-007-9048-4 [DOI] [PubMed] [Google Scholar]
- Lee D. -S., Park S. H., Yang H., Chung K. -H., Yoon T. H., Kim S. -J., Kim K., and Kim Y. T., Lab Chip 4, 401 (2004). 10.1039/b313547k [DOI] [PubMed] [Google Scholar]
- Lee D. -S., Wu M. -H., Ramesh U., Lin C. -W., Lee T. -M., and Chen P. -H., Sens. Actuators B 100, 401 (2004). 10.1016/j.snb.2004.02.012 [DOI] [Google Scholar]
- Bhattacharyya A. and Klapperich C., Biomed. Microdevices 9, 245 (2007). 10.1007/s10544-006-9026-2 [DOI] [PubMed] [Google Scholar]
- Carlo D. D., Wu L. Y., and Lee L. P., Lab Chip 6, 1445 (2006). 10.1039/b605937f [DOI] [PubMed] [Google Scholar]
- Geissler M., Roy E., Diaz-Quijada G. A., Galas J. -C., and Veres T., ACS Applied Materials & Interfaces 1, 1387 (2009). 10.1021/am900285g [DOI] [PubMed] [Google Scholar]
- Geissler M. and Xia Y., Adv. Mater. (Weinheim, Ger.) 16, 1249 (2004). 10.1002/adma.200400835 [DOI] [Google Scholar]
- Chin C. D., Linder V., and Sia S. K., Lab Chip 7, 41 (2007). 10.1039/b611455e [DOI] [PubMed] [Google Scholar]
- Duffy D. C., McDonald J. C., Schueller O. J. A., and Whitesides G. M., Anal. Chem. 70, 4974 (1998). 10.1021/ac980656z [DOI] [PubMed] [Google Scholar]
- Li X., Tian J., and Shen W., Cellulose 17, 649 (2010). 10.1007/s10570-010-9401-2 [DOI] [Google Scholar]
- Martinez A. W., Phillips S. T., and Whitesides G. M., Proc. Natl. Acad. Sci. U.S.A. 105, 19606 (2008). 10.1073/pnas.0810903105 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cheng C. M., Martinez A. W., Gong J., Mace C. R., Phillips S. T., Carrilho E., Mirica K. A., and Whitesides G. M., Angew. Chem., Int. Ed. Engl. 49, 4771 (2010). [DOI] [PubMed] [Google Scholar]
- Coltro W. K. T., de Jesus D. P., da Silva J. A. F., do Lago C. L., and Carrilho E., Electrophoresis 31, 2487 (2010). 10.1002/elps.201000063 [DOI] [PubMed] [Google Scholar]
- Li X., Tian J., and Shen W., ACS Appl. Mater. Interfaces 2, 1 (2010). 10.1021/am9006148 [DOI] [PubMed] [Google Scholar]
- Reches M., Mirica K. A., Dasgupta R., Dickey M. D., Butte M. J., and Whitesides G. M., ACS Appl. Mater. Interfaces 2, 1722 (2010). 10.1021/am1002266 [DOI] [PubMed] [Google Scholar]
- Grimes A., Breslauer D. N., Long M., Pegan J., Lee L. P., and Khine M., Lab Chip 8, 170 (2008). 10.1039/b711622e [DOI] [PubMed] [Google Scholar]
- Nguyen D., Taylor D., Qian K., Norouzi N., Rasmussen J., Botzet S., Lehmann M., Halverson K., and Khine M., Lab Chip 10, 1623 (2010). 10.1039/c001082k [DOI] [PubMed] [Google Scholar]
- Carrilho E., Martinez A. W., and Whitesides G. M., Anal. Chem. 81, 7091 (2009). 10.1021/ac901071p [DOI] [PubMed] [Google Scholar]
- Focke M., Kosse D., Muller C., Reinecke H., Zengerle R., and von Stetten F., Lab Chip 10, 1365 (2010). 10.1039/c001195a [DOI] [PubMed] [Google Scholar]
- Lu Y., Shi W., Jiang L., Qin J., and Lin B., Electrophoresis 30, 1497 (2009). 10.1002/elps.200800563 [DOI] [PubMed] [Google Scholar]
- Lu Y., Shi W., Qin J., and Lin B., Anal. Chem. 82, 329 (2010). 10.1021/ac9020193 [DOI] [PubMed] [Google Scholar]
- Cheung Y. K., Gillette B. M., Zhong M., Ramcharan S., and Sia S. K., Lab Chip 7, 574 (2007). 10.1039/b700869d [DOI] [PubMed] [Google Scholar]
- Wang W., Zhao S., and Pan T., Lab Chip 9, 1133 (2009). 10.1039/b816287e [DOI] [PubMed] [Google Scholar]
- Thomas M. S., Millare B., Clift J. M., Bao D., Hong C., and Vullev V. I., Ann. Biomed. Eng. 38, 21 (2010). 10.1007/s10439-009-9831-x [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yang C. W. T., Ouellet E., and Lagally E. T., Anal. Chem. 82, 5408 (2010). 10.1021/ac902926x [DOI] [PubMed] [Google Scholar]
