Abstract
Recent studies have indicated that nutrient deprivation particularly glucose may play a major role in tumor cell tolerance to a generally oxidative stress environment in solid tumors. Here, we studied the impact of glucose deprivation on the response of human colon (HT29)0020and prostate (DU145) cancer cells to γ radiation. A significant decrease in intracellular glucose level was observed in glucose deprived cells as measured by bioreductive assay. The survival of HT29 and DU145 were increased by 30 and 100% respectively when these cells were exposed to γ radiation in the absence of glucose compared to that in the presence of glucose. In glucose depleted medium, glutathione (GSH), a free radical scavenger, content remained the same, and showed no correlation with the radiation resistance induced by glucose deprivation. GRP78, a stress response survival protein, was not significantly increased in cells deprived of glucose for 4 hours compared to those cells in glucose. DNA repair protein Ku, which is known to play a major role in cellular resistance to radiation, was significantly increased in glucose deprived cancer cells that showed enhanced radiation resistance. These results have demonstrated, for the first time, that glucose deprivation mediated stress increased the expression of nuclear Ku and resistance to radiation induced oxidative stress in human cancer cells. The additional resistance caused by glucose deprivation in cancer cells has clinical significance since solid tumors are known to have low level of glucose due to diffusion limited blood supply and higher metabolic activity.
Keywords: Tumor microenvironment, oxidative stress, Ku protein, glutathione, glucose deprivation, GRP78
INTRODUCTION
Tumor microenvironment is believed to play a major role in the response of cancer cells to therapy (1). It has been suggested that genetic alterations resulting from tumor microenvironment may be responsible for the resistance of cancer cells to DNA damaging agents (2, 3). In addition, p53 mutation, which occurs mostly in cancer cells, plays a significant role in the resistance of cancer cells to DNA damaging agents (4–6). Cancer cells with other molecular changes also respond differently to DNA damaging agents. BCL-2, BCL-x and Bax genes, which affect apoptosis, have been shown to have a direct effect on the survival of cancer cells after exposure to DNA damaging agents (7, 8). A number of cell lines with mutations in genes that are relevant to apoptosis are resistant to radiation (4–7, 9–11).
Radiation sensitivity of mammalian cells is also dependent on DNA repair genes (12–14). DNA damaging agents are supposed to induce the same amount of initial DNA damage (target damage) in different cells provided their molecular targets are exposed/reacted to the same amount of damaging species. However, certain types of cells require a larger dose for similar cell death suggesting an intrinsic resistance (4–7, 9–12). A direct correlation between DNA double strand break (DSB) repair deficiency and increased response to radiation induced cell death has been demonstrated in Chinese hamster ovary (CHO) cell lines with deficiency in DNA repair proteins (12–14). Recent studies have demonstrated that targeting of Ku and DNA dependent protein kinase (DNAPK), major DNA DSB repair proteins, by small interfering RNA in human cancer cells enhanced their sensitivity to DNA damaging agents (15, 16).
Glutathione (GSH) plays a major role in the cellular response to oxidative stress. Several reports have demonstrated a direct correlation between GSH, GSH associated enzymes and cytotoxicity (17–20). GSH is a small molecular weight non protein thiol found ubiquitously in all mammalian cells. This tripeptide acts as a free radical scavenger and repairs oxidized molecules through enzyme catalyzed reactions. Studies have shown that buthionine sulfoximine (BSO) mediated depletion of GSH have increased oxidative stress and cisplatin mediated cell death (21, 22). It has been demonstrated that glutathione may be an important determinant of a cell's ability to repair DNA damage and resist cell death (21, 22).
The microenvironment related changes, which may influence the outcome of therapy, are mainly hypoxia, nutrient deprivation and hypoxia related gene expression (23). Recent studies have indicated that nutrient deprivation, particularly glucose, may play a major role in tumor cell tolerance to a generally oxidative stress environment in solid tumors (23, 24). It has been demonstrated that chronic exposure to low glucose have caused photo dynamic therapy (PDT) resistance in breast cancer cells (25). Glucose regulated protein GRP78 is believed to play a major role in the survival of cancer cells during hypoxia, low pH and stress. Glucose deprivation has also been known to induce GRP78 protein in mammalian cells (23, 24). GRP78 is considered a marker for endoplasmic reticulum stress. However, GRP78 induction has been attributed to cancer cells tolerance to certain DNA damaging agents (26, 27). Its protective ability is believed to be due to its calcium-binding properties and inhibition of caspase activation (26, 28, 29). Endoplasmic reticulum stress response element (ERSE) mediates transcriptional activation of GRP78 (28, 30).
Glucose associated molecular changes are clinically relevant since glucose deprivation is common in human solid tumors (31–33). Although the role of prolonged low glucose exposure in PDT resistance has been demonstrated in breast cancer cells (25), the impact of short-term low glucose exposure has not been studied for any kind of cancer therapy. It is important to determine the impact of short-term exposure to low glucose prior to therapy since it is likely that short term exposure to low glucose and low oxygen may also occur in solid tumors. The radiation resistance induced by short-term low oxygen exposure has been extensively studied in cells and purified DNA (34–36). However, the impact of either short- or long- term glucose deprivation on the response of human cancer cells to γ radiation induced oxidative stress has not been demonstrated. In this investigation, we have examined the effects of short-term (4hrs) glucose deprivation on the response of human colon HT29 and prostate DU145 cancer cells to γ radiation. Our results, for the first time, demonstrated that glucose deprivation increased radioresistance of human cancer cells with a concomitant increase in nuclear Ku, a DNA repair protein. The additional resistance caused by glucose deprivation in cancer cells has clinical significance since solid tumors are known to have low level of glucose due to diffusion limited blood supply and higher metabolic activity (31–33).
METHODS
Cells
Colon (HT29) and prostate (DU145) human cancer cells were primarily used in these studies. All these cancer cells, which were originally obtained from ATCC, USA, were provided by Drs. Cameron Koch and Gary Kao, University of Pennsylvania.
Cell plating and glucose deprivation
Cells (0.8 × 106) plated in six well plates were replenished with fresh Dulbecco’s Modified Eagle Medium (DMEM) containing glucose, HEPES and 2% dialyzed Fetal Bovine Serum (FBS). For glucose deprivation, cells were replenished with glucose free DMEM, 2% dialyzed FBS and 25 mM HEPES. These Cells were incubated in 5% CO2 at 37°C for 4 hours before measuring glucose level, metabolic activity and radiation response.
Extracellular Glucose Assay
Glucose levels in medium before and after glucose starvation were quantified by following the reduction of nicotinamide adenine dinucleotide phosphate (NADP) to NADPH in reaction mixture catalyzed by Adenosine Triphosphate (ATP), hexokinase (HK) and glucose 6- phosphate dehydrogenase (G6PD). Briefly, 25µl of sample was mixed with 175µl of 0.1M sodium acetate buffer (pH 4.6). Fifty µl of this sample mixture was mixed with 50µl of ATP/NADP solution (455 mg ATP-Na2H2.3H2O/50mg NADP-Na2H2 in 5 ml of Trizma (0.3 M), MgSO4 (0.3 M) buffer, pH 7.5) and 20µl of G6PD/HK (1:10 in 3.2 M ammonium sulfate). The optical density (OD) was continuously measured at 340 nm every min for up to10 minutes and the concentration was calculated using a glucose standard curve.
Intracellular Glucose as measured by glucose dependent hydroxyethyl disulfide (HEDS) bioreduction
A stock solution of 0.1M HEDS was prepared fresh in glucose free DMEM growth medium. HEDS exposure (1hr) was effected by replenishing cells with 1 (six well plate) and 1.5ml (60mm dishes) of fresh DMEM growth medium with or without glucose, 2% dialyzed FBS, HEPES and desired concentrations of HEDS. The amount of mercaptoethanol (ME) produced by bioreduction of HEDS was estimated by High Performance Liquid Chromatography/ electrochemical detection (HPLC/EC), and 5, 5-dithiobis 2-nitrobenzoic acid (DTNB) assays (37).
Cells treated with and without HEDS were cooled on ice. To quantify the bioreduction/ME production, 0.5 ml of extracellular medium was mixed with 0.5 ml of 100 mM sulfosalicyclic acid (SSA) lysis buffer in microfuge tubes and centrifuged at high speed in a Fisher 59A microfuge. For 5, 5-dithiobis 2-nitrobenzoic acid (DTNB) assay, 150µl of the extract was mixed with 1200 µl of phosphate buffer and 150 µl of 10 mM DTNB. The optical density of this reaction mixture was measured at 412 nm and the concentration was calculated using an extinction coefficient of 1.36 × 104 for reduced DTNB.
