Abstract
A regulatory mechanism is introduced whereupon the catalytic activity of a given enzyme is controlled by ligand binding to a receptor domain of choice. A small enzyme (barnase) and a ligand-binding polypeptide (GCN4) are fused so that a simple topological constraint prevents them from existing simultaneously in their folded states. The two domains consequently engage in a thermodynamic tug-of-war in which the more stable domain forces the less stable domain to unfold. In the absence of ligand, the barnase domain is more stable and is therefore folded and active; the GCN4 domain is substantially unstructured. DNA binding induces folding of GCN4, forcibly unfolding and inactivating the barnase domain. Barnase–GCN4 is thus a “natively unfolded” protein that uses ligand binding to switch between partially folded forms. The key characteristics of each parent protein (catalytic efficiency of barnase, DNA binding affinity and sequence specificity of GCN4) are retained in the chimera. Barnase–GCN4 thus defines a modular approach for assembling enzymes with novel sensor capabilities from a variety of catalytic and ligand binding domains.
Keywords: molecular switch, GCN4, barnase, protein design, natively unfolded
We previously developed a novel two-domain fusion protein in which the mechanical stress imposed by the folded structure of one domain forces the other to unfold, and vice versa.1 This conformational switching mechanism is achieved by inserting protein X into a surface loop of protein Y, with the condition that the distance between amino and carboxyl ends of X is much greater than the distance between the termini of the loop in Y. If X is more stable than Y, X forcibly stretches and unfolds Y. If Y is more stable, it similarly distorts and denatures X. The fusion protein thus undergoes a thermodynamic tug-of-war in which only one domain can exist in its folded state at any given time. When X is human ubiquitin and Y is the bacterial ribonuclease barnase (Bn), this conformational equilibrium is reversible, cooperative, and controllable by external factors such as temperature, denaturant, and ligand binding.1
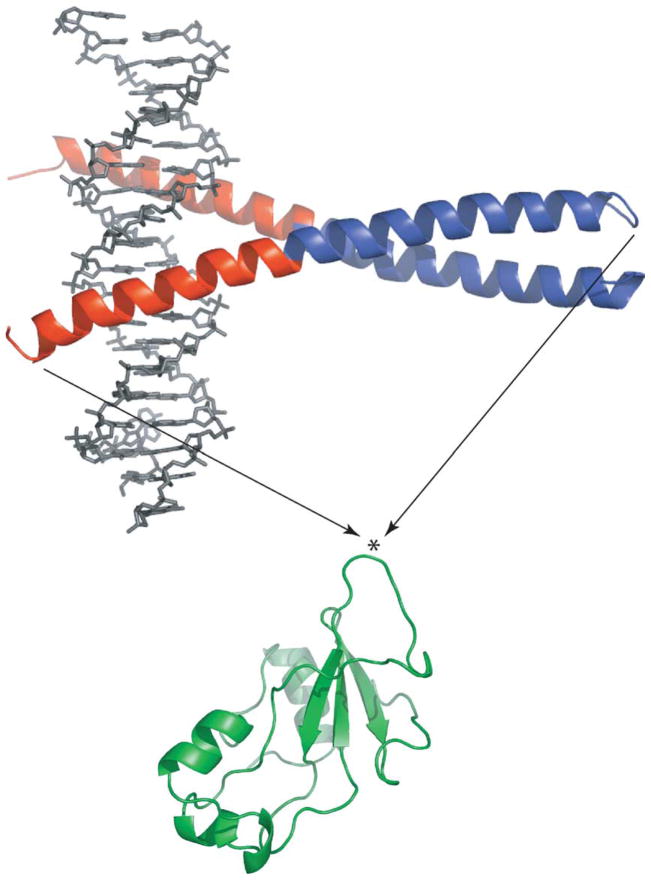
