Abstract
Due to a close genetic relatedness, there is no known antibody that detects Mycobacterium avium subspecies paratuberculosis (MAP), which causes Johne’s disease in cattle and sheep, and does not cross-react with other M. avium subspecies. In the present study, a monoclonal antibody (MAb; 17A12) was identified from mice immunized with a cell membrane fraction of MAP strain K-10. This antibody is 100% specific as it detected a 25-kDa protein in all 29 MAP whole cell lysates, but did not bind to any of the 29 non-paratuberculosis strains tested in immunoblot assays. However, the antibody revealed variable reactivity levels in MAP strains as it detected higher levels in bovine isolates but comparably lower levels in ovine isolates of MAP. In order to identify the target binding protein for 17A12, a lambda phage expression library of MAP genomic fragments was screened with the MAb. Four reactive clones were identified, sequenced and all shown to be overlapping. Further analysis revealed all four clones expressed an unknown protein encoded by a sequence that is not annotated in the K-10 genome and overlapped with MAP3422c on the opposing DNA strand. The epitope of 17A12 was precisely defined to seven amino acids and was used to query the K-10 genome. Similarity searches revealed another protein, encoded by MAP1025, possessed a similar epitope (one-amino acid mismatch) that also reacted strongly to the antibody. A single nucleotide polymorphism (SNP) in MAP1025 was then identified by comparative sequence analysis, which results in a Pro28His change at residue 28, the first amino acid within the 17A12 epitope. This SNP is present in all MAP strains but absent in all non-MAP strains and accounts for the specificity of the 17A12 antibody. This new antibody is the first ever isolated that binds only to the paratuberculosis subspecies of M. avium and opens new possibilities for the specific detection of this significant ruminant pathogen.
Keywords: Mycobacterium paratuberculosis, Johne’s disease, antigens, antibodies, detection and diagnostics
Introduction
There are inherent diagnostic difficulties when a bacterial pathogen is closely related to ubiquitous environmental microorganisms. In these situations, it is difficult to detect the pathogen, but not the environmental bacteria, which would lead to false positive results. Yet this is the case for Mycobacterium avium subsp. paratuberculosis (hereafter referred to as MAP), a veterinary pathogen that causes Johne’s disease in cattle, sheep, and other ruminants (Harris and Barletta, 2001). It belongs to a group of closely related mycobacteria that comprise the Mycobacterium avium complex (MAC), and the remaining members of this complex play the role of environmental contaminants in a veterinary context. The MAC group historically has consisted of Mycobacterium intracellulare and all M. avium subspecies, including avium, hominissuis, paratuberculosis, and silvaticum (Turenne et al., 2007); however, recently species have been added to this complex based on multispacer sequence typing analysis (Cayrou et al., 2010). The M. avium subspecies in particular are closely related as determined long ago by DNA–DNA hybridization studies (Yoshimura and Graham, 1988) which led to the initial proposal to include them as an avium subspecies (Thorel et al., 1990). More recent genome scale studies have demonstrated that hominissuis and paratuberculosis subspecies share greater than 98% genetic similarity among the sequenced strains (Bannantine et al., 2003) and show only a few small differences by competitive genomic DNA hybridizations to microarrays (Paustian et al., 2008).
Methods that include subtractive hybridization and comparative genomics enable identification of sequences specific to a particular bacterium under study. For MAP, these approaches have identified large sequence polymorphisms (LSPs), which appear to represent the main source of genomic diversity. These LSPs and their evolutionary implications are nicely summarized by Marcel Behr and co-workers (Alexander et al., 2009). Single nucleotide polymorphisms (SNPs) and nucleotide repeats have also contributed to the genomic diversity and have resulted in excellent strain typing methods (Bull et al., 2003; Amonsin et al., 2004; Sevilla et al., 2008; Thibault et al., 2008; Castellanos et al., 2009). Thus far, these differences in the DNA sequences of MAC organisms have yet to fully explain the phenotypic differences inherent in each MAC member.
While a monoclonal antibody (MAb) specific to M. avium subsp. avium has long been identified that does not cross-react with MAP (Abe et al., 1989), no such antibodies specific to only MAP have been described. This is to be expected because the MAP genome, at 4.8 Mb, is believed to be the smallest in the MAC group and therefore specific gene targets are more likely available in the other subspecies with larger genomes and thus more coding potential. In spite of this, MAbs were previously developed against a whole cell extract of MAP in an effort to obtain a specific detection reagent. When positive hybridomas secreting MAbs were screened against mycobacterial species in specificity studies, nearly all cross-reacted with M. avium subspecies isolates (Bannantine et al., 2007b). The one antibody that did not, 14D4, surprisingly cross-reacted with more distantly related mycobacterial species such as M. phlei, M. kansasii, and M. bovis. The protein target that 14D4 binds to remains unknown.
An alternative strategy to obtain a specific antibody is to search the LSP regions in MAP for genes that encode proteins with high predicted antigenicities and then make antibodies to recombinant proteins representing those gene products. However, there are only a total of 32 genes that qualify using this criteria (Paustian et al., 2010) and in most instances when a MAb can be obtained, it reacts well to the recombinant protein, but does not react with the native protein produced by MAP (Bannantine, unpublished observations). Therefore, it is ideal to start with the native protein when screening for such antibodies.
It has been shown previously that a surface extraction of MAP has increased specificity and can be used to distinguish this pathogen from other MAC members (Eda et al., 2006). Therefore, we hypothesized that similar surface protein extracts might contain specific components. We prepared a membrane extraction of MAP and used that as antigen for MAb production. We discovered a specific antibody that binds to a protein encoded by MAP1025. Although the protein encoded by this gene is not specific to MAP, sequence analysis revealed a SNP, present only in MAP strains, that alters the epitope.
Materials and Methods
Bacterial strains
A total of 58 Mycobacterium species and strains were used in this study. These include several members of the MAC group and TB complex as well as some saprophytic mycobacteria. They are listed in Table A1 in Appendix. Escherichia coli strains used for cloning and expression are described previously (Bannantine et al., 2010).
Production of monoclonal antibodies
Monoclonal antibodies were produced using standard methods (Harlow and Lane, 1988). Briefly, BALB/c mice were immunized three times intraperitoneally with a membrane-enriched protein extract of MAP K-10 (100 μg per injection) suspended in 0.5 mL of phosphate-buffered saline (PBS; 150 mM NaCl, 10 mM NaPO4, pH 7.2) at 14-day intervals. The membrane-enriched extract was prepared as described previously (Radosevich et al., 2007) and emulsified in Freund’s incomplete adjuvant (Sigma-Aldrich, St. Louis, MO, USA) for all immunizations. Humoral immune responses of each mouse were evaluated by preparative immunoblot analysis using the membrane-enriched extract. Cell fusions with splenic lymphocytes and myeloma cells were performed on the best responder mouse. Positive antibody secreting hybridomas were identified by immunoblot screening with culture supernatant. The 17A12 antibody was immunotyped using isotype kit I from Thermo Scientific (Rockford, IL, USA). The same procedure was subsequently used to generate MAbs against the purified recombinant target protein (termed UP1 for unknown protein 1) expressed from clone #23 (Table A2 in Appendix), which generated MAb 10D11 along with 13 other MAbs.
Production of recombinant protein
The full length MAP1025, MAP3422c, and truncated UP1 fragments were constructed in the pMAL-c2 expression vector as previously described (Bannantine et al., 2010). These constructs were transformed and expressed as previously described (Bannantine et al., 2010). The primers used to construct these clones are listed in Table A2 in Appendix.