For HPLC/EC assay, the medium extract was diluted by 400 times with SSA buffer. The sample was analyzed using HPLC system consisting of a single pump, autosampler, guard cell, 5010 analytical cell and Colouchem III (ESA, USA). Diluted sample (10µl) was loaded onto a C18 column and run in an isocratic mode using a mobile phase with 50mM phosphate, pH 2.7, 0.05 mM octane sulfonic acid and 2.2% acetonitrile.
Quantification of intracellular non-protein thiols (NPSH)
Intracellular non-protein thiol (NPSH/GSH) was determined by DTNB assay as described by Ayene et al (37). Cells were rinsed twice with cell rinse and mixed with 1ml of ice-cold 50mM SSA buffer. After 10 min on ice, cells were scraped with a teflon spatula and centrifuged at high speed in a Fisher 59A microfuge. The supernatant (300µl) was mixed with 1050 µl of phosphate buffer and 150 µl of 10 mM DTNB. The optical density of this reaction mixture was measured at 412 nm and the concentration was calculated using an extinction coefficient of 1.36 × 104 for reduced DTNB.
Western blot analysis of GRP78
Total cellular GRP78 was quantified using Western blot and NIH image analysis. Whole cell extracts were prepared by using nuclear extract kit from Active Motif, CA, USA. Cells grown in tissue culture dishes were cooled on ice and rinsed with 2.5 ml ice-cold phosphate buffered (0.01 M phosphate buffer, pH 7.5, 0.15 M NaCl, 2.7mM KCl) saline (PBS). These cells were detached gently by teflon cell scraper in the presence of ice cold PBS (1.5 ml) and transferred to a 2 ml microfuge tubes. Each sample was centrifuged for 5 minutes at 500 rpm and cell pellet was used for the preparation of whole cell extract.
Cell pellet was resuspended in 100 µl 1X complete lysis buffer (Active Motif) by pipetting up and down. This cell suspension was vortexed for 10 seconds at highest setting and incubated for 10 minutes on ice on a rocking platform set at 150 rpm. The cell suspension was again vortexed for 30 seconds at highest speed and centrifuged for 20 minutes at 14,000 g in a microcentrifuge at 4°. The supernatant (whole cell extract) was transferred to a pre-chilled microcentrifuge tube and stored at −80°. Protein concentration in the supernatant was measured by Bio-Rad protein assay.
Protein extracts (5 µg) were incubated in NUPAGE sample buffer at 70°C for 10 minutes. These samples were then electrophoresed on a 10% BIS/TRIS precast gel (NUPAGE) at 200 V for 60 min in MOPS SDS running buffer (NUPAGE). Proteins in the gel were transferred to nitrocellulose paper by electrophoresis in NUPAGE transfer buffer at 30 V for 60 min. The nitrocellulose paper was incubated with 10 ml blocking buffer (5 g BSA in 100ml PBS/0.1%Tween 20) for 1.5 h at RT on a rocker and stored in the cold room overnight. It was washed five times with PBS containing 0.1% Tween 20 (PBST) and incubated with 100 µl of primary anti GRP78 antibody (Neomarkers) in 10 ml blocking buffer for 2 h at RT. It was washed five times with PBST before incubation with secondary peroxidase-labeled anti-mouse Ab for 1 h and the bands were detected using a standard ECL kit (Amersham, NJ).
Quantification of Ku by Enzyme Linked Immunosorbent Assay (ELISA)
Nuclear extracts were prepared by using the nuclear extract kit from Active Motif, CA, USA. Cell pellet prepared as described above for Western was used for the preparation of nuclear extract. Cell pellet was gently resuspended in 250 µl 1X hypotonic buffer (20 mM HEPES, pH 7.5, 5 mM NaF, 10 µM Na2MoO4, 0.1 mM EDTA) and incubated for 15 minutes on ice. It was mixed with 12.5 µl detergent, vortexed for 10 seconds at highest speed and centrifuged for 30 seconds at 14, 000g. Nuclear pellet in the microfuge tube was resuspended in 25 µl complete lysis buffer (Active Motif), vortexed for 10 seconds at highest setting and incubated for 30 minutes on ice in a rocker set at 150 rpm. Nuclear pellet suspension was again vortexed for 30 seconds at highest speed and centrifuged for 10 minutes at 14,000 g in a microcentrifuge. The supernatant (nuclear fraction) was stored at −80°C. Protein concentration was quantified by Biorad Reagent.
Nuclear Ku was quantified by Ku DNA repair kits from Active Motif, USA as followed by Ayene et al. (38). Forty microliter of AM6 binding buffer was added to each well of an eight-well strip. Nuclear extract (0.5µg) in a total volume of 10 µl complete lysis buffer was then added to each well and incubated for 1 h at room temperature on a orbital shaker at 100 rpm. After incubation, each well was washed three times with 200 µl of 1X washing buffer. Ku70 antibody at a 1:1000 dilution in 1X anti body binding buffer was added to each well and incubated for 1 h at room temperature on an orbital shaker at 100 rpm. After incubation, the wells were washed 4 times with 200 µl 1X washing buffer. For colorimetric reaction, 100 µl developing solution was added to each well and incubated for 4 minutes at room temperature in dark. The reaction was stopped by mixing it with 100 µl of stop solution. The O.D was read in a microplate reader at 450nm with a reference wavelength of 655 nm.
Irradiation Treatment
Cells plated in 60mm dishes with and without glucose after desired incubation time were exposed to 4Gy of radiation (Dose rate- 9.63Gy/min) at room temperature in air using a J.L. Shepherd Mark I 137Cs irradiator.
Determination of Cell Viability
Immediately after irradiation, the cells were replenished with fresh DMEM growth medium containing glucose, 15%FBS and 25 mM HEPES. Each dish containing approximately 1.4 million cells was incubated at 37°C in a humidified 5% CO2 incubator. After 18 hours incubation, the attached cells in these dishes were trypsinized, mixed with medium and counted using a Coulter counter. Approximately 20, 000 cells from each sample were plated in a new six well plate with complete growth medium (glucose,15% FCS, 25 mM HEPES) and incubated in a humidified 5% CO2 incubator at 37°C for 5 days before measuring survival. The survival was measured by counting total number of cells in each dish as described above using a Coulter counter. The survival fraction was calculated using the following formula:
Statistical Analysis
Each experiment was repeated at least three times. Values were presented as Means + Standard Error (SE). The statistical significance of the differences between the groups was determined by Analysis of Variance (ANOVA) with the ‘p’ values presented in the legend of each figure.
RESULTS
Extracellular Glucose in glucose and glucose free medium
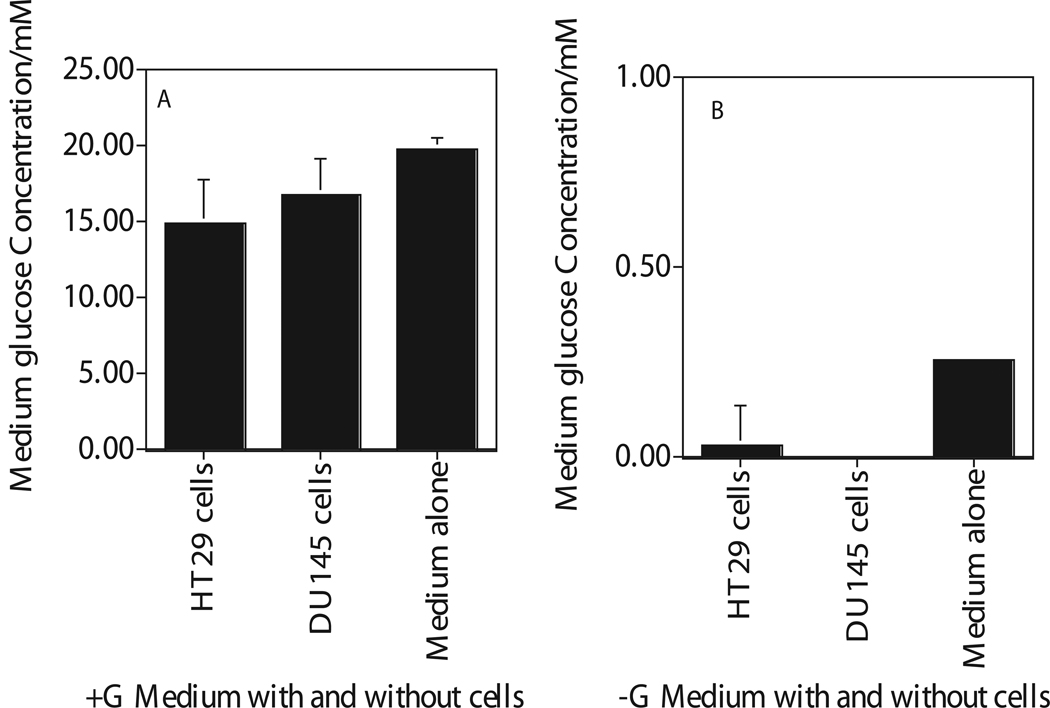
The concentration of extracellular glucose was determined after 4hr incubation of cells in glucose- or glucose free-medium by enzymatic assay that uses hexokinase (HK) to oxidize glucose (Figures 1). Glucose concentration in DMEM medium with glucose was 19 mM (Figure 1a). HK assay showed almost undetectable level of glucose in glucose free DMEM medium with 2% dialyzed FBS measured after 4 hours incubation with these cells (Figure 1b). These results suggested that addition of 2% dialyzed fetal calf serum did not cause significant increase in glucose level in glucose free medium (Figure 1b).
