Abstract
Solid-state NMR spectroscopy is an efficient tool for following conformational dynamics of membrane proteins at atomic resolution. We used this technique for the site-specific detection of light-induced hydrogen-deuterium exchange in the lipid-embedded heptahelical transmembrane photosensor Anabaena sensory rhodopsin to pinpoint the location of its conformational changes upon activation. We show that the light-induced conformational changes result in a dramatic, but localized, increase in the exchange in the transmembrane regions. Most notably, the cytoplasmic half of helix G and the cytoplasmic ends of helices B and C exchange more extensively, probably as a result of their relative displacement in the activated state, allowing water to penetrate into the core of the protein. These light-induced rearrangements must provide the structural basis for the photosensory function of Anabaena sensory rhodopsin.
Solid-state NMR (SSNMR) is rapidly evolving into a powerful tool for studies of structure and dynamics of membrane proteins in their native lipid environment (1–7). Multidimensional SSNMR methods are sufficiently sensitive to determine chemical shifts of individual atoms and provide detailed structural information (8–11). They can be combined with hydrogen-deuterium (H/D) exchange (12–14) to study, in a site-specific manner, how membrane proteins interact with water. In this letter we use magic angle spinning (MAS) SSNMR detection of H/D exchange of individual amino acids to study conformational changes in Anabaena sensory rhodopsin (ASR) upon light activation. ASR is a unique cyanobacterial photosensor, believed to be responsible for chromatic adaptation (15). It differs from other microbial rhodopsins structurally (it has a water-filled cavity on the cytoplasmic side (16)), photochemically (it undergoes photochromic transitions between the two stable states instead of a conventional photocycle (16,17)), and functionally (it interacts with the unique soluble cytoplasmic transducer (15,18)). Recently, we obtained spectroscopic assignments for the majority of ASR resonances and found that many residues on both cytoplasmic and extracellular surfaces interact with the solvent, as follows from the fast exchange of their amide protons (11). In this study, we show that significant localized light-induced conformational changes occur in the transmembrane core of ASR, most probably correlated to its photosensory function.
To probe the changes of the water-accessible surface that occur upon illumination, we compared 1), dark-adapted ASR in H2O; 2), dark-adapted ASR incubated for 24 h in D2O; 3), ASR incubated in D2O for 1.5 h under illumination; and 4), ASR incubated in D2O for an additional 3 h under illumination. Equal amounts of sample were packed in a 3.2-mm rotor, and 2D NCA and 3D NCACX chemical shift correlation experiments were recorded. Short H/N cross-polarization (CP) (19) times of 300 μs ensured that nitrogen spins are excited primarily from their directly bonded protons, as is evident, for example, from the disappearance of most proline NCA correlations in the 2D NCA correlation spectra (Fig. S2 in the Supporting Material). Proline nitrogen atoms do not carry protons and are not affected by H/D exchange, so the reduction or absence of their correlations can serve as a control. From proline crosspeak intensities, we estimate that although CP excitation from remote protons (e.g., Hα) is still possible, its contribution is small and does not exceed 25% of the original signal. Thus, site-specific signal attenuation provides an average measure of the light-modulated degree of accessibility of amide nitrogen atoms to the solvent.
The results of detailed site-specific analysis of the 3D NCACX correlation spectra are shown in Fig. 1 (see also Fig. S3 for representative 2D NC planes and Fig. S1 for the identities of the exchangeable residues). When H/D exchange is performed in the dark, most of the affected residues are confined to the solvent-exposed cytoplasmic and extracellular sides, e.g., the B-C loop (D57-Y70). Under continuous illumination, ASR shuttles between two stable states and undergoes conformational transitions through a series of intermediates with different lifetimes and water accessibilities of the hydrophobic core (16,17). The H/D exchange provides an integral picture of these changes, which mostly saturate in the first 90 min of the illumination. The most significant light-induced exchange occurs in the cytoplasmic ends of helices B (V39–I42) and C (L83–A91), and in the whole cytoplasmic half of helix G (K210–G220), with further enhancement in the exchange of helix F (Figs. 1 and 2). Some residual signals in these parts are likely due to long-range CP effects, which are expected even in fully exchanged fragments. On the extracellular side, we observe a 3.5- to 4-fold reduction of peak intensities in the immediate vicinity of the B-C loop in helix C, and in helix E (Figs. 1 and 2, and Fig. S1).