Here we introduce a new class of enzymes whose activity is regulated by binding of the appropriate ligand to a regulatory domain of choice. Ligand binding induces folding of the regulatory domain and thus unfolds the catalytic domain by the mutually exclusive folding mechanism. We have chosen Bn (110 amino acid residues) and the GCN4 DNA binding domain (56 amino acid residues) as the catalytic and regulatory subunits, respectively (Figure 1). GCN4 was inserted between residues 66 and 67 of Bn, which lie at the tip of a solvent-exposed loop, to generate the fusion protein BG. The Cα–Cα distance between the ends of the loop is approximately 10 Å. When bound to the AP-1 consensus DNA oligonucleotide, GCN4 adopts the homodimeric, parallel coiled-coil structure depicted in Figure 1.2 The end-to-end distance of 75 Å ensures that DNA binding will split the Bn domain in two, thereby inactivating it. In the absence of DNA, GCN4 can still dimerize via the C-terminal coiled-coil region (shown in blue) with a dissociation constant (Kd) of 6–9 nM.3–5 The 25 N-terminal residues that comprise the DNA binding region (shown in red), however, are largely unstructured.6,7 These residues are predicted to uncouple folding/unfolding of the Bn domain and the coiled-coil region of GCN4 by acting as a long, flexible linker. Bn is consequently expected to be folded and active if no DNA is present.
Figure 1.
Creation of the BG fusion protein from GCN4 (top) and Bn (bottom). The DNA binding and coiled-coil regions of GCN4 are colored red and blue, respectively. The bound DNA oligonucleotide is shown in grey. The asterisk indicates the point where GCN4 was inserted (between residues 66 and 67 of Bn). KpnI and NheI restriction sites were created to fuse the Bn and GCN4 genes. The extra nucleotides introduced Gly-Thr and Ala-Ser at the junction points. These dipeptides serve as short linkers. Images were generated by the Pymol program (DeLano Scientific).
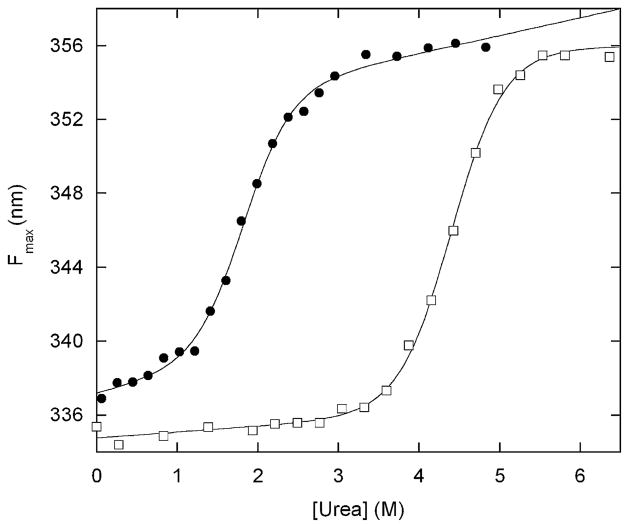
To test that hypothesis, we characterized the structure and stability of BG by fluorescence spectroscopy. All three Trp residues are in the Bn sequence, so Trp fluorescence reports primarily on the structure of the Bn domain. Free Bn exhibits fluorescence emission maxima (Fmax) of 335 nm and 356 nm in native and unfolded states (6 M urea), respectively (Figure 2). Fmax of BG is 337 nm in the absence of denaturant, suggesting that the Bn domain is folded in pH 7 buffer. Addition of urea unfolds both free Bn and the Bn domain of BG in a cooperative and reversible manner. Fitting these data to the linear extrapolation equation8 yields folding free energies of 7.4 kcal mol−1 and 3.7 kcal mol−1, respectively. It is apparent that insertion of the GCN4 domain, when it is largely disordered in the absence of DNA, does not unfold Bn.
Figure 2.