Figure 1.
Enzymatic quantification of extracellular glucose in the medium removed after 4 hours incubation of human cancer cells (HT29, DU145) in glucose (Figure A) and glucose free (Figure B) medium. Medium with Glucose, +G; Medium with zero glucose, −G. Each data point plotted was the mean ± standard error (SE) of at least three experiments with SE as shown unless smaller than points plotted. Statistical comparisons for glucose levels in glucose free medium relative to glucose medium was significant (P<0.01) for both the cells and the medium.
Intracellular Glucose for cells incubated for 4 hours in glucose and glucose free medium
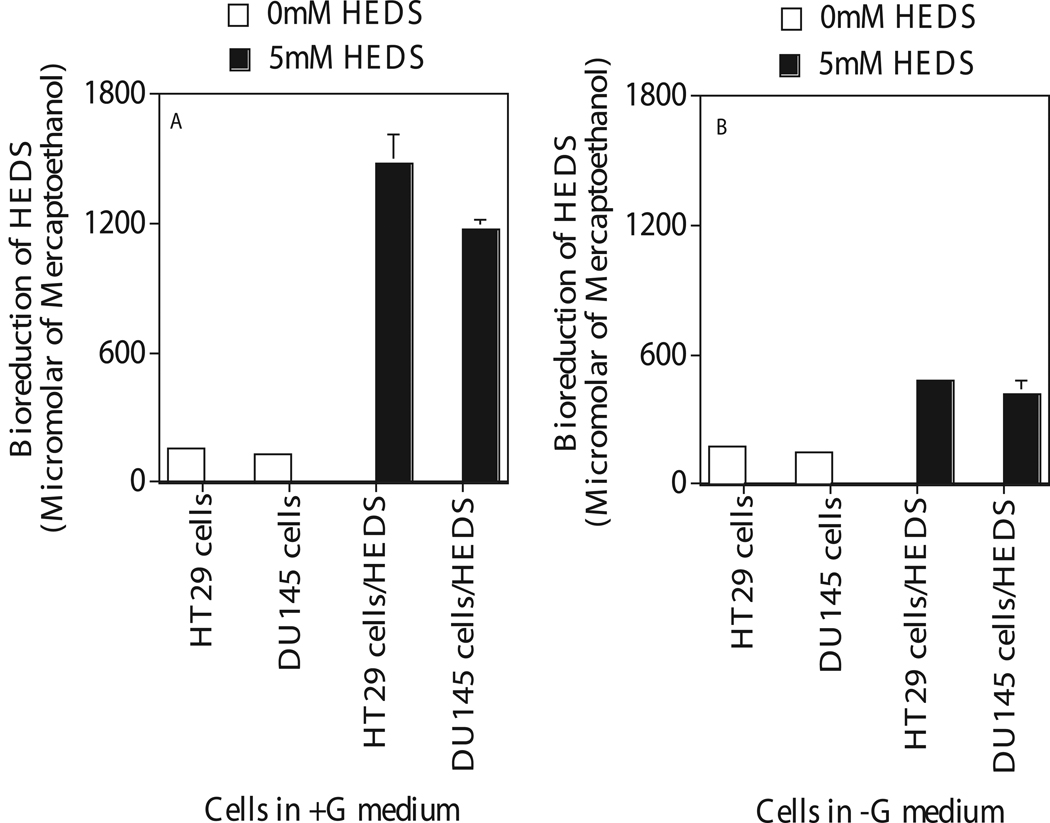
Intracellular glucose was measured by measuring the bioreduction of HEDS (Figure 2). All mammalian cells can convert HEDS into ME by oxidative pentose phosphate cycle using glucose as the substrate (37, 38). Most of the intracellular ME produced by this mechanism is released into the medium. Therefore, extracellular medium can be used to quantify ME produced by the bioreduction of HEDS. All thiols including ME will react with DTNB and this reaction can be quantified by spectrophotometer. In glucose medium, HT29 and DU145 cells converted 5 mM HEDS into 1600 µM of mercaptoethanol in one hour (Figure 2a). In glucose free medium, these cancer cells showed a 70% decrease in bioreduction compared to that measured in glucose medium suggesting a lack of glucose mediated intracellular metabolic activity in these cells (Figure 2b).
Figure 2.
Quantification of intracellular glucose after 4 hours incubation of human cancer cells (HT29, DU145) in glucose (Figure A) and glucose free (Figure B) medium. Medium with Glucose, +G; Medium with zero glucose, −G. Glucose was measured by medium based bioreduction/DTNB assay that quantifies the glucose mediated cellular metabolic activity. Each data point plotted was the mean ± standard error (SE) of at least three experiments with SE as shown unless smaller than points plotted. Statistical comparisons for mercaptoethanol (ME) levels in glucose free medium relative to glucose medium was significant (P<0.001) for both cells.
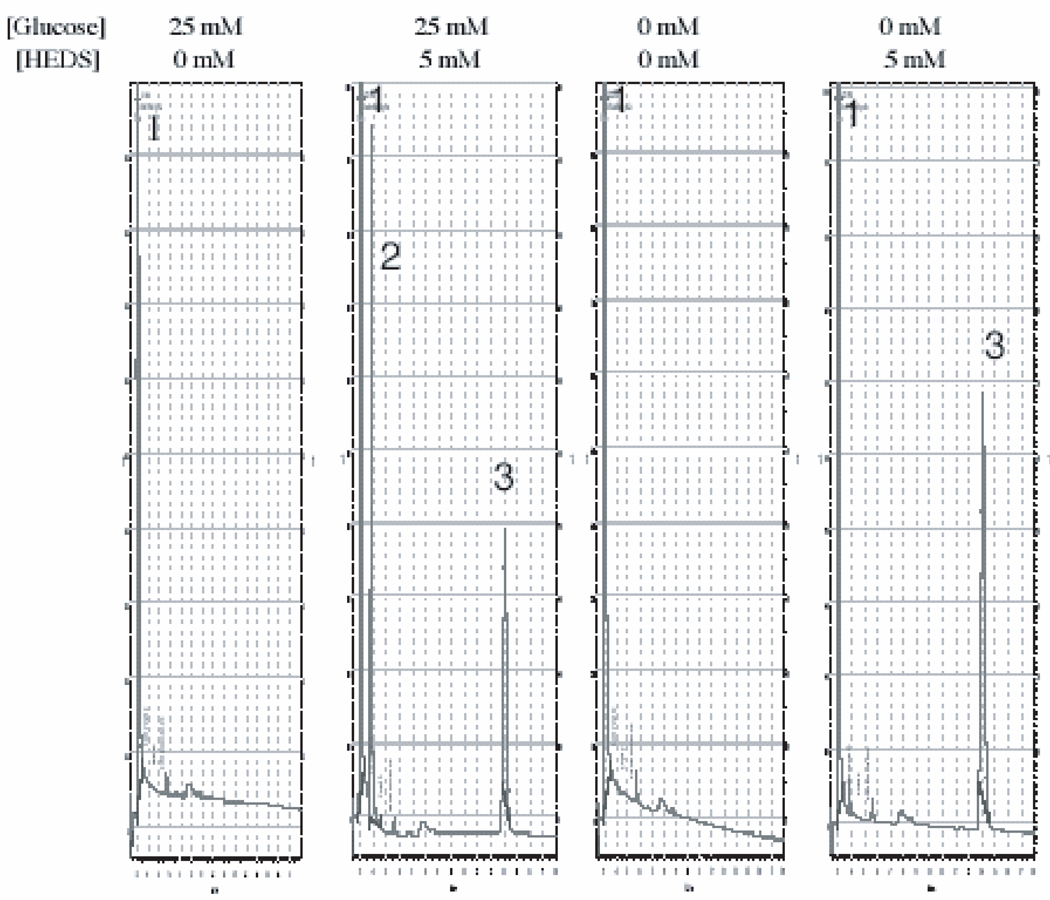
Although DTNB assay has demonstrated glucose dependent bioreduction, this assay measures all DTNB reactive thiols in the medium. The specificity of this DTNB based assay for bioreduction of HEDS was also determined in these cancer cells. An HPLC/electrochemical method was used to determine whether the data obtained by DTNB assay specifically measures the conversion of HEDS into ME (Figure 3).
