Abstract
HIF-2α promotes von Hippel-Lindau (VHL) deficient renal clear cell carcinoma (RCC) tumorigenesis, while HIF-1α inhibits RCC growth. As HIF-1α antagonizes c-Myc function, we hypothesized that HIF-2α might enhance c-Myc activity. We demonstrate here that HIF-2α promotes cell cycle progression in hypoxic RCCs and multiple other cell lines. This correlates with enhanced c-Myc promoter binding, transcriptional effects on both activated and repressed target genes, and interactions with Sp1, Mizl and Max. Finally, HIF-2α augments c-Myc transformation of primary mouse embryo fibroblasts (MEFs). Enhanced c-Myc activity likely contributes to HIF-2α mediated neoplastic progression following loss of the VHL tumor suppressor, and influences the behavior of hypoxic tumor cells.
Introduction
Low oxygen (O2) levels are frequently encountered in solid tumors, and hypoxic stress responses have important effects on the natural history of disease (Pugh and Ratcliffe, 2003). Tumor growth, angiogenesis, invasion and metastasis are all regulated by hypoxia stimulated gene expression, largely mediated by Hypoxia Inducible Factors (HIFs). HIFs function as heterodimers in which the O2-labile α-subunits form a complex with a stable β-subunit (also known as ARNT) and bind hypoxia response elements (HREs) throughout the genome. Two α-subunits, HIF-1α and HIF-2α, have been demonstrated to increase target gene transcription in hypoxic cells. When complexed with ARNT, they activate genes such as those encoding glycolytic enzymes, vascular endothelial growth factor (VEGF), matrix metalloproteinase-2 and erythropoeitin (Hu et al., 2003; Semenza, 2003). The HIFs can alter cell cycle progression through putative transcriptional targets such as Cyclin D1 (Baba et al., 2003) and indirect modulation of p21 and p27 (Gardner et al., 2001; Green et al., 2001; Koshiji et al., 2004).
While initial characterization of hypoxia-induced transcription focused on HIF-1α, HIF-2α has recently been shown to regulate unique genes and physiologic functions (Covello et al., 2006; Hu et al., 2003; Tian et al., 1998). The HIF-α subunits differ in expression profiles, with HIF-1α expressed ubiquitously and HIF-2α limited to endothelium, kidney, heart, lungs and small intestine (Ema et al., 1997; Tian et al., 1997; Wiesener et al., 2003). HIF-1α uniquely activates glycolytic enzyme genes, while HIF-2α preferentially activates VEGF, transforming growth factor-α (TGFα), lysyl oxidase, Oct4 and Cyclin D1 (Baba et al., 2003; Covello et al., 2006; Erler et al., 2006; Gunaratnam et al., 2003; Hu et al., 2003; Wang et al., 2005). Targeted disruption of the HIF-2α locus in different mouse strains results in distinct phenotypes, including embryonic lethality due to bradycardia and vascular defects, perinatal lethality due to impaired lung maturation, and embryonic and post-natal lethality caused by multi-organ failure and mitochondrial dysfunction (Compernolle et al., 2002; Peng et al., 2000; Scortegagna et al., 2003; Tian et al., 1998). Each of these is quite different from the E10.5 lethality caused by cardiac and vascular defects restating from HIF-1α disruption (Carmeliet et al., 1998; Iyer et al., 1998; Ryan et al., 1998).
HIF-2α, but not HIF-1α, promotes tumor growth in RCC xenograft models. Overexpression of stable HIF-2α in 786-O RCC cells expressing pVHL restores xenograft growth to the level of parental VHL null cells (Kondo et al., 2003; Kondo et al., 2002; Raval et al., 2005), whereas overexpression of stable HIF-1α inhibits tumor growth (Maranchie et al., 2002; Raval et al., 2005). Studies of pre-neoplastic lesions from the kidneys of patients with VHL disease also suggest a role for HIF-2α in the transformation of dysplastic cells, as HIF-2α expression increased with the degree of dysplasia, whereas HIF-1α expression decreased (Mandriota et al., 2002; Raval et al., 2005). HIF-2α also appears to have a more general role in promoting tumorigenesis. Subcutaneous teratomas generated from ES cells with the HIF-2α cDNA “knocked in” to the HIF-1α locus exhibit four-fold greater mass than WT controls, largely due to increased proliferation (Covello et al., 2005). This is not simply the result of HIF-1α loss, as multiple studies with Hif-1α−/− ES cell-derived teratomas failed to demonstrate a consistent growth advantage (Carmeliet et al., 1998; Hopfl et al., 2002; Ryan et al., 1998). A recent study in neuroblastoma has shown HIF-2α stabilization under prolonged, but mild hypoxia, raising the intriguing possibility that HIF-2α may promote angiogenesis even in tumors experiencing minimal hypoxic stress (Holmquist-Mengelbier et al., 2006).