Figure 1.
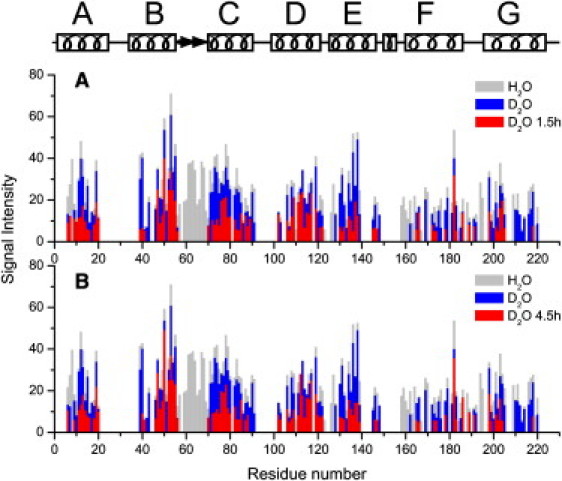
(A) Comparison of crosspeak intensities (in units of RMS of noise) in the nonexchanged sample (gray), the sample exchanged in the dark for 24 h (blue), and the sample exchanged under illumination for 90 min (red). (B) Same as in A, but with the illuminated sample exchanged for an additional 180 min (red). The secondary structure of ASR derived from x-ray and SSNMR data (11) is shown on top.
Figure 2.
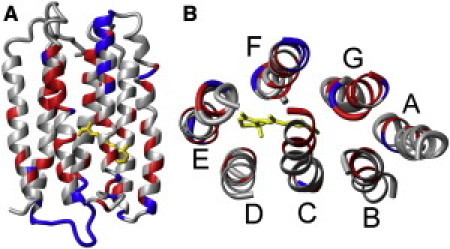
(A) Side view of ASR (structural model derived from SSNMR and x-ray data (11)), with helices A, G, and F facing the viewer; the cytoplasmic side is on top. Residues colored in red show strong enhancement of H/D exchange by light. Residues colored in blue are exchangeable in the dark. (B) Top view of the same model from the cytoplasmic side, with loops removed. Transmembrane helices are marked by letters.
Several residues, especially conspicuous in helices A, C, and E, show a 1.5- to 2.5-fold reduction in the signal intensity. It was surprising to find that longer exposure to D2O under illumination (270 min vs. 90 min) does not cause further signal decrease, which cannot be explained in the framework of a simple open/closed two-state model.
In the 3D structural model of ASR, helices B, C, F, and G form polar semichannels inside the protein core on both sides of the Schiff base (11,16). The x-ray structure (16) reveals a hydrogen-bonded network in the cytoplasmic half, mainly involving helices B, C, and G, as well as a few water molecules. Our data suggest that light-induced conformational changes open a cleft between these helices on the cytoplasmic side (Fig. 2), which allows water to penetrate into the protein core. Significant light-induced conformational changes (outward tilt) of the cytoplasmic half of helix F occur in other microbial rhodopsins (20). Although the observed H/D exchange pattern supports the possibility that a similar tilt of helix F occurs in ASR, we believe that the conformational changes involve other helices as well. The large light-induced increase in the exchange extent of the cytoplasmic half of helix G (along with the ends of helices B and C) can in fact be caused by its own displacement, which may occur without the movement of helix F. This unique conformational change must be related to the unique function of ASR, which interacts with the cytoplasmic soluble transducer.