Figure 3.
A representative HPLC/EC separation of HEDS and ME demonstrating the specificity of medium based glucose dependent bioreductive assay determined by DTNB assay in Figure 2. (peaks 1-SSA; 2-ME; 3-HEDS). Each experiment was repeated at least three times. The area of the individual peak measured and normalized to standard ME by Agilent EZChrom Elite application was used to calculate the concentration of ME. The mercaptoethanol levels in glucose and glucose free medium are 1825±62 and 396±17µM respectively. Statistical comparisons for mercaptoethanol (ME) levels in glucose free medium relative to glucose medium was significant (P<0.0001).
Preliminary experiments showed that thiol standards cysteine, mercaptoethanol (ME), glutathione, mercaptopropionyl glycine, glutathione disulfide and HEDS are eluted at 2.1, 3.8, 4.1, 10.8 and 14.4 minutes (results not shown). These results demonstrated that HPLC/EC method could be used to quantify the bioreduction of HEDS i.e. conversion of HEDS into mercaptoethanol (ME) without interference from other small molecular weight thiol compounds in the medium. HPLC tracing with ME peak (peak 2) observed from the extract of extracellular medium of a human colon cancer cell treated with HEDS is shown in Figure 3. The extracellular medium from cells either in the presence and absence of glucose showed only a solvent peak (peak 1). However, the medium from cells treated with HEDS for three hours in the presence of glucose showed a ME peak (peak 2) and HEDS peak (peak 3). This suggested that the extracellular medium did not contain any other DTNB reactive thiols. Further, the HPLC data demonstrated a glucose dependent bioreduction of HEDS similar to that observed for DTNB assay. These results suggested that the data obtained by medium based DTNB assay (Figure 2) is specific for cellular bioreduction of HEDS, which is an indirect measure of intracellular glucose depletion.
Radiation response of cancer cells incubated for 4 hours in glucose and glucose free medium
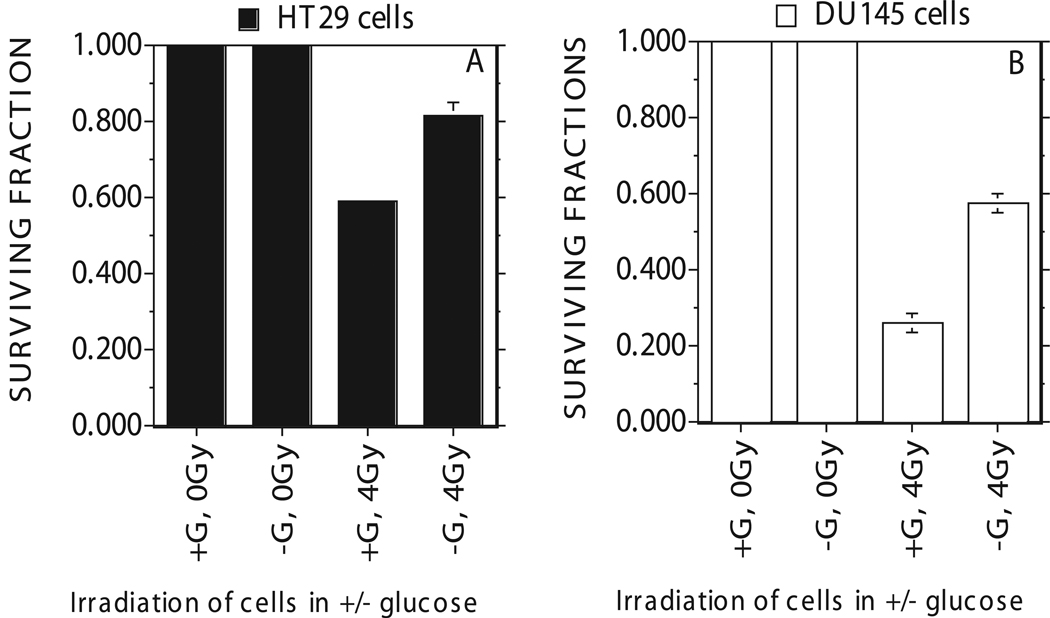
For studies to determine the impact of glucose deprivation on radiation response, these cancer cells were exposed to a clinically relevant dose of 4 Gy (Figures 4a,b). The surviving fraction calculated for cells exposed to 4Gy radiation showed a surviving fraction of 0.6 for HT29 cells (Figure 4a) in glucose containing DMEM medium with 2%FCS. In glucose free DMEM medium, HT29 cancer cells showed a significantly higher surviving fraction of 0.8 compared to 0.6 in the presence of glucose (Figure 4a).
Figure 4.
Radiation induced cell death measured for HT29 human colon cancer cells (Figure A) and DU145 prostate cancer cells (Figure B) after four hours of glucose starvation in DMEM growth medium consisting of 2% dialyzed FBS and 25 mM HEPES. Medium with Glucose, +G; Medium with zero glucose, −G. Each data point plotted was the mean ± standard error (SE) of at least three experiments with SE as shown unless smaller than points plotted. Statistical comparisons for cell survival (Surviving Fraction) in glucose free medium relative to glucose medium was significant (P<0.01) for both cells.
These assays also showed a surviving fraction of 0.25 for DU145 cells in glucose containing DMEM medium after 4 Gy radiation (Figure 4b). In glucose free DMEM medium, DU145 cancer cells showed a significant increase in survival after 4 Gy radiation with a surviving fraction of 0.6 compared to 0.25 in the presence of glucose (Figure 4b).
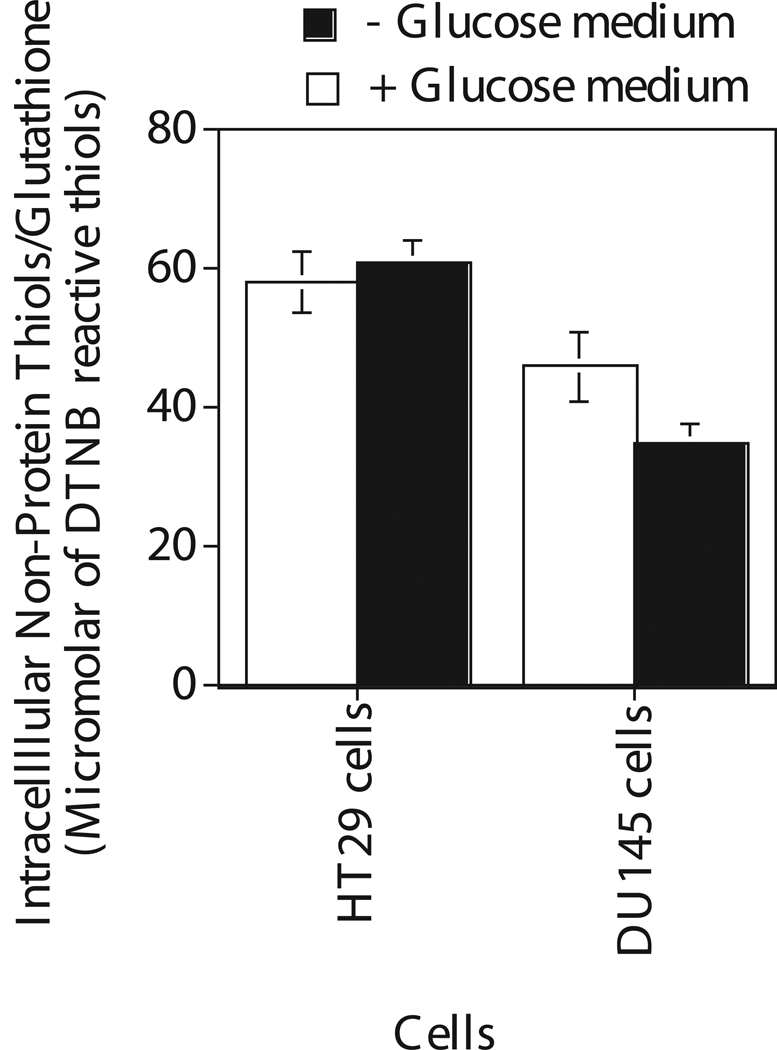
Intracellular non-protein thiols in cells incubated for 4 hours in glucose and glucose free medium
Intracellular non protein thiol (NPSH) was also estimated in these cancer cells to determine any correlation between NPSH and glucose regulated radiation response since NPSH is known to modulate cellular toxicity (Figure 5). This assay measures mostly GSH in cells since 90% of the total intracellular NPSH is glutathione (37). In glucose medium, DTNB reactive intracellular NPSH in HT29 and DU145 cells were almost the same. In glucose free medium, GSH levels in these cells were slightly decreased or remained the same as compared to that in glucose medium.