Lambda ZAP expression library screening
A MAP strain ATCC19698 expression library was constructed in the lambda ZAP phage vector (Agilent technologies-Stratagene, La Jolla, CA, USA) using size selected fragments in the 3–6 kb range as described previously (Bannantine and Stabel, 2001). The recombinant phage were seeded on lawns of E. coli XL1-Blue MRF’ cultured on 150-mm Petri plates containing NZY media made with agarose as the solidifying agent according to the manufacturer’s guidelines. The phage were diluted and plated on E. coli XL1-Blue such that approximately 600–700 plaques per plate were obtained. After plaque formation was barely visible, the plate was overlaid with 0.01 M IPTG-coated Protran® nitrocellulose filters (Sigma-Aldrich) and allowed to incubate for an additional hour. Filters containing the lifted plaques were placed in blocking solution consisting of PBS with 2% bovine serum albumin (BSA; PBS–BSA) overnight and screened with the 17A12 antibody (diluted 1:300 in blocking solution) the following day. Positive plaques were picked and processed according to the manufacturer’s guidelines (Agilent technologies-Stratagene). Subcloning of phage inserts into the pBK-CMV vector for sequencing was also performed according to the manufacturer’s guidelines.
Epitope mapping of 17A12 and other monoclonal antibodies
To localize the epitope of MAbs obtained in this study, several truncated fragments of the target protein (termed UP1) were constructed and expressed in pMAL-c2 similar to that described before for MAP1242 (Wu et al., 2009). High-resolution epitope mapping was preformed using a spot array from JPT peptide (Berlin, Germany). Ten peptides were synthesized directly on a cellulose-β-alanine membrane (5 nmol per peptide spot). The peptides that reacted with MAbs 17A12 and 10D11 were identified by standard immunoblot procedures as described immediately below. Bound antibody was completely removed between experiments using the regeneration protocol I described by the manufacturer (JPT peptide). The membrane was exposed to film for a protracted period of time between experiments to confirm no bound antibody remained.
Electrophoresis, immunoblot, and preparative immunoblot assays
Sodium dodecyl sulfate-polyacrylamide gel electrophoresis (SDS-PAGE) was performed using 12% (w/v) polyacrylamide gels. Electrophoretic transfer of proteins onto pure nitrocellulose was accomplished with the Bio-Rad Trans Blot Cell (Bio-Rad Laboratories, Richmond, CA, USA) with sodium phosphate buffer (25 mM, pH 7.8) at 0.8 A for 90 min. After transfer, filters were blocked with PBS–BSA and 0.1% Tween 20, termed PBS–BSA–T. Culture supernatants containing MAbs were diluted in PBS–BSA and exposed to the blot at room temperature for 2 h. After three washes in PBS–BSA–T, blots were incubated for 1.5 h in goat anti-mouse-peroxidase (Thermo Scientific) diluted 1:20,000 in PBS–BSA. Nitrocellulose blots were again washed three times as described above and developed for chemiluminescence using SuperSignal detection reagents (Thermo Scientific).
Confocal microscopy of M. avium subspecies infected monocyte-derived macrophages (MDM) and MAC-T cells
Bovine MDMs and MAC-T cells were seeded separately at a concentration of 2.0 × 104 cells/mL in a 24-well plate containing No. 1.5 thickness glass coverslips. All cells were incubated at 37°C in a humidified chamber containing 5% CO2 until confluent. Prior to cell infection, MAH 7337 was stained with 0.25 μg/mL of 6-carboxyfluorescein diacetate (Sigma-Aldrich, St. Louis, MO, USA) for 1 h at 37°C and immediately washed 3× with PBS. MAP K-10 (pWes4) GFP expression strain and MAH 7337 infection of bovine MDMs and MAC-T cells were conducted in a similar fashion as described above with the exception of using phenol red free media to prevent fluorescence quenching in MAH invasion. All time points were conducted in triplicate. For immunostaining, cells were washed 3× with PBS at defined time points and fixed using 2% paraformaldehyde at 37°C for 5 min. Cells were immediately washed 2× with PBS containing 1% BSA and permeabilized with ice-cold methanol for 5 min at −20°C. Next cells were blocked with PBS containing 1% BSA for 1 h at room temperature, washed twice with PBS and incubated with 1:300 dilution of either 17A12 or 8G2 MAbs in PBS–Tween 20 overnight at 4°C. After incubation, cells were washed twice with PBS containing 1% BSA, incubated with goat anti-mouse IgG conjugated to Alexa Fluor 350 (1:500; Invitrogen, Carlsbad, CA, USA) for 1 h at room temperature in the dark, rewashed, and counter-stained with CellMask Deep Red plasma membrane stain (2.5 μg/mL; Invitrogen, Carlsbad, CA, USA). A final wash step was conducted and coverslips were mounted on glass slides using prolong gold anti-fade reagent (Invitrogen, Carlsbad, CA, USA). Coverslips were sealed using nail polish and stored at 4°C until visualization. All slides were imaged using the Olympus Fluoview upright confocal microscope and software (Olympus, Center Valley, PA, USA). Slide images were taken using the following lasers: Alexa Fluor 405 or DAPI, FITC, and Cy5. Z-series was collated for all images using a 1.0 μm step size and a Kalman average of 2 acquisitions. Three fields per slide were recorded.
Densitometry analysis and statistics
Protein levels were indirectly measured by antibody binding on immunoblots. Chemiluminescent images were captured on Kodak BioMax MR film and scanned to obtain a digital image. Reactive bands within the images were then analyzed by densitometry using Photoshop CS5 extended software’s measurement tool. Protein levels in ovine and bovine strains were evaluated by unpaired t test. The results are reported as a P value where <0.05 is considered significant.
Results
Monoclonal antibody 17A12 is specific to M. avium subsp. Paratuberculosis
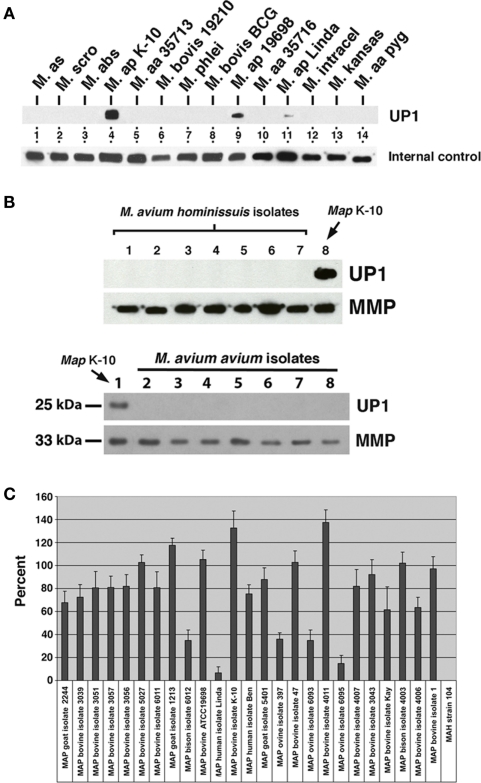
Nine positive, stable hybridomas were obtained following immunization of mice with a membrane-enriched fraction of MAP as described in the materials and methods section. MAbs present in hybridoma culture supernatants were tested for reactivity against several whole cell extracts of mycobacteria. One of these newly obtained MAbs, designated 17A12, reacted only with MAP and not with other mycobacteria (Figure 1A). The isotype for this antibody is IgG1 kappa. While no reactivity with this antibody was observed for other species and subspecies of mycobacteria, including members of the MAC group, reactivity varied among the different strains of MAP. The bovine isolate K-10 showed strong reactivity in lane 4 whereas weak reactivity was observed for the human isolate shown in lane 11. MAb 4B6, which detects an unknown but conserved mycobacterial protein (Bannantine et al., 2007b), shows the relative amounts of protein loaded in each of those lanes. The type strain of MAP, which is another bovine isolate, also reacted with the antibody and is shown in lane 9.
Figure 1.