Urea-induced denaturation of BG (filled circles) and free Bn (open squares) monitored by Trp fluorescence maximum. Lines are best fit of the data to the linear extrapolation equation. Solution conditions are 200 nM protein (monomer concentration), 25 mM Hepes (pH 7.0), 100 mM NaCl at 25 ºC. Data were collected on a Fluoromax-3 fluorometer (Jobin-Yvon/SPEX) with an excitation wavelength of 280 nm. Emission maxima were calculated using the Datamax software package (Jobin-Yvon/SPEX). BG was expressed in Escherichia coli BL21(DE3) and purified using the same protocol developed for barnase–ubiquitin.1 One notable difference is that BG is found completely in inclusion bodies and is thus protected from proteolysis; the yield of BG is correspondingly much higher than that of barnase–ubiquitin.
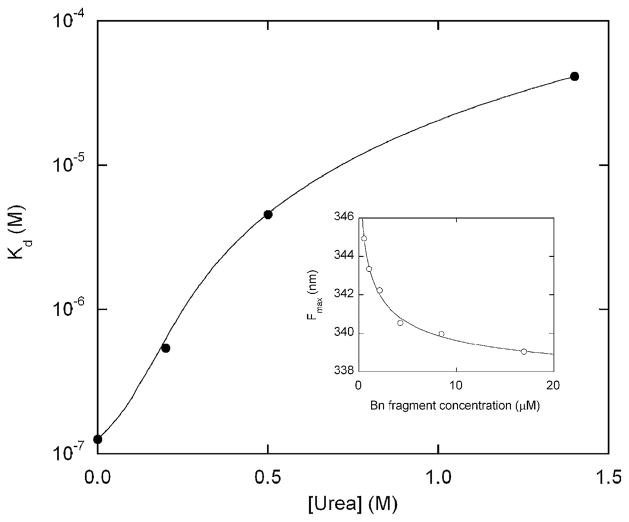
To test whether DNA binding to the GCN4 domain unfolds the Bn domain, we monitored the Fmax of BG as a function of AP-1 concentration. However, it was first necessary to establish conditions that minimize intermolecular complementation of the Bn fragments that are generated upon GCN4/DNA binding and folding. Intermolecular complementation is a direct consequence of the mutually exclusive folding mechanism. It occurs when the N-terminal Bn fragment binds with the C-terminal Bn fragment from another molecule. The resulting complex can refold to a species that exhibits native-like fluorescence spectra (Figure 3), enzymatic activity (not shown), and NMR spectra.9 To determine the apparent Kd for complementation, we dissolved various concentrations of Bn fragments 1–67 and 68–110 in 6 M urea and refolded them by dilution into buffer. Formation of the native complex was monitored by a shift in Fmax. The data are well fit by the simple 1:1 binding equation (Figure 3, inset), which yields an apparent Kd value for complementation of ~100 nM (Figure 3). As expected, binding weakens with increasing urea concentration, reflecting the coupling between binding and folding. We chose to perform the DNA binding experiments in 1.4 M urea because it disrupts intermolecular complementation while allowing the Bn domain of BG to remain largely folded (Figure 2). We point out that destabilizing Bn by mutation should in principle produce a similar effect and eliminate the need for urea.
Figure 3.