Figure 5.
Quantification of intracellular non-protein thiols as measured by dithiobis nitrobenzoic acid in human cancer cells HT29 and DU145 after 4 hours incubaton in the medium with and without glucose. Each data point plotted was the mean ± standard error (SE) of at least three experiments with SE as shown unless smaller than points plotted. Statistical comparisons for non-protein thiol (NPSH) levels in glucose free medium relative to glucose medium was not significant.
Intracellular GRP78 in cells incubated for 4 hours in glucose and glucose free medium
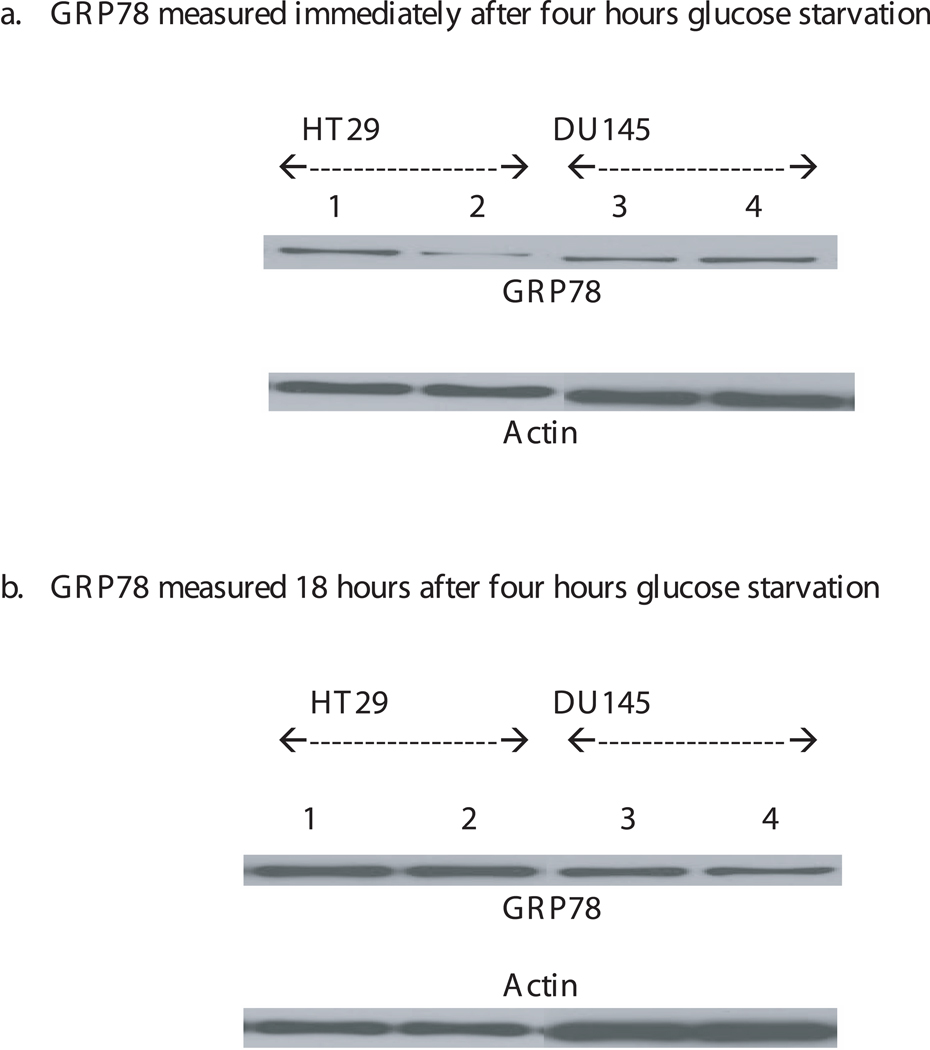
GRP78 levels were measured in these cells to determine its role in the radiation resistance induced by glucose depletion (Figure 6a,b). Western blot analysis of these cancer cells for GRP78 did not show any significant increase in GRP78 in any of these cells after 4 hours incubation in the absence of glucose as compared to that in the presence of glucose (Figure 6a). We have also checked the level of GRP78 in these cells at 18 hours after 4 hours glucose starvation by replacing the glucose free medium with glucose medium right after 4 hours glucose starvation (Figure 6b). The GRP78 levels did not increase in these cells grown in glucose free medium.
Figure 6.
A representative Western blot analysis of GRP78 expression measured at 0 and 18hrs after 4 hours glucose starvation in HT29 (lanes 1 & 2) and DU145 (lanes 3 & 4) cells grown in the presence (lanes 1 & 3) and absence of glucose (lanes 2 & 4). Each experiment was repeated at least three times. The intensity of the protein bands in the western blot was calculated by NIH image analysis and the GRP78 protein band was normalized to actin. The normalized GRP78 values was used to calculate the percentage of GRP78 in glucose starved cells compared to that in cells with glucose. The percentages of GRP78 levels measured in HT29 and DU145 cells immediately after 4hrs glucose starvation are 101 ± 4.5 and 92.53 ± 10.05 respectively. The percentages of GRP78 levels measured in HT29 and DU145 cells 18 hours after 4hrs glucose starvation are 128 ± 22 and 75.40 ± 8.7 respectively. Statistical comparisons for GRP78 protein levels in glucose free medium relative to glucose medium was not significant.