Monoclonal antibody 17A12 detects an unknown protein (UP1) present only in M. avium subsp. paratuberculosis whole cell extracts. Shown are immunoblots of mycobacterial whole cell extracts exposed to MAbs. (A) The top blot was exposed to 17A12, which detects only the three MAP strains present in lanes 4, 9, and 11. The lower blot, labeled internal control, was exposed to MAb 4B6, which detects an unknown but highly conserved mycobacterial protein (Bannantine et al., 2007b) and shows the relative amounts of protein loaded in each of those lanes. The mycobacterial whole cell antigen prep used is indicated. Abbreviations: M, M. avium subsp. silvaticum; M. scro, M. scrofulaceum; M. abs, M. abcessus; M. ap, M. avium subsp. paratuberculosis; M. aa, M. avium subsp. avium; M. intracel, M. intracellulare. (B) UP1 is not present in M. avium subsp. avium or M. avium subsp. hominissuis isolates. The control blot labeled MMP in each image was exposed to a MAb previously developed in our laboratory that binds to the major membrane protein, which is present in all MAC species (Bannantine et al., 2007a). The upper blots are loaded with M. avium subsp. avium isolates and the lower blots are loaded with M. avium subsp. hominissuis isolates not analyzed in (A) (see Table A1 in Appendix for these strains). (C) Quantitative densitometry was performed on several MAP strains and the M. avium subsp. hominissuis strain 104. The results are expressed as a percent of 17A12 divided by the internal control (MMP). Error bars indicate standard deviations of the means. Data are representative of three independent culture replicates and error bars are standard deviation of the mean.
The specificity of this antibody was a fantastic result considering the genetic similarity of these subspecies and was never observed with previously developed antibodies raised against MAP proteins (Leid et al., 2002; Bannantine et al., 2007a,b; Malamo et al., 2007). Therefore, additional isolates of M. avium subsp. hominissuis and M. avium subsp. avium were collected and analyzed including an isolate from endangered pygmy rabbits in the Columbia basin (Harrenstien et al., 2006). None of those isolates produced any protein target detected by the MAb (Figure 1B). When more extensive analysis of additional MAP isolates from different hosts were analyzed, the variable reactivity was confirmed by densitometry analysis (Figure 1C). This variability did not depend on the total protein as antibody to the major membrane protein (MMP) encoded by MAP2121c was used to normalize protein loaded. The results further show that MAb 17A12 detection levels of the target protein from ovine strains were lower compared to bovine strains (P < 0.0003). This strain-to-strain difference was reproducible and is either a result of epitope changes or level of protein expression. To distinguish between these possibilities, the target protein as well as the reactive epitope must first be identified.
Expression library screening for the target protein
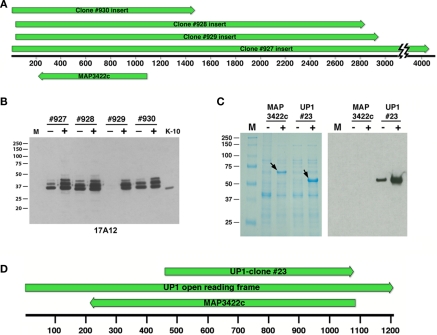
Four immunoreactive plaques were obtained when screening a phage lambda expression library with the 17A12 antibody. Sequence analysis of the subcloned plaque inserts demonstrated they were all overlapping with a 1440-bp segment common to all four clones (Figure 2A). The only annotated gene in this 1440-bp region, MAP3422c, was present in the opposite strand relative to the lacZ promoter for all four library clones. E. coli subclones from these original phage clones expressed a protein appearing as three bands and slightly larger than the native protein in MAP (Figure 2B). The entire coding sequence of MAP3422c, a pseudouridine synthase, which catalyzes the isomerization of specific uridines in an RNA molecule to pseudouridine, was cloned into an expression vector and the resulting protein did not react with 17A12 by immunoblot analysis (Figure 2C). The unannotated ORF on the opposite strand was next cloned and expressed. This protein, termed UP1, did react strongly with the 17A12 antibody (Figure 2C).
Figure 2.
Sequence and immunological analysis of reactive library clones. (A) Alignment of the four positive library clones. The alignment is drawn to scale showing the overlap of clones #927, #928, #929, and #930. Shown beneath the base pair scale bar is the position of the only annotated gene, MAP3422c, common to all four clones but on the opposite DNA strand relative to the lacZ promoter. Arrows indicate direction of transcription. (B) Immunoblot of uninduced and IPTG-induced E. coli lysates harboring the positive library clones was exposed to MAb 17A12. Protein size markers are indicated in the left margins and the clone number and induction status are indicated across the top. K-10 is the MAP whole cell extract. Lane M is the protein size markers. (C) The full length MAP3422c and a truncated section of MAP3422 (UP1-#23) were cloned and expressed in E. coli. Whole cell extracts of these recombinant clones were induced with IPTG analyzed by SDS-PAGE and immunoblot analysis with 17A12. Only the recombinant UP1 reacted with the antibody. Arrows indicate the location of the induced protein and “M” represents protein size markers. The induction status is indicated by a positive or negative symbol beneath the label. (D) Schematic sequence alignment showing the positions of MAP3422c and UP1-clone #23 relative to the full length UP1 open reading frame. Arrows indicate direction of transcription and scale is in base pairs.
The UP1 ORF was only partially present in the genomic phage clones. The size of this partial ORF is 1.2-kb and it completely overlaps with MAP3422c on the opposing or complementary DNA strand (Figure 2D). The calculated size of the translated product from this partial ORF is 42 kDa, which is much larger than the 25-kDa size observed on immunoblots (Figure 1). Similarity searches of the nucleotide sequence showed the UP1 ORF was present in other mycobacteria; however, the translated protein showed no significant similarity to any proteins in the public databases including NCBI’s GenBank and SwissProt. Furthermore, there are no motifs for UP1. Finally, analysis of the UP1 sequence using the conserved domain database (CDD) search detected no conserved domains, indicating the unique nature of this putative protein.
Production of additional MAbs to the UP1 protein
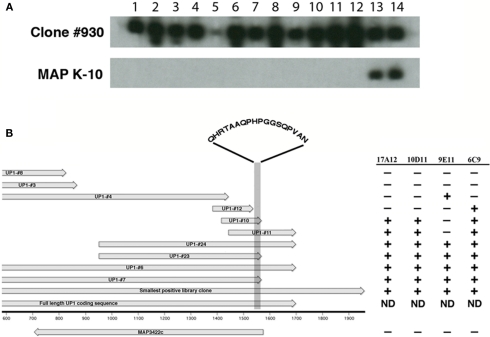
To further determine if the UP1 protein is indeed produced by MAP and that 17A12 binding is not due to a non-specific reaction, hybridomas were screened from mice immunized with the truncated recombinant UP1 protein expressed from clone #23 (Figure 2D; Table A2 in Appendix). This screen identified 13 secreting hybridomas that reacted with the truncated version of UP1. However, only one of these antibodies, 10D11, also reacted with the native protein produced in MAP (Figure 3A).
Figure 3.
Production of monoclonal antibodies to the recombinant UP1 protein. Thirteen new MAbs were obtained after immunization with purified recombinant UP1. (A) Shown are two preparative slot blots, one containing UP1 expressed from phage clone #930 and the other MAP K-10 whole cell extract. Only the 17A12 (slot 14) and 10D11 (slot 13) antibodies reacted with both the native protein in K-10 and the E. coli expressed UP1. Slot assignments: 1 = 14A5, 2 = 5H4, 3 = 14G5, 4 = 10A4, 5 = 14B7, 6 = 14D7, 7 = 14G7, 8 = 3G12, 9 = 3G2, 10 = 6F7, 11 = 9C9, 12 = 10D2, 13 = 10D11, and 14 = 17A12. (B) Alignment of truncated recombinant clones expressing UP1 along with a MAb reactivity table. The alignment shows the relative sizes and position of recombinant clones expressing fragments of the UP1 protein. Arrows indicate direction of transcription. The reactivity table indicates that 17A12 and 10D11 both bind to an 18-amino acid section of the UP1 fragment (shaded area). The amino acids present within this shaded region are indicated. The remaining antibodies all showed either the 9E11 or 6C9 reactivity patterns. ND, not determined.