Moderate levels of hypoxia (0.5% – 3% O2) can cause cell cycle arrest through a HIF-1α dependent increase in cyclin dependent kinase inhibitor (CKI) p21 and p27 expression (Gardner et al., 2001; Goda et al., 2003; Green et al., 2001). p21 and p27 are not direct HIF targets; their expression changes as a result of HIF-1α inhibition of the proto-oncogene c-Myc and consequent relief of transcriptional repression at their promoters (Koshiji et al., 2004; Koshiji et al., 2005; Mack et al., 2005). HIF-1α may antagonize c-Myc activity by altering interaction with Sp1, one of its co-factors (Koshiji et al., 2004; Koshiji et al., 2005). There are no published data as yet on HIF-2α effects on cell cycle progression.
c-Myc is a basic helix-loop-helix/leucine zipper (bHLH/LZ) transcription factor that controls the G1-S cell cycle transition and is overexpressed in many human tumors (Adhikary and Eilers, 2005; Nilsson and Cleveland, 2003). c-Myc activates the transcription of growth promoting genes such as Cyclin D2, ornithine decarboxylase, and E2F1 by binding to a conserved E box (CACGTG) with its binding partner Max, and inhibits the expression of multiple genes, notably p21 and p27, by binding to the transcription initiator element (Inr) in a complex with Max and either Sp1 or Miz1 (Coller et al., 2000; Fernandez et al., 2003). As c-Myc is normally labile and expressed at low levels, it competes for Max binding with antagonistic transcription factors such as Mad, Mnt and Mga. Interestingly, Mad/Max complexes repress expression via E boxes, while Miz1 functions as a transcriptional activator when bound to an Inr without c-Myc (Adhikary and Eilers, 2005).
Given that HIF-1α and HIF-2α may have opposite effects on renal tumor growth and the established inhibitory effect of HIF-1α on c-Myc (Koshiji et al., 2004; Koshiji et al., 2005), we hypothesized that HIF-2α actually promotes c-Myc transcriptional activity. This would result in enhanced cell cycle progression and increased tumor growth. We have examined 786-O and RCC4 RCCs with restored pVHL expression, embryonic epithelial cells (ECs), and NIH3T3 cells, and observed HIF-2α mediated enhancement of c-Myc activity and cell cycle progression. As a control, HIF-1α effects were also assessed in each context, and HIF-1α exhibited the anticipated inhibition of proliferation. HIF-2α also has a role in tumor initiation, promoting c-Myc/RasV12G induced transformation in primary MEFs.
Results
HIF-2α enhances cell cycle progression while HIF-1α inhibits it
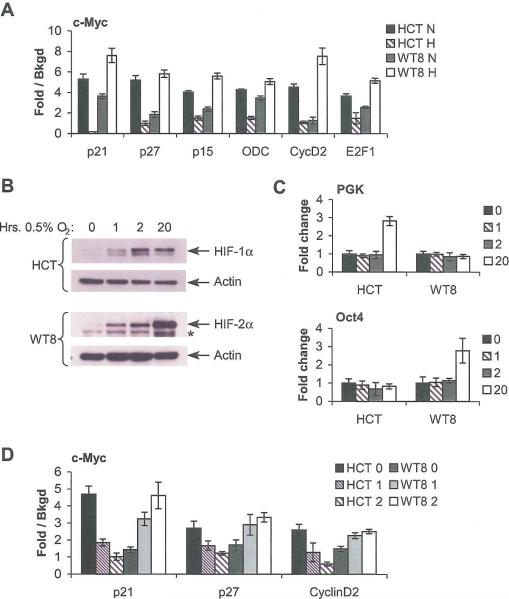
To study differential effects of HIF-α subunits on tumor cell cycle progression, we selected cell lines expressing predominantly HIF-1α (HCT116 colon carcinoma cells) or HIF-2α (WT8, 786-O RCC cells expressing pVHL). We confirmed published data concerning HIF-α subunit expression and localization in these cells (Koshiji et al., 2004; Maxwell et al., 1999). As expected, HCT116 cells expressed HIF-1α in the nuclear fraction whereas WT8 cells did not, even upon long exposures of radiographic films. HIF-2α expression in hypoxic WT8 cells was detected predominantly in the nuclear fraction. HIF-2α was also detected in HCT116 cells, but at approximately 10% the levels observed in WT8 cells (Figure 1A). To correlate these data with transcriptional targets, mRNA was extracted from cells grown at 21% or 0.5% O2 for 24, 48, and 72 hrs., and analyzed by quantitative real time PCR (QRT-PCR). As expected, both cell lines increased expression of the HIF-1α/HIF-2α shared target VEGF (Supplemental Figure 1A), whereas only HCT116 cells showed elevated levels of the HIF-1α-specific target phosphoglycerate kinase (PGK; Supplemental Figure 1B; Hu et al., 2003) and only WT8 cells increased Levels of the HIF-2α-specific target Oct4 (Supplemental Figure 1C; Covello et al., 2006). Of note, while HCT116 cells exhibited basal Oct4 expression, hypoxic induction of HIF-2α in these cells had no further effect on transcript levels, likely due to low HIF-2α protein levels. We therefore concluded that HIF-1α is the functional HIF-α subunit in HCT116 cells, and HIF-2α the functional HIF-α subunit in WT8 cells.