Urea dependence of the apparent dissociation constant for intermolecular complementation (Bn fragments 1–67 and 68–110). The line is meant to guide the eye only. Various concentrations of 1–67 and 68–110 Bn fragments (always present at a 1:1 ratio) were unfolded in 6 M urea then rapidly diluted to the urea concentration indicated. Refolding of the complex was monitored by shift in Trp fluorescence maximum (inset; data obtained in 0.2 M urea). Kd values were obtained by fitting fluorescence maxima to the simple 1:1 binding equation (continuous line in the inset indicates curve fit). Solution conditions are the same for Figure 2. Bn fragments 1–67 and 68–110 were prepared by digesting purified S67M mutant with CNBr.18
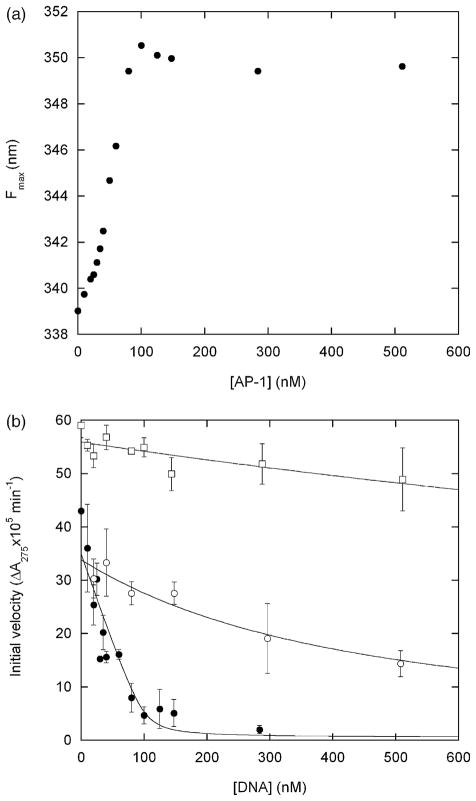
AP-1 binding by the GCN4 domain induces a large shift in fluorescence of the Bn domain (Figure 4(a)). The Fmax value of the fully bound species is 350 nm. The Fmax value of the urea-unfolded state of BG extrapolates to 353 nm at 1.4 M urea (Figure 2), suggesting that DNA binding induces nearly complete unfolding of the Bn domain. The binding curve is linear and breaks sharply at the point where the AP-1 concentration equals that of the BG dimer (100 nM). DNA binding is therefore stoichiometric and Kd is too low to be determined accurately; we estimate the upper limit to be ~5 nM. This value is consistent with the minimal mechanism of Scheme 1:
| (1) |
Figure 4.
(a) DNA binding-induced unfolding of the Bn domain of BG monitored by Trp fluorescence maximum. Samples were incubated with for 2 h with AP-1 oligonucleotide (5′-AGTGGAGATGACTCATCTCGTGC-3′) prior to measurements. (b) Inhibition of RNase activity by DNA binding. Filled and open circles designate BG incubated with AP-1 and with the non-consensus oligonucleotide 5′-CAGGGTGCTATGAACAAATGCCTCGAGCTGTTCCG T-3′, respectively. Open squares represent free Bn incubated with AP-1. Lines are best fits of the data to the simple binding equation. Fitted Kd values are ~2 nM (filled circles), 360 nM (open circles) and ~3 μM (open squares). Error bars represent standard deviations of three measurements. Samples were prepared as for (a) and assayed for RNase activity by addition of 20 μM guanylyl(3′-5′)uridine 3′-monophosphate.19 Substrate transesterification was monitored by absorbance at 275 nm on a Cary 100 spectrophotometer (Varian Instruments). Initial velocities were obtained from least-squares fits of the linear portion of the data. The concentration of free Bn was reduced to 60 nM to lower the initial velocity to measurable levels. Conditions are identical to those for Figure 2 except 1.4 M urea is present in all samples, and samples for enzyme assays contain 0.1 mg ml−1 bovine.
The presence and absence of an underscore indicates that the domain is folded and unfolded, respectively, and BG is the dimeric coiled-coil form of the protein. The observed Kd for DNA binding is equal to K1(1+K2), where K1 is the intrinsic dissociation constant for the GCN4–DNA interaction in the absence of a structured Bn domain, and K2 is the equilibrium constant for Bn folding when the DNA binding region of GCN4 is unstructured. K1 has been reported to be 2–20 nM for free GCN4.3,10 Extrapolation of ΔG to 1.4 M urea yields K2=4.2 (Figure 2). The observed Kd value for the BG–DNA complex is thus predicted to be 10–100 nM, in reasonable agreement with Figure 3. The higher than expected affinity of BG compared to free GCN4 may be due to the use of Hepes buffer in the present experiments and phosphate buffer in the GCN4 studies. Nevertheless, it is evident that BG retains the tight binding affinity of the parent GCN4 protein.