Epitope mapping of monoclonal antibodies
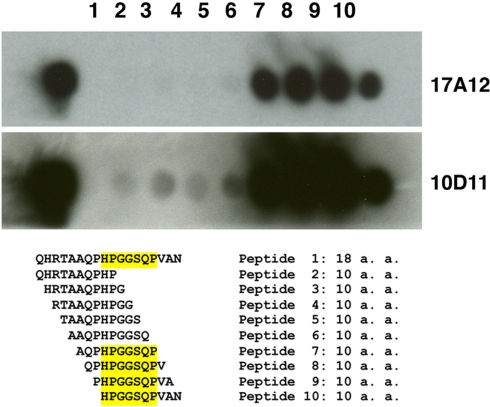
A series of truncated UP1 peptides were produced from recombinant expression clones (Table A2 in Appendix). All MAbs obtained in this study were mapped to these recombinant UP1 fragments. They bound to at least three distinct epitopes as determined by immunoblot analysis (data not shown) and shown schematically (Figure 3B). Note that both 10D11 and 17A12 had the same reactivity pattern and also were the only two antibodies that reacted with the native protein expressed by MAP. Analysis of the aligned recombinant UP1 peptides showed the epitope was contained on an 18-amino acid region near the C-terminal end of the ORF (Figure 3B). Using this information, the epitopes for these two antibodies were further mapped to determine if they are identical and also to provide an anchor point on the protein. A set of 10 peptides was used to more precisely map the epitope, each with one-amino acid extension (Figure 4). Analysis of both antibodies using these peptides identified the seven amino acid linear epitope as HPGGSQP (Figure 4). These data demonstrated that only this epitope appears to react with the native protein and all other MAbs with epitopes distinct from HPGGSQP failed to react with the native protein.
Figure 4.
Epitope mapping of 17A12 using synthetic peptides. An overlapping peptide array was constructed and used to precisely map the 17A12 binding site. Peptides were synthesized onto cellulose membranes as either the full 18-amino acid peptide identified from Figure 3 or decapeptides with one amino acid overlap. The cellulose membrane was exposed to 17A12 or 10D11 and then processed as a standard immunoblot. The reactive peptides, which comprise the epitope, are highlighted. The antibody used is indicated in the right margin.
The lack of reactivity to the native MAP protein for most of the MAbs was a cause of concern. Therefore, the epitope was used to query the MAP K-10 genome using BLAST analysis. No identical matches were discovered; however, MAP1025, encoding an RDD family protein, possessed a similar epitope with a single amino acid difference (Ser32Gln) and its calculated size is 25.0 kDa, which is within range of that observed by immunoblot of MAP sonicated extracts. A recombinant protein to MAP1025 was already available from a previous study (Bannantine et al., 2010) so it was immediately used in an immunoblot assay with 17A12. The antibody bound very strongly to this protein (Figure 5). Collectively, these data indicate that a mimotope was present in UP1 and the real epitope is present in MAP1025.
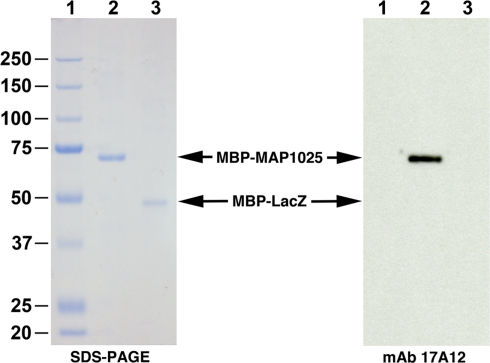
Figure 5.
MAP1025 reacts with MAb 17A12. SDS-PAGE and immunoblot analysis of the MAP1025 recombinant fusion protein. The MBP-MAP1025 fusion protein migrates at approximately 70 kDa, which agrees with predicted sizes (25 kDa for MAP1025 plus 42 kDa for MBP). The MBP-LacZ control was loaded identically for both the SDS-PAGE and immunoblot; however, MBP-MAP1025 was diluted 1:2,000 for the immunoblot and 1:2 for SDS-PAGE. Note that the antibody only reacts with MAP1025 and not the MBP affinity tag. Kilodalton size standards are located in the left margin. Lanes: 1, protein size standards; 2, MBP-MAP1025; 3, MBP-LacZ.
The monoclonal antibody specificity is due to a SNP within MAP1025
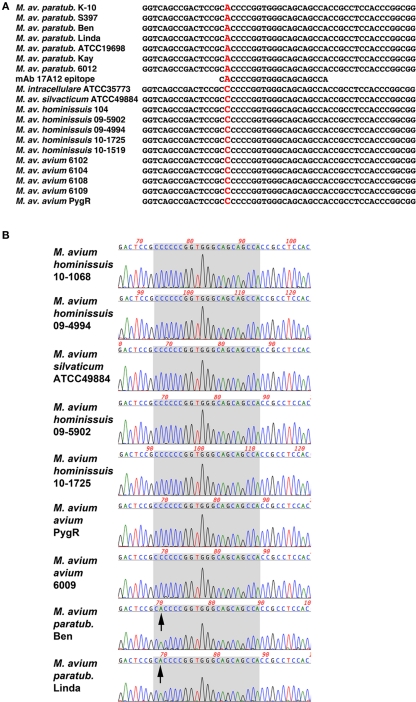
Nucleotide similarity searches showed that MAP1025 was also present in the genome of M. avium subspecies hominissuis strain 104. There are six SNPs in this 720 bp gene; all are positioned within the first 240 bp. Only two of these SNPs result in an amino acid change and one of these was within the epitope. Sequence alignments show that a SNP (C → A) changes the codon of the first amino acid in the epitope from His-28 in MAP to Pro-28 in strain 104. Additional isolates were PCR amplified and sequenced in this region. They also contained the same SNP (Figure 6; Table 1). In total, 12 MAP isolates and 18 non-MAP MAC isolates were sequenced. The identical SNP was present in all MAC strains tested. These data suggest that the reason for the 17A12 specificity is due to this non-synonymous SNP.
Figure 6.
Sequence alignment of the 17A12 epitope region reveals a non-synonymous SNP in genomic DNA. (A) Alignment of 19 amplified products from mycobacterial genomic DNA were compared with the 17A12 epitope. The polymorphic nucleotide is shown in red, which results in an amino acid change from proline to histidine. (B) Sequence chromatogram of selected mycobacterial templates reveals sequence quality at the site of the C → A polymorphism. The seven codons that encode the 17A12 epitope are shaded in gray. Arrows point to the adenine nucleotide SNP present in the first codon of the epitope.
Table 1.
Epitopes from this study.
| Epitope sequence | Amino acid position | Amino acid change | Source | Presence in mycobacteria? | Reactivity with 17A12? |
|---|---|---|---|---|---|
| HPGGSQP | Not appl. | Gln32Ser | UP1 | Not expressed | Yes |
| HPGGQQP | 28–34 | Pro28His | MAP1025 | MAP only | Yes |
| PPGGQQP | 28–34 | His28Pro | MAV_1202 | Non-MAP only | No |
The N-terminal 75 amino acids of MAP1025 are strongly biased to proline and glycine amino acids. The first 75 amino acids consist of 46% proline and 18% glycine residues. After amino acid 75 is an RDD family domain. This family of proteins contain three conserved amino acids: one arginine and two aspartates, hence the name RDD family. This region also contains two predicted transmembrane domains.