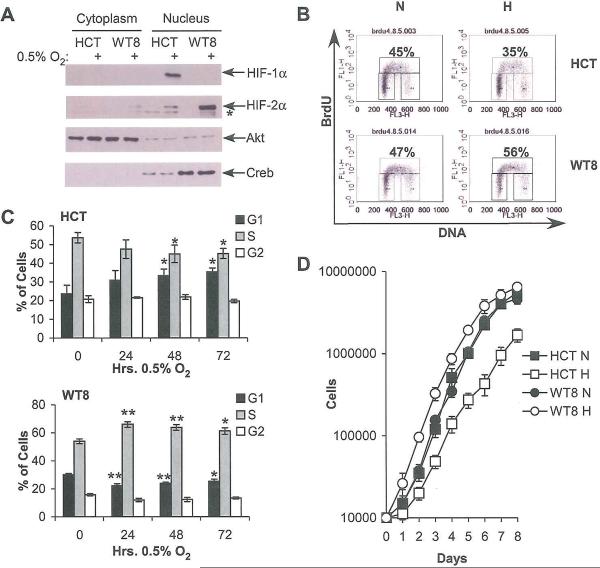
Figure 1.
Differential expression of HIF-1α and HIF-2α in HCT116 and WT8 cells correlates with differential cell cycle progression under hypoxia. A. Western blot of HIF-α expression in HCT116 (“FICT”) and WT8 cells following 4 hrs. incubation at 0.5% O2 shows differential expression of HIF-α subunits. Asterisk denotes a background band. Akt and Creb immunoblots assess loading and the efficiency of cellular fractionation; Akt is present in both the nucleus and cytoplasm, while Creb is exclusively nuclear. B. Representative BrdU incorporation plots from HCT116 and WT8 cells grown at 21% or 0.5% O2 for 48 hrs. C. Summary of changes in BrdU incorporation in HCT116 and WT8 cells after 24, 48 and 72 hrs. hypoxia. Results averaged from 3 experiments, error bars ±1 SEM, * p < 0.05, ** p < 0.01. D. Proliferation measured by serial cell counts under normoxia (N) or hypoxia (H); data from one representative experiment. Error ±1 SD.
Given that these two cell lines exhibited differential HIF-α expression and activity, we assessed their cell cycle progression by BrdU incorporation. When the HCT116 cells were grown under hypoxia, they accumulated in G1 phase, with a corresponding decrease in the percentage of cells in S phase. Representative FACS plots (Figure 1B, upper panel) and the summary of three experiments (Figure 2C, upper panel) are shown. These changes were statistically significant following 48 or 72 hrs. hypoxia (p < 0.05). Conversely, when the WT8 cells were grown at 0.5% O2, statistically significant increases in the percentage of cells in S phase and decreases in the percentage in G1 phase after 24, 48 (p < 0.01) or 72 hrs. (p < 0.05) were observed. Again, both representative FACS plots (Figure 1B, lower panel) and summary data (Figure 1C, lower panel) are shown. Changes in cell cycle profiles correlated with proliferation as measured by serial cell counts under normoxic or hypoxic conditions (Figure 1D), with increased WT8 cell numbers and decreased HCT116 cell numbers under hypoxia. These effects are most apparent at early time points, becoming attenuated after 6 days in culture due to confluency. Thus, when HIF-1α and HIF-2α are expressed separately they have opposite effects on cell cycle progression and proliferation that are stable over long time courses, where HIF-2α promotes both processes.