DNA repair protein Ku in cells incubated for 4 hours in glucose and glucose free medium
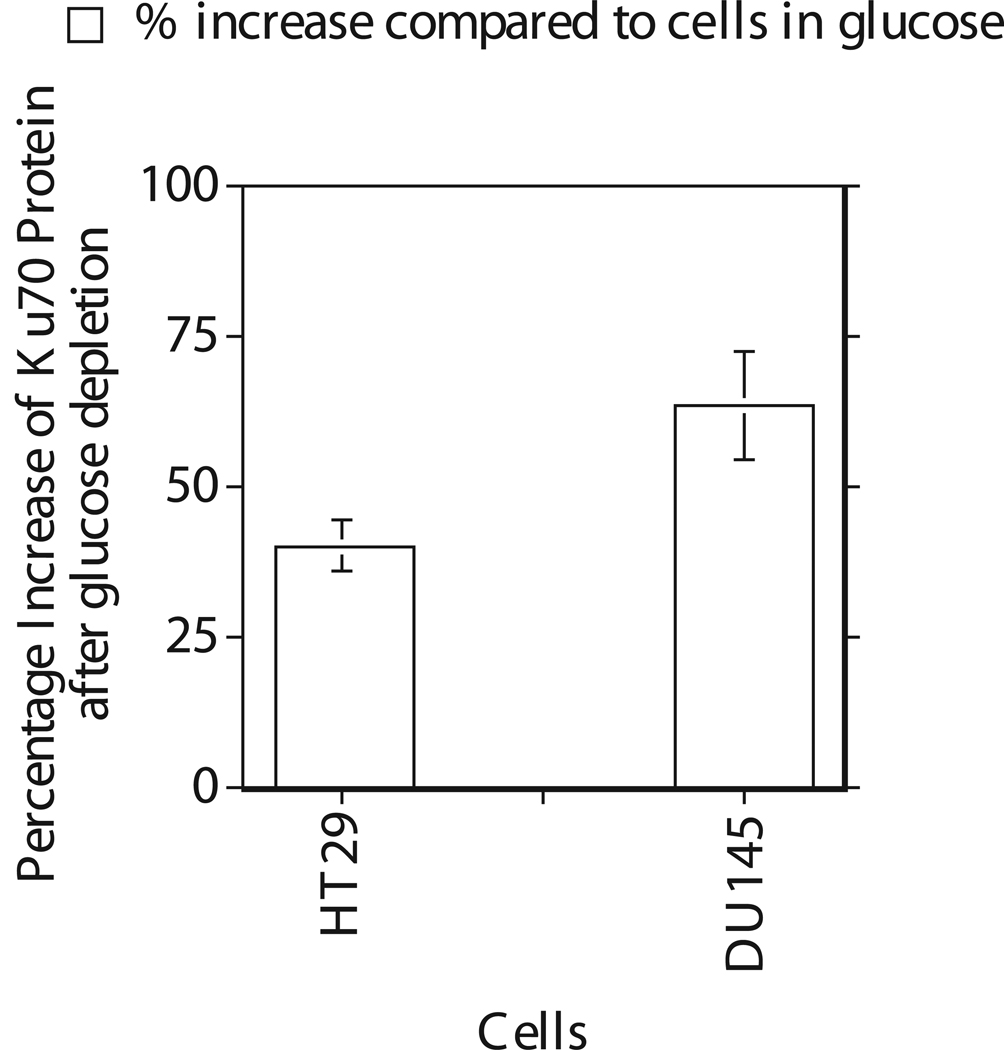
A direct correlation between increased response to radiation induced cell death and Ku mutation has been demonstrated in CHO cells (12–14). Studies have also demonstrated that targeting of Ku by small interfering RNA in human cancer cells enhanced their sensitivity to radiation and etoposide (18, 20). Therefore, Ku was quantified in the nuclei since nuclear Ku is responsible for the repair of DNA double strand breaks induced by γ radiation. ELISA analysis of nuclear extract from glucose deprived HT29 and DU145 cells showed a 40 to 60% increase in nuclear Ku as compared to that in cells grown in glucose medium (Figure 7).
Figure 7.
Over expression of Ku as measured by ELISA in human cancer cells HT29 and DU145 after 4 hours incubation in the medium with and without glucose. Each data point plotted was the mean ± standard error (SE) of at least three experiments with SE as shown unless smaller than points plotted. Statistical comparisons for Ku protein levels in glucose free medium relative to glucose medium was significant (P<0.001) for both cells.
DISCUSSION
Several studies have indicated that the therapeutic response of tumors may be modulated by intrinsic molecular factors such as over expression of oncogenes, repair genes and/or mutation of tumor suppressor genes (4–14). In addition, the microenvironmental factors such as hypoxia, hypoxia related gene expression and nutrient deprivation might also contribute to the overall outcome of cancer therapy (1, 23). Although the radiation response of HT29 and to some extent DU145 have been well established, the impact of glucose deprivation on the response of these human cancer cells to γ radiation has not been demonstrated. In this report, we have studied the impact of glucose deprivation on the response of human colon (HT29) and prostate (DU145) cancer cells to γ radiation since glucose depletion is associated with diffusion -limited hypoxia and higher metabolic activity (31–33).
Microenvironment in solid tumors expands from hypoxia to cytokines, stress, nutrient deprivation, etc. In order to overcome these adversities, cancer cells express or over express genes, which enable them to survive and grow. A recent report had shown that tumors survive and grow by taking advantage of complementary metabolic pathways between cancer cells and the newly formed stroma and vasculature (39). Our results demonstrated, for the first time, that the radiation response of HT29 and DU145 human cancer cells were decreased up to 2.0 fold when these cells were exposed to γ radiation in the absence of glucose. These results demonstrated that glucose depletion mediated stress regulated the response of cancer cells to radiation in both colon and prostate cancer cells. These results suggested that glucose deprivation, which is common in most solid tumors, make human cancer cells more resistant to radiation than that has been reported for cells grown in glucose.
The glucose deprivation in these cells was confirmed by measuring the glucose level in extracellular medium by enzymatic glucose assay. The assay demonstrated that the glucose level in glucose depleted medium is 0.1mM as compared to approximately 15 mM in glucose medium at the end of four hour incubation. In addition, a bioreductive assay was also used to determine the intracellular glucose level under these conditions. The bioreductive assay showed a five fold decrease in bioreductive capacity in glucose deprived cells as compared to cells grown in glucose medium suggesting that the intracellular glucose level also low at the time of irradiation.
Previous studies had shown that depletion of GSH by BSO increased the oxidative stress mediated cell death (21, 22). These studies had also suggested that glutathione may be an important determinant of the cell's ability to repair DNA damage and resist cell death (21, 22). However, in glucose free medium, GSH levels in these cancer cells remained the same and did not show any correlation with radiation response. These results demonstrated that GSH did not play a role in the radiation resistance of these human cancer cells induced by glucose depletion mediated stress since there was no increase in GSH after glucose deprivation.
Studies had looked into the impact of stress on both survival and GRP78 expression of mammalian cells in in vitro (24, 27, 28, 40). Reports on the role of GRP78 in cancer cells response to chemotherapeutic agents varied from increased resistance to sensitivity. Zhang et al demonstrated a correlation between parthenolide depleted intracellular thiols, increase in intracellular reactive oxygen species (ROS), calcium levels and GRP78 protein (41). However, these changes preceded parthenolide-induced cell death in these cells suggesting a correlation between induction of GRP78, which is a marker for endoplasmic reticulum stress, and cell death. However, others had shown that GRP78 small interfering RNA decreased the resistance of GRP78 over expressed breast cancer cells to etoposide, which suggested that GRP78 conferred resistance to etoposide (42). Similarly, studies had reported that cells developed resistance to etoposide with concomitant increase in GRP78 after 6-aminonicotinamide treatment (43). Later studies from the same laboratory showed that the GRP78- over expressing V79 and colon cancer cells are hypersensitive to DNA cross-linking agents melphalan, cisplatin and 1,3-bis(2-chloroethyl)-1-nitrosourea (BCNU) compared to the control cells (44).
Current studies in these cancer cells showed that 4 hours glucose starvation did not induce GRP78 protein expression but caused radiation resistance in two different types of human cancer cells. This is consistent with other reports that had shown no significant increase in GRP78 protein expression and mRNA in certain cells until after 12 hours of glucose starvation (45). This suggested that GRP78 is not involved in the radiation resistance of glucose deprived cancer cells that was observed after 4 hours glucose deprivation. In contrast, results from these studies showed that 4 hours glucose starvation increased nuclear Ku, a DNA repair protein, in HT29 and DU145 cells. These results suggested that glucose depletion mediated stress increased the expression of nuclear Ku, which is known to play a major role in the repair of lethal DNA lesions induced by γ radiation (12–16), and radiation resistance in both prostate and colon cancer cells.
The survival of tumor cells is believed to be regulated by both inherent cellular response and tumor microenvironment (46). It had been shown that inherent cellular response was important in the survival of some tumor cells to hypoxia induced stress (47). Radiation resistance of some primary tumor cells had been shown to be independent of inherent cellular response (48). A recent study had demonstrated that the clinical outcome of patients with classic Hodgkin lymphoma was dependent on genes related to tumor microenvironment, cell growth/apoptosis and regulation of mitosis (49). The radioresistance induced by glucose deprivation irrespective of the differences in inherent cellular responses of two different types of cancer cells is consistent with previous reports that the microenvironment regulated by tumor vasculature may be an important determinant of tumor survival after therapy (46, 48, 49).
Acknowledgments
FUNDING:
This work was supported by grants from the National Institutes of Health (CA 109604) and Pennsylvania Department of Health (SAP#: 4100042735) to ISA. K. M. Ward is a recipient of graduate research assistantship from the Brook J. Lenfest Foundation.
The abbreviations used are
- GSH
glutathione
- G6PD
glucose 6 phosphate dehydrogenase
- HEDS
hydroxyethyldisulfide
- ROS
Reactive oxygen species
- OPPC
oxidative pentose phosphate cycle
- ME
mercaptoethanol
- GRP78
Glucose regulated protein78
- NPSH
non protein thiol
- DTNB
–dithiobisnitrobenzoic acid
Footnotes
This work was presented at the 13th International Congress of Radiation Research, July 8–12, 2007, San Francisco, USA.