- Yuen P. K. and Goral V. N., Lab Chip 10, 384 (2010). 10.1039/b918089c [DOI] [PubMed] [Google Scholar]
- Berkowski K. L., Plunkett K. N., Yu Q., and Moore J. S., J. Chem. Educ. 82, 1365 (2005). 10.1021/ed082p1365 [DOI] [Google Scholar]
- Yuen P. K., Lab Chip 8, 1374 (2008). 10.1039/b805086d [DOI] [PubMed] [Google Scholar]
- Nguyen D., Sa S., Pegan J. D., Rich B., Xiang G. X., McCloskey K. E., Manilay J. O., and Khine M., Lab Chip 9, 3338 (2009). 10.1039/b914091c [DOI] [PubMed] [Google Scholar]
- Chen C. S., Breslauer D. N., Luna J. I., Grimes A., Chin W. C., Lee L. P., and Khine M., Lab Chip 8, 622 (2008). 10.1039/b719029h [DOI] [PubMed] [Google Scholar]
- Taylor D., Dyer D., Lew V., and Khine M., Lab Chip 10, 2472 (2010). 10.1039/c004737f [DOI] [PubMed] [Google Scholar]
- Young E. W. K. and Simmons C. A., Chem. Eng. Educ. 43, 232 (2009). [Google Scholar]
- Brody J. P., Osborn T. D., Forster F. K., and Yager P., Sens. Actuators, A 54, 704 (1996). 10.1016/S0924-4247(97)80042-8 [DOI] [Google Scholar]
- Brody J. P. and Yager P., Sens. Actuators, A 58, 13 (1997). 10.1016/S0924-4247(97)80219-1 [DOI] [Google Scholar]
- Brody J. P., Yager P., Goldstein R. E., and Austin R. H., Biophys. J. 71, 3430 (1996). 10.1016/S0006-3495(96)79538-3 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yager P., Edwards T., Fu E., Helton K., Nelson K., Tam M. R., and Weigl B. H., Nature (London) 442, 412 (2006). 10.1038/nature05064 [DOI] [PubMed] [Google Scholar]
- Jeon N. L., Baskaran H., Dertinger S. K. W., Whitesides G. M., Van De Water L., and Toner M., Nat. Biotechnol. 20, 826 (2002). [DOI] [PubMed] [Google Scholar]
- Zhang Y. W., Barber R. W., and Emerson D. R., Current Analytical Chemistry 1, 345 (2005). 10.2174/157341105774573983 [DOI] [Google Scholar]
- Abramoff M. D., Magelhaes P., and Ram S. J., Biophotonics Int. 11, 36 (2004). [Google Scholar]
- Kim J., Surapaneni R., and Gale B. K., Lab Chip 9, 1290 (2009). 10.1039/b818389a [DOI] [PubMed] [Google Scholar]
- Haubert K., Drier T., and Beebe D., Lab Chip 6, 1548 (2006). 10.1039/b610567j [DOI] [PubMed] [Google Scholar]
- Giddings J. C., Sep. Sci. Technol. 20, 749 (1985). 10.1080/01496398508060702 [DOI] [Google Scholar]
- Giddings J. C., Science 260, 1456 (1993). 10.1126/science.8502990 [DOI] [PubMed] [Google Scholar]
- Giddings J. C., Yang F. J., and Myers M. N., Science 193, 1244 (1976). 10.1126/science.959835 [DOI] [PubMed] [Google Scholar]
- Weibel D. B. and Whitesides G. M., Curr. Opin. Chem. Biol. 10, 584 (2006). 10.1016/j.cbpa.2006.10.016 [DOI] [PubMed] [Google Scholar]
- Wu H., Huang B., and Zare R. N., J. Am. Chem. Soc. 128, 4194 (2006). 10.1021/ja058530o [DOI] [PubMed] [Google Scholar]
- Gorman B. R. and Wikswo J. P., Microfluid. Nanofluid. 4, 273 (2008). 10.1007/s10404-007-0169-0 [DOI] [Google Scholar]
- Keenan T. M. and Folch A., Lab Chip 8, 34 (2008). 10.1039/b711887b [DOI] [PMC free article] [PubMed] [Google Scholar]
- Saadi W., Rhee S., Lin F., Vahidi B., Chung B., and Jeon N., Biomed. Microdevices 9, 627 (2007). 10.1007/s10544-007-9051-9 [DOI] [PubMed] [Google Scholar]
- Heit B. and Kubes P., Sci. STKE 2003, Issue 170, l5 (2003). 10.1126/stke.2003.170.pl5 [DOI] [PubMed] [Google Scholar]
- Chon H., Lim C., Ha S. -M., Ahn Y., Lee E. K., Chang S. -I., Seong G. H., and Choo J., Anal. Chem. 82, 5290 (2010). 10.1021/ac100736t [DOI] [PubMed] [Google Scholar]
- Jiang X., Ng J. M., Stroock A. D., Dertinger S. K., and Whitesides G. M., J. Am. Chem. Soc. 125, 5294 (2003). 10.1021/ja034566+ [DOI] [PubMed] [Google Scholar]
- Dertinger S. K. W., Chiu D. T., Jeon N. L., and Whitesides G. M., Anal. Chem. 73, 1240 (2001). 10.1021/ac001132d [DOI] [Google Scholar]