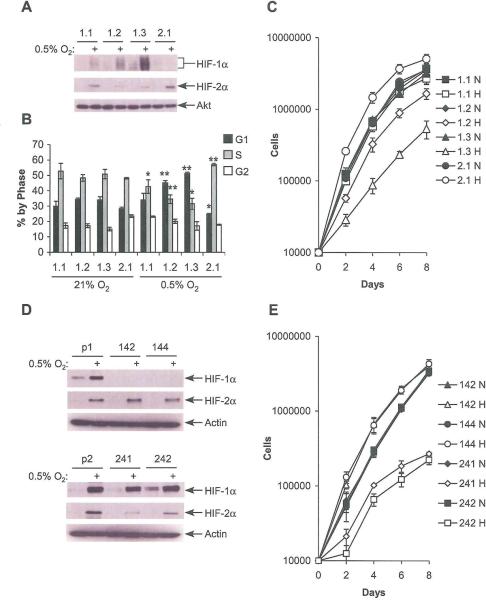
Figure 2.
HIF-1α and HIF-2α have antagonistic effects on cell cycle progression. A. HIF-α subunit expression in WT8 cells stably overexpressing HIF-1α (1.1, 1.2, 1.3) or HIF-2α (2.2) after 24 hrs. at 0.5% O2. B. Summary of changes in BrdU incorporation in WT8 cells overexpressing HIF-1α or HIF-2α after 24 hrs. hypoxia. Results averaged from at least 3 experiments, error bars ±1 SEM, * p < 0.05, ** p < 0.01. C. Proliferation measured by serial cell counts under normoxia (N) or hypoxia (H); data from one representative experiment. Error ±1 SD. D. HIF-α subunit expression in pVHL rescued RCC4 cells transduced with empty vector (p1, p2), shRNA against HFF-1α (142, 144) or HIF-2α (241, 242) after 24 hrs. at 0.5% O2. E. Proliferation in clones with HIF-1α or HIF-2α knockdown, measured by serial cell counts under normoxia (N) or hypoxia (H); data from one representative experiment. Error ±1 SD.
Given that these experiments were performed in tumor cell lines of different tissue origins, it is possible that the observed effects were a result of HIF independent hypoxic responses, or tissue specific differences in responses to O2 deprivation. We therefore used WT8 cells engineered to stably overexpress increasing amounts of HIF-1α (called “1.1,” “1.2” and “1.3”) or HIF-2α, called “2.2” (Figure 2A). Cell cycle progression was evaluated in these cells after growth at 0.5% O2 for 24 hrs. Consistent with observations in HCT116 cells, overexpressed HIF-1α restored the hypoxia-induced increase in G1 phase and decrease in S phase populations (p < 0.05) in WT8 cells, while HIF-2α overexpression continued to elevate the proportion of cells in S phase (Figure 2B; p < 0.01). Proliferation was also assessed in these cells, and found to correlate with hypoxia induced changes in cell cycle profile (Figure 2C). By co-expressing HIF-1α and HIF-2α we confirmed that HIF-1α induces cell cycle arrest in WT8 cells, eliminating the possibility that they exhibit a tissue specific cell cycle response to hypoxia. To further support these data, we used the pVHL rescued RCC4 T3-14 cell line, which expresses both HIF-1α and HIF-2α (Hu et al., 2003) stably transduced with shRNAs targeting either HIF-1α or HIF-2α or an empty vector (Lum et al., manuscript submitted). HIF-α subunit expression was assessed by immunoblot, showing essentially complete knockdown of HIF-1α and >75% knockdown of HIF-2α (Figure 2D). The efficacy of knockdown was confirmed by measuring expression of HIF-α shared and unique targets VEGF, PGK and Oct4 (Supplemental Figure 2A). Next, BrdU incorporation was measured after 0, 24, or 48 hrs. hypoxia. While clones transduced with the empty vector showed a gradual decrease in percentage of cells in S-phase, those with HIF-2α knockdown showed a more precipitous decrease. In direct contrast, pVHL rescued RCC4 cells with HIF-1α knockdown showed increased percentage of cells in S-phase (Supplemental Figure 2B). When cell counts were performed, only a small proliferation decrease was noted under hypoxia in vector transfected cells (Supplemental Figure 2C), whereas a substantial increase was noted with HIF-1α knockdown and a substantial decrease with HIF-2α knockdown. All clones showed indistinguishable proliferation under normoxic conditions (Figure 2E). Thus, HIF-2α enhances cell cycle progression in a fashion that is antagonized in a dose-dependent manner by HIF-1α, suggesting that both HIF-α subunits act on the same pathway. These effects on proliferation in RCC4 are particularly notable, as selective knockdown of each HIF-α disrupts a balanced effect on hypoxic proliferation to either potently enhance it (in the presence of HIF-2α alone) or repress it (with only HIF-1α). This analysis is particularly relevant to RCC development, since most human renal tumors exhibit HIF-1α loss while maintaining HIF-2α during tumor progression (Mandriota et al, 2002; Raval et al, 2005).