To determine whether DNA binding switches off enzymatic function, we measured BG ribonuclease activity under the same conditions as those for Figure 4(a). One equivalent of AP-1 reduces activity to background levels (Figure 4(b)). One explanation is that DNA acts as a competitive inhibitor by binding to the BG active site. This possibility is eliminated by the finding that AP-1 has little effect on the activity of free Bn (Figure 4(b)). Moreover, inhibition is sequence specific: AP-1 inhibits BG≥100-fold more effectively than a non-consensus DNA oligonucleotide (Figure 4(b)). The apparent Kd values for binding the consensus and non-consensus sequences are ~2 nM and 360 nM, respectively.
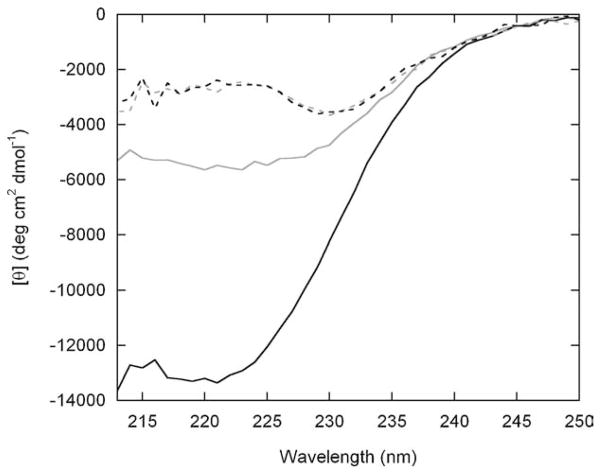
To further characterize the DNA-induced conformational transition, we measured circular dichroism (CD) spectra of BG and free Bn in the presence and absence of AP-1 (Figure 5). CD spectra of Bn are identical with and without AP-1. This result corroborates the enzyme assays of Figure 4(b) and confirms that Bn does not bind the DNA oligonucleotide. Compared to Bn, BG displays enhanced ellipticity with a broad minimum near 222 nm in the absence of AP-1. This finding suggests that the GCN4 domain is partially helical and is consistent with the notion that BG exists as a coiled-coil dimer when no DNA is present. The presence of partial helical structure may be responsible for the lower enzymatic activity of BG relative to Bn (Figure 4(b)). In marked contrast to Bn, BG exhibits a large change in ellipticity upon AP-1 addition. The [θ]222 value of −13,400 deg cm2 dmol−1 corresponds to a helix content of 32%,11 in close agreement with the predicted value of 33% if the GCN4 domain (56 residues of 170 total in BG) is fully helical. Taken together, Figures 4 and 5 demonstrate that: (i) enzymatic activity is regulated exclusively by DNA binding to the GCN4 domain; (ii) binding is both tight and sequence-specific; and (iii) DNA binding unfolds the Bn domain.
Figure 5.
CD spectra of free Bn (broken lines) and BG (continuous lines) in the absence (grey) and presence (black) of AP-1 DNA. Protein and AP-1 concentrations were 0.50 μM and 0.94 μM, respectively. The black traces were generated by subtracting spectra of free AP-1 at the same concentration. Solution conditions are identical to those for Figure 4. Data were collected on a model 202 spectropolarimeter (Aviv Biomedical, Inc.) in a 1 cm × 1 cm cuvette. Wavelengths below 212 nm are not shown due to excessive sample absorbance.