MAP1025 is located primarily in the membrane
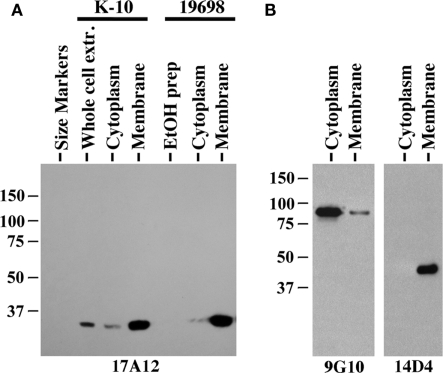
Immunoblot analysis of membrane-enriched fractions for two MAP strains shows the target protein is predominantly present in those fractions (Figure 7A). However, MAP1025 was not detected in the EtOH extract of MAP, which contains lipids and some proteins gently removed from the surface of the bacilli and has been shown to be an effective antigen in ELISA testing for Johne’s disease (Eda et al., 2006). The quality of the membrane and cytoplasm fractions for the ATCC19698 strain was tested with antibodies to proteins known to be present predominantly in one fraction or the other (Figure 7B). Finally, the 17A12 antibody did not react with the Johnin purified protein derivative (PPD) prepared from ATCC19698 at the National Veterinary Services Laboratory (data not shown) indicating that the protein is not secreted in conditions used to prepare PPD. Computational prediction of the subcellular localization of MAP1025 using the PSORTb algorithm (http://www.psort.org/psortb/index.html) identified MAP1025 as a membrane protein (score 9.82) with no signal peptide detected. Collectively, these data suggest the protein is present predominantly in the membrane, is not an easily extracted component of the cell wall, and is not secreted.
Figure 7.
MAP1025 is present primarily in the membrane. (A) Equal amounts of protein obtained from various protein preparations were analyzed by SDS-PAGE and immunoblot analysis. MAP1025 is detected at higher relative abundance in the membrane-enriched fractions by immunoblot. Protein preparations derived from both the K-10 strain and the ATCC type strain are shown. EtOH prep is the ethanol vortex prep. Kilodalton size standards are indicated in the left margins. (B) Control immunoblots demonstrating the integrity of the cytosol and membrane-enriched preps. MAb 9G10 binds to the cytoplasmic protein isocitrate lyase (AceAB) and 14D4 detects an unknown protein present in the membrane as demonstrated previously (Bannantine et al., 2007b).
17A12 binds to MAP infected macrophages and MAC-T cells
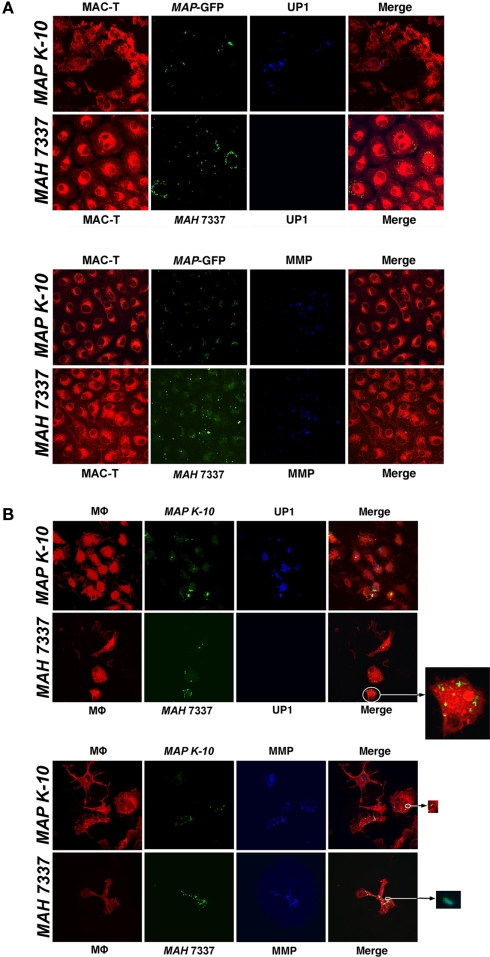
To conclude the study, we tested the utility of 17A12 to specifically label MAP within infected host cells. Both MAP and M. avium subsp. hominissuis infected macrophages and MAC–T epithelial cells were fixed and stained with either 17A12 or 8G2 antibodies. The 17A12 MAb detects MAP in infected MAC–T cells (Figure 8A) and macrophages (Figure 8B), but does not detect M. avium subsp. hominissuis as observed by confocal microscopy. The 8G2 control antibody detected both subspecies in both host cells. Infected cells were examined from 30 min to 48 h postinfection, but only the 1 and 24 h time points are shown in Figure 8. MAP1025 expression was sustained throughout the observed time, including as early as 30 min postinfection (data not shown).
Figure 8.
17A12 detects MAP K-10 but not MAH 7337 in bovine monocyte-derived macrophages (MDMs) and MAC-T epithelial cells. M. avium subsp. infections of MAC-T cells (A) and MDMs (B) at a 10:1 MOI were visualized by confocal microscopy. The MAP K-10 (pWes4) expressing GFP and fluorescein-stained MAH 7337 (green) were used for infections. 17A12 immunostaining (blue) shows only MAP K-10 infection at 24-h in MDMs and 1-h in MAC-T epithelial cells. The control antibody, 8G2, labels MMP (blue) demonstrating infection for MAP and MAH in both cell types. MDMs and MAC-T cells are shown in red. Arrows point to enlarged inserts of intracellular bacteria. Magnification, ×1000.
Discussion
Antibodies are among the most frequently used tools in basic science research and yet no specific antibody had been developed for MAP. This report describes the only known antibody that specifically detects the paratuberculosis subspecies and not other mycobacteria. It has been difficult obtaining such an antibody due to the strong genetic similarity among members of this complex as demonstrated by previous efforts (Leid et al., 2002; Bannantine et al., 2003; Malamo et al., 2007). Development of the 17A12 specific antibody opens new lines of research that utilize immunoblot, immunoprecipitation, ELISA, immunohistochemistry, and flow cytometric procedures. This novel antibody may now be used to specifically enrich for MAP in environmental samples as well as milk samples or used in diagnostic applications. It has already shown utility in labeling MAP within infected cells (Figure 8). Furthermore, studies to determine the presence of MAP in tissue samples from any host species can now be approached with this newly developed tool. Currently used methods to detect the presence of MAP in human tissues include PCR amplification of IS900 (Kirkwood et al., 2009; Sasikala et al., 2009) and in situ hybridization or non-specific acid fast staining (Jeyanathan et al., 2007), but antigen detection with a specific antibody would add a layer of certainty that the organism itself is actually present within tissues.
Efforts to identify the target protein of MAb 17A12 initially led to a non-sense ORF that was not annotated in the MAP genome. It remains unknown as to why the expression library screen did not initially reveal any MAP1025 expression clones. Instead, only the four overlapping UP1 clones were obtained. The library was constructed using Sau3AI partial digestion (Bannantine and Stabel, 2001) and one possible explanation might be a potential lack of Sau3A1 sites in the sequence surrounding MAP1025. However, analysis shows that there is one in-frame Sau3AI site at the beginning of MAP1025 and prior to the sequence encoding the epitope, which precludes this explanation. Nonetheless, all four reactive library clones of UP1 underwent forced expression, under control of the E. coli lac promoter, and thus were detected by the antibody. This UP1 sequence fortuitously had an epitope (HPGGSQP) similar to that present between residues 28 and 34 in MAP1025 (HPGGQQP). Without this defined seven-amino acid epitope from UP1, we would not have located the real binding partner for the 17A12 antibody and thus would have never known the reason for its specificity.
The example provided in this study serves as a word of caution when screening expression libraries. They are artificial systems designed to express cloned inserts regardless of the reading frame or orientation. The fact that four independent clones were obtained using an antibody specific to MAP initially led to the conclusion that UP1 was real, despite the fact that it was not annotated and had no similarity in the sequence databases. Our suspicion was raised after 14 additional MAbs were obtained to the recombinant UP1 protein and only one of these (10D11) bound the same epitope and reacted with a native protein produced in MAP. The predicted size of UP1 also did not agree with that observed with the native protein. Finally, the UP1 ORF was 100% conserved in all subspecies of MAC. These factors prompted the continued search for the target protein.