HIF-2α enhances the expression of c-Myc target genes in a c-Mye dependent fashion
To investigate the role of c-Myc in these cell cycle changes, we assessed hypoxic expression of c-Myc targets p21 and p27 (repressed by c-Myc), as well as Cyclin D2 and E2F1 (activated by c-Myc). Although additional factors also regulate these genes, we will refer to them as “c-Myc targets” to differentiate them from direct HIF targets such as PGK and Oct4. Under the conditions described above, we observed increased p21 and p27 mRNA expression in hypoxic HCT116 cells, consistent with attenuated repression by c-Myc. Conversely, hypoxic WT8 cells exhibited decreased p21 and p27 mRNA expression, consistent with enhanced repression by c-Myc (Figure 3A). We observed the opposite effect on c-Myc activated targets: Cyclin D2 and E2F1 mRNA levels were decreased in hypoxic HCT116 cells and increased in hypoxic WT8 (Figure 3B). These results were confirmed by immunoblot assays on extracts from ceils grown at 21% O2 or 0.5% O2 for 48 hrs. (Figure 3C). While the data demonstrate relatively subtle changes in expression, they are consistent with those commonly observed for c-Myc (Bouchard et al., 1999; Coller et al., 2000). To confirm the antagonism between HIF-1α and HIF-2α on c-Myc target genes, p27 and Cyclin D2 expression was also assessed in stable WT8 lines overexpressing HIF-1α and HIF-2α. Consistent with the cell cycle analysis, increased HIF-1α expression correlated with dose-dependant increases in p27 induction and Cyclin D2 repression, while increased HIF-2α expression correlated with decreased p27 expression and increased Cyclin D2 expression, as observed in the parental cell line (Figure 3D).
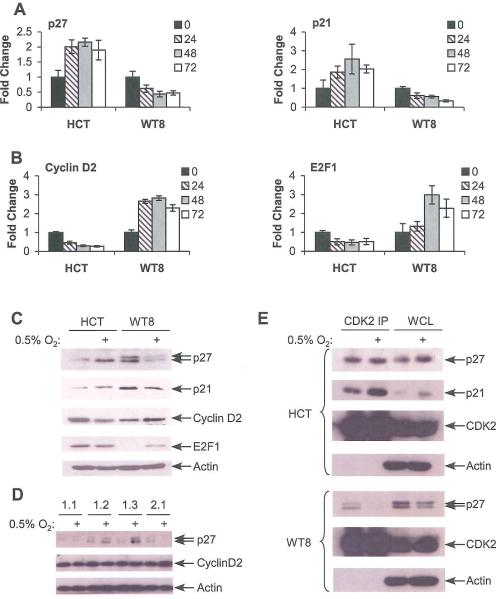
Figure 3.
Tumor cell lines expressing HIF-1α or HIF-2α exhibit differential hypoxic effects on cell cycle regulators. A. Expression of c-Myc repressed targets p21 and p27 in HCT116 and WT8 following 24, 48 or 72 hrs. at 0.5% O2. Results measured by QRT-PCR and averaged from 4 experiments, error bars ±1 SEM. As described in the text, HIF-1α expressing HCT116 cells and HIF-2α expressing WT8 cells exhibit opposite responses to hypoxia with respect to c-Myc target expression. B. Expression of c-Myc activated targets Cyclin D2 and E2F1 in HCT116 and WT8 as above. C. Western blot analysis of c-Myc target expression in HCT116 and WT8 following 48 hrs. hypoxia. D. Expression of c-Myc targets in WT8 cells overexpressing HIF-1α or HIF-2α following 24 hrs. at 0.5% O2. E. Change in p21 and p27 interaction with CDK2 assessed by CDK2 IP following 48 hrs. hypoxia in HCT116 and WT8.