The mutually exclusive mechanism can be proven by demonstrating that DNA binding affinity and Bn stability are coupled in an inverse fashion. Since AP-1 binds stoichiometrically to BG (Figure 3), a straightforward test consists of stabilizing the Bn domain and determining whether Kd increases to a measurable value. At least a 20-fold increase in Kd would be required, corresponding to K2≥100 and ΔΔG≥2.8 kcal mol−1. We attempted to stabilize Bn by binding it to the mononucleotide inhibitor 3′-guanylic acid (3′-GMP)12,13 in 1.4 M urea. The 3′-GMP affinity is too low, however, to generate appreciable amounts of the complex at the highest nucleotide concentration permissible in the assay (~200 μM; limited by excessive absorbance at 280 nm). Similarly, phosphate has been shown to bind free Bn,14 but 50 mM phosphate stabilizes the Bn domain of BG by only 1.0 kcal mol−1 under the conditions used for Figure 2 (data not shown). An alternate approach is to stabilize the Bn domain by introducing mutations. A Bn variant harboring substitutions at six positions was found to be 3.0 kcal mol−1 more stable than wild-type.15 It remains to be tested whether the increase in stability is enough to perturb DNA binding affinity to a measurable extent.
BG and its cousin, barnase-ubiquitin,1 serve as a platform for the design of enzymes that possess novel sensor capabilities. The main requirement is that the end-to-end length of the inserted protein must be longer than the distance between termini of the surface loop of the target protein. The ratio of these distances is ~7.5 for BG and ~4.0 for barnase–ubiquitin. The minimum value has not been determined. Another consideration is that the stabilities of the two domains should be roughly comparable. If the catalytic domain is very stable, the affinity of the binding domain will be weakened and large concentrations of ligand will be required to trigger unfolding. Our experiments suggest that the optimal condition is when the catalytic domain is only marginally stable (e.g. ΔG=0.9 kcal mol−1 in 1.4 M urea; Figure 2), so that it is catalytically active in the absence of ligand but unfolded by low concentrations of ligand. The switching mechanism is highly responsive under these circumstances.
Bn is a particularly useful catalytic domain because it offers the potential for therapeutic applications. It is highly toxic when introduced into both prokaryotic and mammalian cells.16,17 By linking Bn to an appropriate binding domain, the fusion protein could have the ability to selectively kill cell types depending on whether a specific ligand is present or not. Stabilizing or destabilizing mutations can be introduced into either domain to fine-tune the position and sensitivity of the conformational switch to match the needs of the application.
Acknowledgments
We thank Tom Cutler for providing barnase samples and for discussions. This work was supported by NIH grant GM069755 (to S.N.L.).
Abbreviations used
- Bn
barnase
- BG
fusion protein barnase–GCN4
- 3′-GMP
3′-guanylic acid
References
- 1.Radley TL, Markowska AI, Bettinger BT, Ha JH, Loh SN. Allosteric switching by mutually exclusive folding of protein domains. J Mol Biol. 2003;332:529–536. doi: 10.1016/s0022-2836(03)00925-2. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.O’Shea EK, Klemm JD, Kim PS, Alber T. X-ray structure of the GCN4 leucine zipper, a two-stranded, parallel coiled coil. Science. 1991;254:539–544. doi: 10.1126/science.1948029. [DOI] [PubMed] [Google Scholar]
- 3.Cranz S, Berger C, Baici A, Jelesarov I, Bosshard HR. Monomeric and dimeric bZIP transcription factor GCN4 bind a the same rate to their target DNA site. Biochemistry. 2004;43:718–727. doi: 10.1021/bi0355793. [DOI] [PubMed] [Google Scholar]