The elements required to produce a specific epitope is another interesting feature of this study. The 17A12 antibody was obtained using a membrane prep of MAP as the immunizing antigen whereas 10D11 was obtained using recombinant UP1. Although each antigen possessed a slightly different epitope, HPGGQQP for MAP1025, and HPGGSQP for UP1, both antibodies bound to both proteins. However, the MAP1025 epitope also differed by a single amino acid when comparing the paratuberculosis subspecies with all other subspecies in the MAC (Pro28His), yet that change resulted in specificity of 17A12. This suggests that His-28 is required for 17A12 binding. Furthermore, the epitope mapping experiment (Figure 4) shows that Pro-34 must also be present for 17A12 recognition. Thus the beginning (His-28) as well as the end (Pro-34) of the epitope are well defined. Residue changes within this epitope may not be as important since the Gln32Ser change did not affect 17A12 binding. Finally, it should be noted that 10D11 was not tested further in these studies because it was raised to the epitope from UP1 and not the native epitope present in MAP1025.
In general, the bovine isolates had stronger reactivity with 17A12 compared to the ovine isolates of MAP. While the cause of the variable reactivity among these two MAP lineages was never identified, the reason is not due to changes in the epitope itself. Sequence analysis of MAP1025 in several MAP strains, including those isolated from bovine, human and ovine hosts, showed 100% conservation within the epitope (Figure 6). Therefore, reactivity may be due to expression levels in those isolates and is a subject for further study. The fact that the protein has an RDD family motif does not suggest any obvious reason for potential expression differences.
It has recently been discovered that the surface molecules of MAP enable the specific detection of this organism in both a flow cytometric assay (Eda et al., 2005) as well as an ELISA format (Speer et al., 2006). This antigen prep consists of a gentle vortex of logarithmically growing bacilli in 80% ethanol. The extracted surface components are then easily dried down and used as the antigen in these assays. The components of this preparation have not yet been determined, but initial studies suggest that diagnostic proteins, carbohydrates, or lipids are more predominant in membrane fractions or surface extractions as opposed to a whole cell extract of the bacterium. In this study, MAP1025 was shown to be present in membrane fractions; however, it was not detected in the EtOH prep suggesting that it is not easily extracted from the surface of the bacterium with this solvent.
SNPs have been used to distinguish ovine from bovine strains using molecular subtyping techniques (Marsh et al., 1999; Castellanos et al., 2009). However, no study to date has uncovered a SNP that affects MAb binding as has been describe here. This SNP-sensitive MAb has so far sharply divided MAP from all other mycobacterial species. The importance of having this reagent for research and detection of MAP cannot be underestimated and opens new avenues of research with this pathogen. Finally, the discovery of a novel SNP that defines MAb specificity shows the power of non-synonymous SNPs in relation to the immunological response in the host.
Conflict of Interest Statement
The authors declare that the research was conducted in the absence of any commercial or financial relationships that could be construed as a potential conflict of interest.
Acknowledgments
The technical assistance of Janis Hansen and Brad Criswell was critical to this study’s success. This study was supported by the USDA-Agricultural Research Service intramural funds. Portions of this study were also supported by the USDA-NIFA-CAP program entitled the Johne’s disease integrated program (JDIP). Finally, we gratefully acknowledge the Iowa State University Hybridoma Facility for hybridoma production.
Appendix
Table A1.
Mycobacterial strains and isolates used in this study.
| Isolate | Organism | Host | Location | Reference or source |
|---|---|---|---|---|
| K-10 | M. avium subsp. paratuberculosis | Bovine | Feces | NADC, ATCC BAA-968 |
| 19698 | M. avium subsp. paratuberculosis | Bovine | Feces | ATCC 19698 |
| 187 | M. avium subsp. paratuberculosis | Bovine | Ileum | Recent clinical isolate, NADC |
| 523 | M. avium subsp. paratuberculosis | Bovine | Ileum | NADC, Ames, Iowa |
| 803 | M. avium subsp. paratuberculosis | Bovine | Ileum | NADC, Ames, Iowa |
| 3039 | M. avium subsp. paratuberculosis | Bovine | Feces | NADC, Ames, Iowa |
| 3051 | M. avium subsp. paratuberculosis | Bovine | Feces | NADC, Ames, Iowa |
| 3057 | M. avium subsp. paratuberculosis | Bovine | Feces | NADC, Ames, Iowa |
| 3056 | M. avium subsp. paratuberculosis | Bovine | Feces | NADC, Ames, Iowa |
| 5027 | M. avium subsp. paratuberculosis | Bovine | Mesenteric LN | NADC, Ames, Iowa |
| 6011 | M. avium subsp. paratuberculosis | Bovine | Ileum | Robert Whitlock, U of Penn |
| 47 | M. avium subsp. paratuberculosis | Bovine | Ileum | NADC, Ames, Iowa |
| 4011 | M. avium subsp. paratuberculosis | Bovine | IC lymph node | NADC, Ames, Iowa |
| 4007 | M. avium subsp. paratuberculosis | Bovine | IC lymph node | NADC, Ames, Iowa |
| 3043 | M. avium subsp. paratuberculosis | Bovine | Feces | NADC, Ames, Iowa |
| Kay | M. avium subsp. paratuberculosis | Bovine | Feces | NADC, Ames, Iowa |
| 4006 | M. avium subsp. paratuberculosis | Bovine | IC lymph node | NADC, Ames, Iowa |
| 1003 | M. avium subsp. paratuberculosis | Bovine | Lymph node | NADC, Ames, Iowa |
| 6012 | M. avium subsp. paratuberculosis | Bison | Ileum | Robert Whitlock, U of Penn |
| 4003 | M. avium subsp. paratuberculosis | Bison | Seminal vesicles | NADC, Ames, Iowa |
| Linda | M. avium subsp. paratuberculosis | Human | Ileum | ATCC 43015 |
| Ben | M. avium subsp. paratuberculosis | Human | Intestine | ATCC 43544 |
| 2244 | M. avium subsp. paratuberculosis | Goat | ||
| 1213 | M. avium subsp. paratuberculosis | Goat | ||
| 5401 | M. avium subsp. paratuberculosis | Goat | ||
| S397 | M. avium subsp. paratuberculosis | Ovine | Ileum | Recent clinical isolate, NADC |
| 6093 | M. avium subsp. paratuberculosis | Ovine | Ileum | NADC |
| 6094 | M. avium subsp. paratuberculosis | Ovine | Mesenteric LN | NADC |
| 6095 | M. avium subsp. paratuberculosis | Ovine | Ileum | NADC |
| 724 | M. avium subsp. avium | Chicken | Liver | ATCC 25291 |
| 6003 | M. avium subsp. avium | Chicken | ATCC 35713 (TMC702) | |
| 801 | M. avium subsp. avium | Chicken | ATCC 35719 | |
| 6009 | M. avium subsp. avium | Bovine | ATCC 35716 (TMC715) | |
| 6102 | M. avium subsp. avium | Deer | USDA-APHIS, Ames, Iowa | |
| 6104 | M. avium subsp. avium | Gazelle | USDA-APHIS, Ames, Iowa | |
| 6106 | M. avium subsp. avium | Avian | CDC, Atlanta, GA | |
| 6107 | M. avium subsp. avium | Avian | USDA-APHIS, Ames, Iowa | |
| 6108 | M. avium subsp. avium | Swine | USDA-APHIS, Ames, Iowa | |
| 6109 | M. avium subsp. avium | Human | CDC, Atlanta, GA | |
| 6110 | M. avium subsp. avium | Human | CDC, Atlanta, GA | |
| PygR | M. avium subsp. avium | Pygmy rabbit | (17) | |
| 104 | M. avium subsp. hominissuis | Human | Blood | Luiz E. Bermudez |
| 09-4407 | M. avium subsp. hominissuis | Elk | NVSL | |
| 09-4994 | M. avium subsp. hominissuis | Swine | NVSL | |
| 09-5902 | M. avium subsp. hominissuis | Swine | NVSL | |
| 10-1519 | M. avium subsp. hominissuis | Dog | NVSL | |
| 10-1068 | M. avium subsp. hominissuis | Bovine | NVSL | |
| 10-1725 | M. avium subsp. hominissuis | Bovine | NVSL | |
| 10-2173 | M. avium subsp. hominissuis | Bovine | NVSL | |
| 6006 | M. avium subsp. silvaticum | Roe deer | Vi-72 | |
| 6409 | M. avium subsp. silvaticum | Wood pigeon | Liver and spleen | ATCC 49884 |
| L948 | M. abscessus | ATCC 19977 | ||
| 19210 | M. bovis | Bovine | Lymph node | ATCC 19210 |
| 1011 | M. bovis BCG Pasteur | Bovine | Milk | ATCC 35734 (TMC1011) |
| 6081 | M. kansasii | Human | ATCC 12478 | |
| 6010 | M. intracellulare | Swine | ATCC 35773 | |
| 6083 | M. phlei | ATCC 11758 | ||
| 6077 | M. scrofulaceum | Human | Lymph node | ATCC 19981 |
Abbreviations: LN, lymph node; IC, ileal cecal; NADC, National Animal Disease Center; CDC, Center for Disease Control; APHIS, Animal Plant Health Inspection Service; ATCC, American Type Culture Collection.