To further extend these observations, two other model systems were analyzed. First, we measured mRNA and protein changes in p27 and Cyclin D2 in the RCC4 cell lines described above. In empty vector transduced cells, little change was detected in mRNA or protein levels of these targets. In clones with HIF-1α knockdown decreased p27 and increased Cyclin D2 were observed, while in clones with HIF-2α knockdown increased p27 and decreased Cyclin D2 were observed (Supplemental Figure 2E, 2F). These findings correlate with the cell cycle changes described above, though a minor decrease in S-phase percentage is noted in the vector transduced cells with no change in c-Myc targets. This is likely to be a result of HIF-independent effects of the stringent level of hypoxia (0.5% O2) used in this study. Second, to use a defined genetic system that is relatively undifferentiated and unlikely to have the mutational load of a human tumor cell line, we produced embryonic epithelial cells (ECs; Mansfield et al, 2005) from day 8.5 embryos harboring targeted deletions of Hif-1α (Carmeliet et al., 1998) or Hif-2α (Compernolle et al., 2002). Six lines were analyzed in total, with two Hif-1α and two Hif-2α null lines, and a WT littermate control for each. Results from one representative cell line from each genotype are shown. First, HIF-α expression and target gene induction were confirmed (Supplemental Figure 3A, 3B). Next, we measured mRNA expression of c-Myc targets p27 and Cyclin D2. No change in p27 was detected in hypoxic WT lines, whereas p27 mRNA levels increased in hypoxic cells expressing only HIF-1α, and decreased in cells expressing only HIF-2α (Supplemental Figure 3C, upper panel). For Cyclin D2, we observed that all HIF-2α expressing cell lines (i.e., WT or Hif-1α−/−) exhibited increased mRNA expression under hypoxia, while only the Hif-2α−/− cells exhibited decreased Cyclin D2 mRNA levels (Supplemental Figure 3C, lower panel). Western blot analysis confirmed the observed mRNA changes (Supplemental Figure 3D). Thus, consistent HIF-α mediated modulation of c-Myc influenced cell cycle regulators was observed in multiple cell systems.
As the biological effects of c-Myc are mediated through coordinated regulation of multiple factors influencing the G1-S phase transition, cyclin dependant kinase 2 (CDK2) immunoprecipitations (IPs) were performed on hypoxic HCT116 and WT8 cells at 48 hrs., and tested for co-precipitated p21 and p27. We observed substantially increased levels of p21 associated with CDK2 in hypoxic HCT116 cells, while less change was observed for p27. However, it should be noted that HCT116 cells express higher levels of p21 than p27. In direct contrast, p27 association with CDK2 was almost completely abrogated in hypoxic WTS cells, while minimal p21 was detected (Figure 3D and data not shown). Therefore, changes in c-Myc target expression correlate with more dramatic differences in CDK2 complex components, consistent with the hypoxic effects on cell cycle progression observed above.
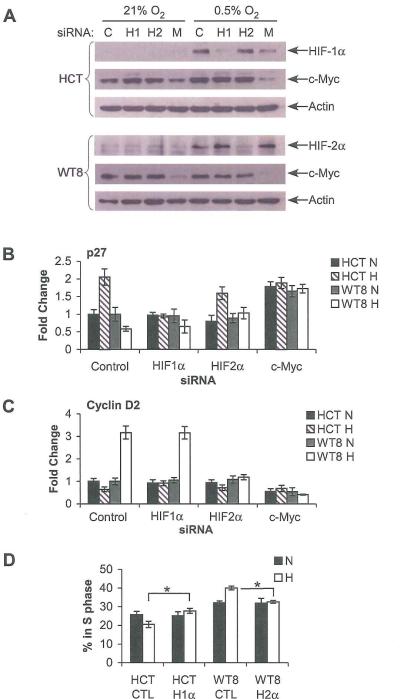
The data suggested a correlation between HIF-α effects on cell cycle progression and c-Myc target gene expression. To confirm that gene expression and cell cycle changes were a direct result of hypoxic activation of the HIF-α subunits and test a requirement for c-Myc, HCT116 and WT8 cells were transfected with siRNA against HIF-1α, HIF-2α, and c-Myc, or a control. siRNA. Each siRNA achieved greater than 60% knockdown of its target (Figure 4A). c-Myc knockdown sometimes correlated with less HIF-1α or HIF-2α expression. We believe this to be a direct result of c-Myc knockdown, as it occurred in a dose-dependent fashion, and with two separate siRNAs against c-Myc (data not shown). HIF effects on p27 were not altered by the control siRNA at 20 hrs. hypoxia (Figure 4B). When treated with siRNA against HIF-1α, hypoxic HCT116 cells failed to induce p27 mRNA, and when treated with siRNA against HIF-2α, the WT8 cells no longer exhibited decreased p27 expression. Finally, c-Myc siRNA resulted in high p27 mRNA levels, with no effect of hypoxia on either cell type. Similarly, Cyclin D2 levels were not decreased in hypoxic HCT116 cells treated with HIF-1α siRNA or increased in hypoxic WT8 cells treated with HIF-2α siRNA. With c-Myc siRNA, low Cyclin D2 levels were observed in all conditions. Furthermore, when cells were tested by BrdU incorporation after 48 hrs. hypoxia, knockdown of the HIF-1α in HCT116 and HIF-2α in WT8 abrogated the effect of hypoxia on cell cycle progression (Figure 4D). These data indicate that HIF-1α and HIF-2α are required to alter hypoxic cell cycle progression and modulate c-Myc target gene expression in a c-Myc dependent manner.