- 4.Zitzewitz JA, Bilsel O, Luo J, Jones BE, Matthews CR. Probing the folding mechanism of a leucine zipper peptide by stopped-flow circular dichroism spectroscopy. Biochemistry. 1995;34:12812–12819. doi: 10.1021/bi00039a042. [DOI] [PubMed] [Google Scholar]
- 5.Zitzewitz JA, Ibarra-Molero B, Fishel DR, Terry KL, Matthews CR. Preformed secondary structure drives the association reaction of GCN4-p1, a model coiled-coil system. J Mol Biol. 2000;286:1105–1116. doi: 10.1006/jmbi.2000.3507. [DOI] [PubMed] [Google Scholar]
- 6.Saudek V, Pasley HS, Gibson T, Gausepohl H, Frank R, Pastore A. Solution structure of the basic region from the transcriptional activator GCN4. Biochemistry. 1991;30:1310–1317. doi: 10.1021/bi00219a022. [DOI] [PubMed] [Google Scholar]
- 7.Bracken C, Carr PA, Cavanagh J, Palmer AG. Temperature dependence of intramolecular dynamics of the basic leucine zipper of GCN4: implications for the entropy of association with DNA. J Mol Biol. 1999;285:2133–2146. doi: 10.1006/jmbi.1998.2429. [DOI] [PubMed] [Google Scholar]
- 8.Santoro MM, Bolen DW. Unfolding free energy changes determined by the linear extrapolation method. 1 Unfolding of phenylmethanesulfonyl alpha chymotrypsin using different denaturants. Biochemistry. 1988;27:8063–8068. doi: 10.1021/bi00421a014. [DOI] [PubMed] [Google Scholar]
- 9.Neira JL, Vázquez E, Fersht AR. Stability and folding of the protein complexes of barnase. Eur J Biochem. 2000;267:2859–2870. doi: 10.1046/j.1432-1327.2000.01290.x. [DOI] [PubMed] [Google Scholar]
- 10.Hollenbeck JJ, Oakley MG. GCN4 binds with high affinity to DNA sequences comtaining a single half-site. Biochemistry. 2000;39:6380–6389. doi: 10.1021/bi992705n. [DOI] [PubMed] [Google Scholar]
- 11.Scholtz JM, Barrick D, York EJ, Stewart JM, Baldwin RL. Urea unfolding of peptide helices as a model for interpreting protein unfolding. Proc Natl Acad Sci USA. 1995;92:185–189. doi: 10.1073/pnas.92.1.185. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Buckle AM, Fersht AR. Subsite binding in an RNase: structure of a barnase–tetranucleotide complex at 1.76-Å resolution. Biochemistry. 1994;33:1644–1653. doi: 10.1021/bi00173a005. [DOI] [PubMed] [Google Scholar]
- 13.Guillet V, Lapthorn A, Mauguen Y. Three-dimensional structure of a barnase-3′GMP complex at 2.2 Å resolution. FEBS Letters. 1993;330:137–140. doi: 10.1016/0014-5793(93)80259-w. [DOI] [PubMed] [Google Scholar]
- 14.Meiering EM, Bycroft M, Fersht AR. Characterization of phosphate binding in the active site of barnase by site-directed mutagenesis and NMR. Biochemistry. 1991;30:11348–11356. doi: 10.1021/bi00111a022. [DOI] [PubMed] [Google Scholar]
- 15.Serrano L, Day AG, Fersht AR. Stepwise mutation of barnase to binase: a procedure for engineering increased stability of proteins and an experimental analysis of the evolution of protein stability. J Mol Biol. 1993;233:305–312. doi: 10.1006/jmbi.1993.1508. [DOI] [PubMed] [Google Scholar]
- 16.Leuchtenberger S, Perz A, Gatz C, Bartsch JW. Conditional cell ablation by stringent tetra-cycline-dependent regulation of barnase in mammalian cells. Nucl Acids Res. 2001;29:e76. doi: 10.1093/nar/29.16.e76. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Bi YM, Rothstein SJ, Wildeman AG. A novel strategy for regulated expression of a cytotoxic gene. Gene. 2001;279:175–179. doi: 10.1016/s0378-1119(01)00742-9. [DOI] [PubMed] [Google Scholar]
- 18.Matsudaira P. Limited N-terminal sequence analysis. Methods Enzymol. 1990;182:602–613. doi: 10.1016/0076-6879(90)82047-6. [DOI] [PubMed] [Google Scholar]
- 19.Day AG, Parsonage D, Ebel S, Brown T, Fersht AR. Barnase has subsites that give rise to large rate enhancements. Biochemistry. 1992;31:6390–6395. doi: 10.1021/bi00143a005. [DOI] [PubMed] [Google Scholar]