REFERENCES
- 1.Helmlinger G, Yuan F, Dellian M, Jain RK. Interstitial pH and pO2 gradients in solid tumors in vivo: high-resolution measurements reveal a lack of correlation. Nat Med. 1997;3:177–182. doi: 10.1038/nm0297-177. [DOI] [PubMed] [Google Scholar]
- 2.Brown JM, Giaccia AJ. The unique physiology of solid tumors: opportunities (and problems) for cancer therapy. Cancer Res. 1998;58:1408–1416. [PubMed] [Google Scholar]
- 3.Izuishi K, Kato K, Ogura T, Kinoshita T, Esumi H. Remarkable tolerance of tumor cells to nutrient deprivation: possible new biochemical target for cancer therapy. Cancer Res. 2000;60:6201–6207. [PubMed] [Google Scholar]
- 4.Blaszyk H, Hartmann A, Cunningham JM, Schaid D, Wold LE, Kovach JS, Sommer SS. A prospective trial of midwest breast cancer patients: a p53 gene mutation is the most important predictor of adverse outcome. Int J Cancer. 2000;89:32–38. doi: 10.1002/(sici)1097-0215(20000120)89:1<32::aid-ijc6>3.0.co;2-g. [DOI] [PubMed] [Google Scholar]
- 5.Pirollo KF, Bouker KB, Chang EH. Does p53 status influence tumor response to anticancer therapies? Anticancer Drugs. 2000;11:419–432. doi: 10.1097/00001813-200007000-00002. [DOI] [PubMed] [Google Scholar]
- 6.Bristow RG, Benchimol S, Hill RP. The p53 gene as a modifier of intrinsic radiosensitivity: implications for radiotherapy. Radiother Oncol. 1996;40:197–223. doi: 10.1016/0167-8140(96)01806-3. [DOI] [PubMed] [Google Scholar]
- 7.Cartron PF, Juin P, Oliver L, Meflah K, Vallette FM. Impact of proapoptotic proteins Bax and Bak in tumor progression and response to treatment. Expert Rev Anticancer Ther. 2003;3:563–570. doi: 10.1586/14737140.3.4.563. [DOI] [PubMed] [Google Scholar]
- 8.Kyprianou N, King ED, Bradbury D, Rhee JG. bcl-2 over-expression delays radiation-induced apoptosis without affecting the clonogenic survival of human prostate cancer cells. Int J Cancer. 1997;70:341–348. doi: 10.1002/(sici)1097-0215(19970127)70:3<341::aid-ijc16>3.0.co;2-i. [DOI] [PubMed] [Google Scholar]
- 9.Ayene IS, Bernhard EJ, McKenna WG, Muschel RJ, Krisch RE, Koch CJ. DNA as an important target in radiation-induced apoptosis of MYC and MYC plus RAS transfected rat embryo fibroblasts. Int J Radiat Biol. 2000;76:343–354. doi: 10.1080/095530000138682. [DOI] [PubMed] [Google Scholar]
- 10.Geng L, Walter S, Melian E, Vaughan AT. Transfection of a vector expressing wild-type p53 into cells of two human glioma cell lines enhances radiation toxicity. Radiat Res. 1998;150:31–37. [PubMed] [Google Scholar]
- 11.Minagawa Y, Kigawa J, Itamochi H, Kanamori Y, Shimada M, Takahashi M, Terakawa N. Cisplatin-resistant HeLa cells are resistant to apoptosis via p53-dependent and -independent pathways. Jpn J Cancer Res. 1999;90:1373–1379. doi: 10.1111/j.1349-7006.1999.tb00722.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Kemp LM, Sedgwick SG, Jeggo PA. X-ray sensitive mutants of Chinese hamster ovary cells defective in double-strand break rejoining. Mutat Res. 1984;132:189–196. doi: 10.1016/0167-8817(84)90037-3. [DOI] [PubMed] [Google Scholar]
- 13.Koberle B, Masters JR, Hartley JA, Wood RD. Defective repair of cisplatin-induced DNA damage caused by reduced XPA protein in testicular germ cell tumours. Curr Biol. 1999;9:273–276. doi: 10.1016/s0960-9822(99)80118-3. [DOI] [PubMed] [Google Scholar]
- 14.Rosenberg E, Taher MM, Kuemmerle NB, Farnsworth J, Valerie K. A truncated human xeroderma pigmentosum complementation group A protein expressed from an adenovirus sensitizes human tumor cells to ultraviolet light and cisplatin. Cancer Res. 2001;61:764–770. [PubMed] [Google Scholar]
- 15.Ayene IS, Ford LP, Koch CJ. Ku protein targeting by Ku70 small interfering RNA enhances human cancer cell response to topoisomerase II inhibitor and gamma radiation. Mol Cancer Ther. 2005;4:529–536. doi: 10.1158/1535-7163.MCT-04-0130. [DOI] [PubMed] [Google Scholar]
- 16.Peng Y, Zhang Q, Nagasawa H, Okayasu R, Liber HL, Bedford JS. Silencing expression of the catalytic subunit of DNA-dependent protein kinase by small interfering RNA sensitizes human cells for radiation-induced chromosome damage, cell killing, and mutation. Cancer Res. 2002;62:6400–6404. [PubMed] [Google Scholar]
- 17.Chang M, Shi M, Forman HJ. Exogenous glutathione protects endothelial cells from menadione toxicity. Am J Physiol. 1992;262:L637–L643. doi: 10.1152/ajplung.1992.262.5.L637. [DOI] [PubMed] [Google Scholar]
- 18.DeLeve LD, Wang X. Role of oxidative stress and glutathione in busulfan toxicity in cultured murine hepatocytes. Pharmacology. 2000;60:143–154. doi: 10.1159/000028359. [DOI] [PubMed] [Google Scholar]
- 19.Phimister AJ, Lee MG, Morin D, Buckpitt AR, Plopper CG. Glutathione depletion is a major determinant of inhaled naphthalene respiratory toxicity and naphthalene metabolism in mice. Toxicol Sci. 2004;82:268–278. doi: 10.1093/toxsci/kfh258. [DOI] [PubMed] [Google Scholar]
- 20.Stenius U, Warholm M, Rannug A, Walles S, Lundberg I, Hogberg J. The role of GSH depletion and toxicity in hydroquinone-induced development of enzyme-altered foci. Carcinogenesis. 1989;10:593–599. doi: 10.1093/carcin/10.3.593. [DOI] [PubMed] [Google Scholar]
- 21.Pendyala L, Perez R, Weinstein A, Zdanowicz J, Creaven PJ. Effect of glutathione depletion on the cytotoxicity of cisplatin and iproplatin in a human melanoma cell line. Cancer Chemother Pharmacol. 1997;40:38–44. doi: 10.1007/s002800050622. [DOI] [PubMed] [Google Scholar]
- 22.Yang Z, Faustino PJ, Andrews PA, Monastra R, Rasmussen AA, Ellison CD, Cullen KJ. Decreased cisplatin/DNA adduct formation is associated with cisplatin resistance in human head and neck cancer cell lines. Cancer Chemother Pharmacol. 2000;46:255–262. doi: 10.1007/s002800000167. [DOI] [PubMed] [Google Scholar]
- 23.Tomida A, Tsuruo T. Drug resistance mediated by cellular stress response to the microenvironment of solid tumors. Anticancer Drug Des. 1999;14:169–177. [PubMed] [Google Scholar]
- 24.Lee AS. The glucose-regulated proteins: stress induction and clinical applications. Trends Biochem Sci. 2001;26:504–510. doi: 10.1016/s0968-0004(01)01908-9. [DOI] [PubMed] [Google Scholar]
- 25.Wyld L, Tomlinson M, Reed MW, Brown NJ. Aminolaevulinic acid-induced photodynamic therapy: cellular responses to glucose starvation. Br J Cancer. 2002;86:1343–1347. doi: 10.1038/sj.bjc.6600234. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Reddy RK, Mao C, Baumeister P, Austin RC, Kaufman RJ, Lee AS. Endoplasmic reticulum chaperone protein GRP78 protects cells from apoptosis induced by topoisomerase inhibitors: role of ATP binding site in suppression of caspase-7 activation. J Biol Chem. 2003;278:20915–20924. doi: 10.1074/jbc.M212328200. [DOI] [PubMed] [Google Scholar]