Figure 4.
HIF-α effects on p27 and Cyclin D2 levels require c-Myc. A. Western blot analysis showing siRNA inhibition of HIF-1α (H1), HIF-2α (H2) and c-Myc (M) expression, as well as a luciferase control (C) in HCT116 and WT8 cells. B. QRT-PCR analysis showing expression of p27 in siRNA treated cells after 20 hrs. at 0.5% O2. Results measured by QRT-PCR from 4 experiments ate shown, error bars ±1 SEM. C. QRT-PCR analysis showing expression of Cyclin D2 in siRNA treated cells. Results measured as above. D. Cell cycle progression measured by BrdU incorporation in asynchronous HCT116 cells treated with control or HIF-1α siRNA and WT8 cells treated with control or HIF-2α siRNA at 21% O2 (N) or 0.5% O2 (H) for 48 hrs. Results averaged from 3 experiments are shown, error bars ±1 SEM, * p < 0.05
HIF-α subunits regulate c-Myc promoter binding and interactions with Sp1, Miz1 and Max
To elucidate the mechanism(s) by which HIF-α subunits regulate c-Myc activity, we tested the effects of hypoxia on c-Myc promoter occupancy by chromatin immunoprecipitation (ChIP) in HCT116 and WT8 cells. ChIP was performed for c-Myc, HIF-1α, HIF-2α, and p53 and analyzed with QRT-PCR. The data are represented as the average fold difference between IP with a specific antibody and the background signal of an isotype matched control. Results are averaged from four separate experiments with hypoxic treatment, ChIP and PCR analysis performed independently. For these experiments, 3 c-Myc repressed targets (p21, p27, and p15) were analyzed with primers at the Inr, and 3 c-Myc activated targets (ODC, Cyclin D2 and E2F1) were analyzed, with primers directed at their E box elements, centered at bases −1158, 282 (in intron 1) and −354, respectively. In all cases, signal detected for c-Myc ChIP of normoxic HCT116 cells decreased to near background levels under hypoxia. When c-Myc ChIP was performed in WT8 cells, a substantial increase in the signal from all promoters tested was observed in hypoxic cells (Figure 5A). Previous studies have demonstrated HIF-1α binding to the Inr of p21 and MutSα (Koshiji et al., 2004; Koshiji et al., 2005). We also detected HIF-1α binding to the p21 Inr and the VEGF HRE (centered at base −968) in HCT116 cells and HIF-2α binding to the same sites in WT8 cells. However, neither HIF-α subunit bound to any of the other c-Myc DNA binding regions tested (Supplemental Figure 4A, 4B). These data showed that HIF-2α effects on c-Myc regulated genes correlated with increased levels of c-Myc binding to their promoters, but that HIF-2α DNA binding near c-Myc was not required for hypoxic regulation of c-Myc targets. To confirm the effect of each HIF-α subunit on c-Myc promoter binding in an isogenic cell system, we repeated this analysis in HEK293 cells which expressed Myc-tagged normoxically stable double proline mutants (DPM) of the HIF-α subunits with doxycycline induction (Supplemental Figure 5A; Hu et al., 2003). As before, HIF-1α led to decreased c-Myc DNA binding and HIF-2α led to increased c-Myc DNA binding (Supplemental Figure 5B). Similarly, Myctag IPs (for the HIF-α subunits) showed HIF-α DNA binding to p21 Inr and the VEGF HRE, but no other assayed sites (Supplemental Figure 5C). A control PCR reaction was performed with primers targeting a p53 binding site at −2.3 kB on the p21 promoter. Confirming antibody specificity, signal was detected with p53 ChIP but no other antibody tested (Supplemental Figure 4C, 5D; Koshiji et al., 2004).