Table A2.
Primers used in this study.
| Construct | Forward primer | Reverse primer | Product size (bp) |
|---|---|---|---|
| UP1-#1 | ATCCTCTAGAGGTGATCTCAATCCTGCTGCG | GCGCAAGCTTCTACACCGCCCGGGTGCAGGC | 1182 |
| UP1-#3 | ATCCTCTAGAGGTGATCTCAATCCTGCTGCG | GCGCAAGCTTACCGATCATCGGTGATCCGT | 351 |
| UP1-#4 | ATCCTCTAGAGGTGATCTCAATCCTGCTGCG | GCGCAAGCTTAGTGGTTACCGCCGACGGGGCG | 927 |
| UP1-#6 | ATCCTCTAGAGATCAGCGACTCCCGGGTGCC | GCGCAAGCTTCTACACCGCCCGGGTGCAGGC | 1206 |
| UP1-#7 | ATCCTCTAGAGATCAGCGACTCCCGGGTGCC | GCGCAAGCTTACCGGCGCCGCTGCCGGTTCGC | 1077 |
| UP1-#8 | ATCCTCTAGAGATCAGCGACTCCCGGGTGCC | GCGCAAGCTTACCGATCATCGGTGATCCGT | 375 |
| UP1-#10 | ATCCTCTAGAGTCGACCACCGCCCCGTCGGC | GCGCAAGCTTACCGGCGCCGCTGCCGGTTCGC | 156 |
| UP1-#11 | ATCCTCTAGACACTTCGGCGGCAAGGACTTT | GCGCAAGCTTACACCGCCCGGGTGCAGGC | 258 |
| UP1-#12 | ATCCTCTAGACACACTGGCCCCGGCCGGCAA | GCGCAAGCTTACCCGCCCGGGTGCGGCTGCGC | 156 |
| UP1-#16 | ATCCTCTAGACTCCAGGAAGCTGTGCACGTG | GCGCAAGCTTAGTGGTTACCGCCGACGGGGCG | 1146 |
| UP1-#21 | ATCCTCTAGACGCCAGGATCGTCGAGAGCAC | GCGCAAGCTTACCGATCATCGGTGATCCGT | 465 |
| UP1-#23 | ATCCTCTAGACAACAGCTCCACCTCCGTCAG | GCGCAAGCTTACCGGCGCCGCTGCCGGTTCGC | 621 |
| UP1-#24 | ATCCTCTAGACAACAGCTCCACCTCCGTCAG | GCGCAAGCTTCTACACCGCCCGGGTGCAGGC | 750 |
| MAP1025 | ATCCTCTAGATTGCCCATGACCGATCAACCGC | GCGCAAGCTTCTAGCTCGGCGGGCTTTCGGAG | 726 |
| MAP3422c | ATCCTCTAGACCGGCGCCGCTGCCGGTTCGC | GCGCAAGCTTCATGGACCTGGGTCGTCGAG | 873 |
| UpET1 | CACCATGGATCAGCGACTCCCGGGT | CTACACCGCCCGGGTGCAGGCCGA | 1209 |
| UpET2 | CACCCTGGGCAAGCAGCTGCAGCGG | CTACACCGCCCGGGTGCAGGCCGA | 915 |
| UpET4 | CACCCTGTCATGGACCTGGGTCGTC | CTACACCGCCCGGGTGCAGGCCGA | 999 |
| UpET5 | CACCTTGCCGTCGCACACGCCACCA | CTACACCGCCCGGGTGCAGGCCGA | 1047 |
| UpET6 | CACCGTGCTGGTTCCAATCCAGCAG | CTACACCGCCCGGGTGCAGGCCGA | 1158 |
All primers are listed 5′–3′. Nucleotides specific for cloning purposes are underlined.
References
- Abe C., Saito H., Tomioka H., Fukasawa Y. (1989). Production of a monoclonal antibody specific for Mycobacterium avium and immunological activity of the affinity-purified antigen. Infect. Immun. 57, 1095–1099 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Alexander D. C., Turenne C. Y., Behr M. A. (2009). Insertion and deletion events that define the pathogen Mycobacterium avium subsp. paratuberculosis. J. Bacteriol. 191, 1018–1025 10.1128/JB.01340-08 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Amonsin A., Li L. L., Zhang Q., Bannantine J. P., Motiwala A. S., Sreevatsan S., Kapur V. (2004). Multilocus short sequence repeat sequencing approach for differentiating among Mycobacterium avium subsp. paratuberculosis strains. J. Clin. Microbiol. 42, 1694–1702 10.1128/JCM.42.4.1694-1702.2004 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bannantine J. P., Radosevich T. J., Stabel J. R., Berger S., Griffin J. F., Paustian M. L. (2007a). Production and characterization of monoclonal antibodies against a major membrane protein of Mycobacterium avium subsp. paratuberculosis. Clin. Vaccine Immunol. 14, 312–317 10.1128/CVI.00353-06 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bannantine J. P., Radosevich T. J., Stabel J. R., Sreevatsan S., Kapur V., Paustian M. L. (2007b). Development and characterization of monoclonal antibodies and aptamers against major antigens of Mycobacterium avium subsp. paratuberculosis. Clin. Vaccine Immunol. 14, 518–526 10.1128/CVI.00353-06 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bannantine J. P., Stabel J. R. (2001). Identification of two Mycobacterium avium subspecies paratuberculosis gene products differentially recognised by sera from rabbits immunised with live mycobacteria but not heat-killed mycobacteria. J. Med. Microbiol. 50, 795–804 [DOI] [PubMed] [Google Scholar]
- Bannantine J. P., Stabel J. R., Bayles D. O., Geisbrecht B. V. (2010). Characteristics of an extensive Mycobacterium avium subspecies paratuberculosis recombinant protein set. Protein Expr. Purif. 72, 223–233 10.1016/j.pep.2010.03.019 [DOI] [PubMed] [Google Scholar]
- Bannantine J. P., Zhang Q., Li L. L., Kapur V. (2003). Genomic homogeneity between Mycobacterium avium subsp. avium and Mycobacterium avium subsp. paratuberculosis belies their divergent growth rates. BMC Microbiol. 3, 10. 10.1186/1471-2180-3-10 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bull T. J., Sidi-Boumedine K., Mcminn E. J., Stevenson K., Pickup R., Hermon-Taylor J. (2003). Mycobacterial interspersed repetitive units (MIRU) differentiate Mycobacterium avium subspecies paratuberculosis from other species of the Mycobacterium avium complex. Mol. Cell. Probes 17, 157–164 10.1016/S0890-8508(03)00047-1 [DOI] [PubMed] [Google Scholar]
- Castellanos E., Aranaz A., De Juan L., Alvarez J., Rodriguez S., Romero B., Bezos J., Stevenson K., Mateos A., Dominguez L. (2009). Single nucleotide polymorphisms in the IS900 sequence of Mycobacterium avium subsp. paratuberculosis are strain type specific. J. Clin. Microbiol. 47, 2260–2264 10.1128/JCM.00544-09 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cayrou C., Turenne C., Behr M. A., Drancourt M. (2010). Genotyping of Mycobacterium avium complex organisms using multispacer sequence typing. Microbiology 156, 687–694 10.1099/mic.0.033522-0 [DOI] [PubMed] [Google Scholar]
- Eda S., Bannantine J. P., Waters W. R., Mori Y., Whitlock R. H., Scott M. C., Speer C. A. (2006). A highly sensitive and subspecies-specific surface antigen enzyme- linked immunosorbent assay for diagnosis of Johne’s disease. Clin. Vaccine Immunol. 13, 837–844 10.1128/CVI.00148-06 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Eda S., Elliott B., Scott M. C., Waters W. R., Bannantine J. P., Whitlock R. H., Speer C. A. (2005). New method of serological testing for Mycobacterium avium subsp. paratuberculosis (Johne’s disease) by flow cytometry. Foodborne Pathog. Dis. 2, 250–262 10.1089/fpd.2005.2.250 [DOI] [PubMed] [Google Scholar]