- 27.Shen J, Hughes C, Chao C, Cai J, Bartels C, Gessner T, Subjeck J. Coinduction of glucose-regulated proteins and doxorubicin resistance in Chinese hamster cells. Proc Natl Acad Sci U S A. 1987;84:3278–3282. doi: 10.1073/pnas.84.10.3278. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Rao RV, Peel A, Logvinova A, del Rio G, Hermel E, Yokota T, Goldsmith PC, Ellerby LM, Ellerby HM, Bredesen DE. Coupling endoplasmic reticulum stress to the cell death program: role of the ER chaperone GRP78. FEBS Lett. 2002;514:122–128. doi: 10.1016/s0014-5793(02)02289-5. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Scorrano L, Oakes SA, Opferman JT, Cheng EH, Sorcinelli MD, Pozzan T, Korsmeyer SJ. BAX and BAK regulation of endoplasmic reticulum Ca2+: a control point for apoptosis. Science. 2003;300:135–139. doi: 10.1126/science.1081208. [DOI] [PubMed] [Google Scholar]
- 30.Harding HP, Calfon M, Urano F, Novoa I, Ron D. Transcriptional and translational control in the Mammalian unfolded protein response. Annu Rev Cell Dev Biol. 2002;18:575–599. doi: 10.1146/annurev.cellbio.18.011402.160624. [DOI] [PubMed] [Google Scholar]
- 31.Aronen HJ, Pardo FS, Kennedy DN, Belliveau JW, Packard SD, Hsu DW, Hochberg FH, Fischman AJ, Rosen BR. High microvascular blood volume is associated with high glucose uptake and tumor angiogenesis in human gliomas. Clin Cancer Res. 2000;6:2189–2200. [PubMed] [Google Scholar]
- 32.Rajendran JG, Mankoff DA, O'Sullivan F, Peterson LM, Schwartz DL, Conrad EU, Spence AM, Muzi M, Farwell DG, Krohn KA. Hypoxia and glucose metabolism in malignant tumors: evaluation by [18F]fluoromisonidazole and [18F]fluorodeoxyglucose positron emission tomography imaging. Clin Cancer Res. 2004;10:2245–2252. doi: 10.1158/1078-0432.ccr-0688-3. [DOI] [PubMed] [Google Scholar]
- 33.Schroeder T, Yuan H, Viglianti BL, Peltz C, Asopa S, Vujaskovic Z, Dewhirst MW. Spatial heterogeneity and oxygen dependence of glucose consumption in R3230Ac and fibrosarcomas of the Fischer 344 rat. Cancer Res. 2005;65:5163–5171. doi: 10.1158/0008-5472.CAN-04-3900. [DOI] [PubMed] [Google Scholar]
- 34.Becker D, Summerfield S, Gillich S, Sevilla MD. Influence of oxygen on the repair of direct radiation damage to DNA by thiols in model systems. Int J Radiat Biol. 1994;65:537–548. doi: 10.1080/09553009414550631. [DOI] [PubMed] [Google Scholar]
- 35.Michael BD, Prise KM. A multiple-radical model for radiation action on DNA and the dependence of OER on LET. Int J Radiat Biol. 1996;69:351–358. doi: 10.1080/095530096145913. [DOI] [PubMed] [Google Scholar]
- 36.Prise KM, Folkard M, Davies S, Michael BD. The irradiation of V79 mammalian cells by protons with energies below 2 MeV. Part II. Measurement of oxygen enhancement ratios and DNA damage. Int J Radiat Biol. 1990;58:261–277. doi: 10.1080/09553009014551611. [DOI] [PubMed] [Google Scholar]
- 37.Ayene IS, Stamato TD, Mauldin SK, Biaglow JE, Tuttle SW, Jenkins SF, Koch CJ. Mutation in the glucose-6-phosphate dehydrogenase gene leads to inactivation of Ku DNA end binding during oxidative stress. J Biol Chem. 2002;277:9929–9935. doi: 10.1074/jbc.M111366200. [DOI] [PubMed] [Google Scholar]
- 38.Ayene IS, Biaglow JE, Kachur AV, Stamato TD, Koch CJ. Mutation in G6PD gene leads to loss of cellular control of protein glutathionylation: Mechanism and implication. J Cell Biochem. 2008;103:123–135. doi: 10.1002/jcb.21394. [DOI] [PubMed] [Google Scholar]
- 39.Koukourakis MI, Giatromanolaki A, Harris AL, Sivridis E. Comparison of metabolic pathways between cancer cells and stromal cells in colorectal carcinomas: a metabolic survival role for tumor-associated stroma. Cancer Res. 2006;66:632–637. doi: 10.1158/0008-5472.CAN-05-3260. [DOI] [PubMed] [Google Scholar]
- 40.Kato K, Ogura T, Kishimoto A, Minegishi Y, Nakajima N, Miyazaki M, Esumi H. Critical roles of AMP-activated protein kinase in constitutive tolerance of cancer cells to nutrient deprivation and tumor formation. Oncogene. 2002;21:6082–6090. doi: 10.1038/sj.onc.1205737. [DOI] [PubMed] [Google Scholar]
- 41.Zhang S, Ong CN, Shen HM. Critical roles of intracellular thiols and calcium in parthenolide-induced apoptosis in human colorectal cancer cells. Cancer Lett. 2004;208:143–153. doi: 10.1016/j.canlet.2003.11.028. [DOI] [PubMed] [Google Scholar]
- 42.Dong D, Ko B, Baumeister P, Swenson S, Costa F, Markland F, Stiles C, Patterson JB, Bates SE, Lee AS. Vascular targeting and antiangiogenesis agents induce drug resistance effector GRP78 within the tumor microenvironment. Cancer Res. 2005;65:5785–5791. doi: 10.1158/0008-5472.CAN-05-0754. [DOI] [PubMed] [Google Scholar]
- 43.Chatterjee S, Cheng MF, Berger SJ, Berger NA. Induction of M(r) 78,000 glucose-regulated stress protein in poly(adenosine diphosphate-ribose) polymerase- and nicotinamide adenine dinucleotide-deficient V79 cell lines and its relation to resistance to the topoisomerase II inhibitor etoposide. Cancer Res. 1994;54:4405–4411. [PubMed] [Google Scholar]
- 44.Chatterjee S, Hirota H, Belfi CA, Berger SJ, Berger NA. Hypersensitivity to DNA cross-linking agents associated with up-regulation of glucose-regulated stress protein GRP78. Cancer Res. 1997;57:5112–5116. [PubMed] [Google Scholar]
- 45.Chang SH, Barbosa-Tessmann I, Chen C, Kilberg MS, Agarwal A. Glucose deprivation induces heme oxygenase-1 gene expression by a pathway independent of the unfolded protein response. J Biol Chem. 2002;277:1933–1940. doi: 10.1074/jbc.M108921200. [DOI] [PubMed] [Google Scholar]
- 46.Hazlehurst LA, Landowski TH, Dalton WS. Role of the tumor microenvironment in mediating de novo resistance to drugs and physiological mediators of cell death. Oncogene. 2003;22:7396–7402. doi: 10.1038/sj.onc.1206943. [DOI] [PubMed] [Google Scholar]
- 47.Kiani MF, Fenton BM. Inherent cellular differences may explain the dissimilar survival of RIF-1 and KHT tumour cells under aerobic and hypoxic conditions. Int J Radiat Biol. 1995;67:449–452. doi: 10.1080/09553009514550511. [DOI] [PubMed] [Google Scholar]
- 48.Weichselbaum RR, Epstein J, Little JB, Kornblith P. Inherent cellular radiosensitivity of human tumors of varying clinical curability. AJR Am J Roentgenol. 1976;127:1027–1032. doi: 10.2214/ajr.127.6.1027. [DOI] [PubMed] [Google Scholar]
- 49.Sanchez-Aguilera A, Montalban C, de la Cueva P, Sanchez-Verde L, Morente MM, Garcia-Cosio M, Garcia-Larana J, Bellas C, Provencio M, Romagosa V, de Sevilla AF, Menarguez J, Sabin P, Mestre MJ, Mendez M, Fresno MF, Nicolas C, Piris MA, Garcia JF. Tumor microenvironment and mitotic checkpoint are key factors in the outcome of classic Hodgkin lymphoma. Blood. 2006;108:662–668. doi: 10.1182/blood-2005-12-5125. [DOI] [PubMed] [Google Scholar]