The proposed movement of helix G is consistent with earlier FTIR measurements (21,22), which point at possible light-induced disruption of the hydrogen-bonded network formed by several polar residues from helices B, C, and G in the cytoplasmic half of ASR (16). Furthermore, the presence of a unique Pro-206 in helix G can give this helix an additional flexibility, further enhanced by the disappearance of several important interhelical hydrogen bonds maintained by the homologous superconserved Asp in other microbial rhodopsins. The proposed separation of helix G from B and C, which may occur in the M intermediate upon the transition from the all-trans- to the 13-cis-retinal form, would affect the cytoplasmic side and may change the binding affinity between ASR and its soluble transducer.
In conclusion, we have shown that SSNMR can be used for site-specific detection of H/D exchange in a seven-helix transmembrane protein to reveal and locate conformational changes occurring upon its activation. With SSNMR assignments of many proteins in hand (8–11), our methodology can be easily extended to study their solvent-accessible surfaces, or to indirectly probe invisible or metastable states.
Acknowledgments
We thank Dr. K.-H. Jung for providing the plasmid for ASR expression and Dr. George Harauz for carefully reading the manuscript.
This research was supported by the Natural Science Engineering Research Council of Canada, the Canada Foundation for Innovation, the Ontario Ministry of Research and Innovation, Bruker Canada Ltd, and MEXT of Japan (22770101). V.L. holds the Canada Research Chair in Biophysics. S.W. is supported by the Canadian Institutes of Health Research (CIHR) fellowship. L.S. is a recipient of the MITACS Accelerate Award.
Footnotes
Lichi Shi's present address is Department of Medical Genetics and Microbiology, University of Toronto, Toronto, Ontario, Canada.
Contributor Information
Leonid S. Brown, Email: lebrown@uoguelph.ca.
Vladimir Ladizhansky, Email: vladizha@uoguelph.ca.
Supporting Material
References and Footnotes
- 1.McDermott A. Structure and dynamics of membrane proteins by magic angle spinning solid-state NMR. Annu. Rev. Biophys. 2009;38:385–403. doi: 10.1146/annurev.biophys.050708.133719. [DOI] [PubMed] [Google Scholar]
- 2.Ramamoorthy A. Beyond NMR spectra of antimicrobial peptides: dynamical images at atomic resolution and functional insights. Solid State Nucl. Magn. Reson. 2009;35:201–207. doi: 10.1016/j.ssnmr.2009.03.003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Renault M., Cukkemane A., Baldus M. Solid-state NMR spectroscopy on complex biomolecules. Angew. Chem. Int. Ed. Engl. 2010;49:8346–8357. doi: 10.1002/anie.201002823. [DOI] [PubMed] [Google Scholar]
- 4.Goncalves J.A., Ahuja S., Smith S.O. Structure and function of G protein-coupled receptors using NMR spectroscopy. Prog. Nucl. Magn. Reson. Spectrosc. 2010;57:159–180. doi: 10.1016/j.pnmrs.2010.04.004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Hu F., Luo W., Hong M. Mechanisms of proton conduction and gating in influenza M2 proton channels from solid-state NMR. Science. 2010;330:505–508. doi: 10.1126/science.1191714. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Sharma M., Yi M., Cross T.A. Insight into the mechanism of the influenza A proton channel from a structure in a lipid bilayer. Science. 2010;330:509–512. doi: 10.1126/science.1191750. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Bajaj V.S., Mak-Jurkauskas M.L., Griffin R.G. Functional and shunt states of bacteriorhodopsin resolved by 250 GHz dynamic nuclear polarization-enhanced solid-state NMR. Proc. Natl. Acad. Sci. USA. 2009;106:9244–9249. doi: 10.1073/pnas.0900908106. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Li Y., Berthold D.A., Rienstra C.M. Chemical shift assignment of the transmembrane helices of DsbB, a 20-kDa integral membrane enzyme, by 3D magic-angle spinning NMR spectroscopy. Protein Sci. 2008;17:199–204. doi: 10.1110/ps.073225008. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Etzkorn M., Seidel K., Baldus M. Complex formation and light activation in membrane-embedded sensory rhodopsin II as seen by solid-state NMR spectroscopy. Structure. 2010;18:293–300. doi: 10.1016/j.str.2010.01.011. [DOI] [PubMed] [Google Scholar]
- 10.Shi L., Ahmed M.A.M., Ladizhansky V. Three-dimensional solid-state NMR study of a seven-helical integral membrane proton pump—structural insights. J. Mol. Biol. 2009;386:1078–1093. doi: 10.1016/j.jmb.2009.01.011. [DOI] [PubMed] [Google Scholar]
- 11.Shi L., Kawamura I., Ladizhansky V. Conformation of a seven-helical transmembrane photosensor in the lipid environment. Angew. Chem. Int. Ed. Engl. 2011;50:1302–1305. doi: 10.1002/anie.201004422. [DOI] [PubMed] [Google Scholar]
- 12.Wagner G., Wüthrich K. Amide protein exchange and surface conformation of the basic pancreatic trypsin inhibitor in solution. Studies with two-dimensional nuclear magnetic resonance. J. Mol. Biol. 1982;160:343–361. doi: 10.1016/0022-2836(82)90180-2. [DOI] [PubMed] [Google Scholar]
- 13.Cotten M., Fu R., Cross T.A. Solid-state NMR and hydrogen-deuterium exchange in a bilayer-solubilized peptide: structural and mechanistic implications. Biophys. J. 1999;76:1179–1189. doi: 10.1016/S0006-3495(99)77282-6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.del Amo J.M., Fink U., Reif B. Quantification of protein backbone hydrogen-deuterium exchange rates by solid state NMR spectroscopy. J. Biomol. NMR. 2010;48:203–212. doi: 10.1007/s10858-010-9450-8. [DOI] [PubMed] [Google Scholar]
- 15.Jung K.H., Trivedi V.D., Spudich J.L. Demonstration of a sensory rhodopsin in eubacteria. Mol. Microbiol. 2003;47:1513–1522. doi: 10.1046/j.1365-2958.2003.03395.x. [DOI] [PubMed] [Google Scholar]
- 16.Vogeley L., Sineshchekov O.A., Luecke H. Anabaena sensory rhodopsin: a photochromic color sensor at 2.0 Å. Science. 2004;306:1390–1393. doi: 10.1126/science.1103943. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Kawanabe A., Furutani Y., Kandori H. Photochromism of Anabaena sensory rhodopsin. J. Am. Chem. Soc. 2007;129:8644–8649. doi: 10.1021/ja072085a. [DOI] [PubMed] [Google Scholar]
- 18.Vogeley L., Trivedi V.D., Luecke H. Crystal structure of the Anabaena sensory rhodopsin transducer. J. Mol. Biol. 2007;367:741–751. doi: 10.1016/j.jmb.2006.11.074. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Pines A., Gibby M.G., Waugh J.S. Proton-enhanced NMR of dilute spins in solids. J. Chem. Phys. 1973;59:569–590. [Google Scholar]
- 20.Klare J.P., Bordignon E., Steinhoff H.J. Sensory rhodopsin II and bacteriorhodopsin: light activated helix F movement. Photochem. Photobiol. Sci. 2004;3:543–547. doi: 10.1039/b402656j. [DOI] [PubMed] [Google Scholar]
- 21.Shi L., Yoon S.R., Brown L.S. Cytoplasmic shuttling of protons in Anabaena sensory rhodopsin: implications for signaling mechanism. J. Mol. Biol. 2006;358:686–700. doi: 10.1016/j.jmb.2006.02.036. [DOI] [PubMed] [Google Scholar]
- 22.Kawanabe A., Furutani Y., Kandori H. FTIR study of the L intermediate of Anabaena sensory rhodopsin: structural changes in the cytoplasmic region. Biochemistry. 2008;47:10033–10040. doi: 10.1021/bi800941a. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.