- Harlow E., Lane D. (eds). (1988). Anitbodies: A Laboratory Manual. Cold Spring Harbor. New York: Cold Spring Harbor Laboratory Press [Google Scholar]
- Harrenstien L. A., Finnegan M. V., Woodford N. L., Mansfield K. G., Waters W. R., Bannantine J. P., Paustian M. L., Garner M. M., Bakke A. C., Peloquin C. A., Phillips T. M. (2006). Mycobacterium avium in pygmy rabbits (Brachylagus idahoensis): 28 cases. J. Zoo Wildl. Med. 37, 498–512 10.1638/05-002.1 [DOI] [PubMed] [Google Scholar]
- Harris N. B., Barletta R. G. (2001). Mycobacterium avium subsp. paratuberculosis in veterinary medicine. Clin. Microbiol. Rev. 14, 489–512 10.1128/CMR.14.3.489-512.2001 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jeyanathan M., Boutros-Tadros O., Radhi J., Semret M., Bitton A., Behr M. A. (2007). Visualization of Mycobacterium avium in Crohn’s tissue by oil-immersion microscopy. Microbes Infect. 9, 1567–1573 10.1016/j.micinf.2007.09.001 [DOI] [PubMed] [Google Scholar]
- Kirkwood C. D., Wagner J., Boniface K., Vaughan J., Michalski W. P., Catto-Smith A. G., Cameron D. J., Bishop R. F. (2009). Mycobacterium avium subspecies paratuberculosis in children with early-onset Crohn’s disease. Inflamm. Bowel Dis. 15, 1643–1655 10.1002/ibd.20967 [DOI] [PubMed] [Google Scholar]
- Leid J. G., Hunter D., Speer C. A. (2002). Early diagnosis of Johne’s disease in the American bison by monoclonal antibodies directed against antigen 85. Ann. N. Y. Acad. Sci. 969, 66–72 10.1111/j.1749-6632.2002.tb04352.x [DOI] [PubMed] [Google Scholar]
- Malamo M., Okazaki K., Sakoda Y., Kida H. (2007). Carboxyl terminus of the 34 kDa protein of Mycobacterium paratuberculosis shares homologous B-cell epitopes with Mycobacterium avium and Mycobacterium intracellulare. Vet. Rec. 161, 853–857 [PubMed] [Google Scholar]
- Marsh I., Whittington R., Cousins D. (1999). PCR-restriction endonuclease analysis for identification and strain typing of Mycobacterium avium subsp. paratuberculosis and Mycobacterium avium subsp. avium based on polymorphisms in IS1311. Mol. Cell. Probes 13, 115–126 10.1006/mcpr.1999.0227 [DOI] [PubMed] [Google Scholar]
- Paustian M. L., Bannantine J. P., Kapur V. (2010). “ Mycobacterium avium subsp. paratuberculosis genome,” in Paratuberculosis: Organism, disease, control, eds Behr M. A., Collins D. M. (Oxfordshire: CAB International; ), 73–82 [Google Scholar]
- Paustian M. L., Zhu X., Sreevatsan S., Robbe-Austerman S., Kapur V., Bannantine J. P. (2008). Comparative genomic analysis of Mycobacterium avium subspecies obtained from multiple host species. BMC Genomics 9, 135. 10.1186/1471-2164-9-135 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Radosevich T. J., Reinhardt T. A., Lippolis J. D., Bannantine J. P., Stabel J. R. (2007). Proteome and differential expression analysis of membrane and cytosolic proteins from Mycobacterium avium subsp. paratuberculosis strains K-10 and 187. J. Bacteriol. 189, 1109–1117 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sasikala M., Reddy D. N., Pratap N., Sharma S. K., Balkumar P. R., Sekaran A., Banerjee R., Reddy D. B. (2009). Absence of Mycobacterium avium ss paratuberculosis-specific IS900 sequence in intestinal biopsy tissues of Indian patients with Crohn’s disease. Indian J. Gastroenterol. 28, 169–174 10.1186/1471-2180-8-204 [DOI] [PubMed] [Google Scholar]
- Sevilla I., Li L., Amonsin A., Garrido J. M., Geijo M. V., Kapur V., Juste R. A. (2008). Comparative analysis of Mycobacterium avium subsp. paratuberculosis isolates from cattle, sheep and goats by short sequence repeat and pulsed-field gel electrophoresis typing. BMC Microbiol. 8, 204. 10.1128/CVI.13.5.535-540.2006 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Speer C. A., Scott M. C., Bannantine J. P., Waters W. R., Mori Y., Whitlock R. H., Eda S. (2006). A novel enzyme-linked immunosorbent assay for diagnosis of Mycobacterium avium subsp. paratuberculosis infections (Johne’s Disease) in cattle. Clin. Vaccine Immunol. 13, 535–540 10.1128/JCM.01349-08 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Thibault V. C., Grayon M., Boschiroli M. L., Willery E., Allix-Beguec C., Stevenson K., Biet F., Supply P. (2008). Combined multilocus short-sequence-repeat and mycobacterial interspersed repetitive unit-variable-number tandem-repeat typing of Mycobacterium avium subsp. paratuberculosis isolates. J. Clin. Microbiol. 46, 4091–4094 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Thorel M. F., Krichevsky M., Levy-Frebault V. V. (1990). Numerical taxonomy of mycobactin-dependent mycobacteria, emended description of Mycobacterium avium, and description of Mycobacterium avium subsp. avium subsp. nov., Mycobacterium avium subsp. paratuberculosis subsp. nov., and Mycobacterium avium subsp. silvaticum subsp. nov. Int. J. Syst. Bacteriol. 40, 254–260 10.1128/CMR.00036-06 [DOI] [PubMed] [Google Scholar]
- Turenne C. Y., Wallace R., Jr., Behr M. A. (2007). Mycobacterium avium in the postgenomic era. Clin. Microbiol. Rev. 20, 205–229 10.1016/j.micpath.2009.01.010 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wu C. W., Schmoller S. K., Bannantine J. P., Eckstein T. M., Inamine J. M., Livesey M., Albrecht R., Talaat A. M. (2009). A novel cell wall lipopeptide is important for biofilm formation and pathogenicity of Mycobacterium avium subspecies paratuberculosis. Microb. Pathog. 46, 222–230 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yoshimura H. H., Graham D. Y. (1988). Nucleic acid hybridization studies of mycobactin-dependent mycobacteria. J. Clin. Microbiol. 26, 1309–1312 [DOI] [PMC free article] [PubMed] [Google Scholar]