Figure 5.
ChIP demonstrates altered c-Myc promoter occupancy in hypoxic cells. A. HCT116 and WT8 cells were grown at 21% O2 (N) or 0.5% O2 (H) for 20 hrs., and then assayed by ChIP. Following IP with antibody against c-Myc or isotype control, extracts were assessed by QRT-PCR using SYBR green. The graphs show the fold difference between c-Myc IP and Rabbit IgG control (background) with results from 4 separate experiments, error bars ±1 SEM. B. Time course of HIF-α protein induction in HCT116 and WT8 at 1, 2, and 20 hrs. at 0.5% O2. * indicates a non-specific band. C. QRT-PCR time course of HIF target gene induction at the same time points as above. Results averaged from 3 experiments, error bars ±1 SEM. D. c-Myc promoter binding in HCT116 and WT8 cells incubated at 0.5% O2 for 1 or 2 hrs., then analyzed by ChIP as above. Results from 4 independent experiments are shown, error bars ±1 SEM.
The data strongly suggest a model in which HIF-α subunits alter c-Myc promoter binding, as opposed to its ability to activate transcription. To understand the underlying mechanism(s) for these effects, we determined whether HIF transcriptional activity is required. ChIP was performed as above, but cells were incubated at 0.5% O2 for only 1–2 hrs. At these time points, HIF-α protein induction is observed (Figure 5B), but induction of HIF-α target genes is not (Figure 5C). HCT116 cells exhibited a decrease in c-Myc binding to p21, p27 and Cyclin D2 promoters at 0.5% O2, while enhanced binding was observed in WT8 cells (Figure 5D). As an additional control, ChIP was performed on cells pre-treated with Actinomycin D and similar results were observed (data not shown). Thus, we concluded that the effects of HIF-1α and HIF-2α on c-Myc promoter binding were independent of their respective transcriptional targets.
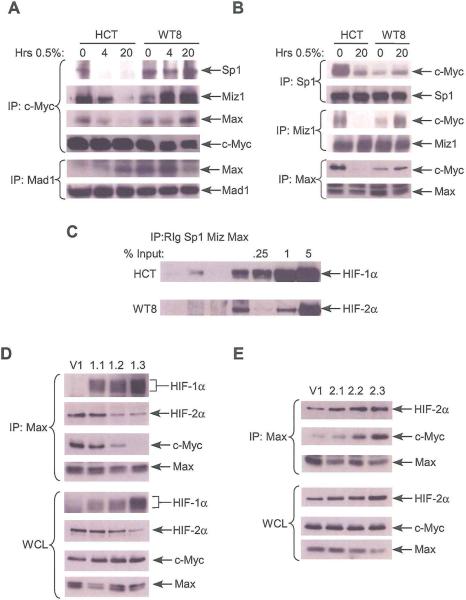
As c-Myc activity is a function of its protein level, we investigated hypoxic effects on c-Myc accumulation, as well as that of its binding partners Sp1, Miz1 and Max, and its competitor Mad1. No consistent changes in protein levels were detected in HCT116 and WT8 cells grown for 4 or 20 hrs. at 0.5% O2 (Supplemental Figure 6A). Assessment of c-Myc mRNA levels and protein stability revealed no consistent changes under hypoxia (data not shown). Of note, hypoxic culture for 4 or 20 hrs. correlated with a substantial decrease in c-Myc co-precipitation with Sp1, Miz1 and Max in HCT116 cells and an increase in c-Myc co-precipitation with these proteins in WT8 cells, albeit less dramatically than the decrease seen in HCT116 cells (Figure 6A). When Mad1 IP was performed the opposite result was observed: increased Mad1/Max association in hypoxic HCT116 cells and decreased association in hypoxic WT8 cells. To confirm these data, IP was also performed for Sp1, Miz1 and Max. As before, we observed decreased co-precipitation of these proteins with c-Myc in hypoxic HCT116 cells and increased co-precipitation in hypoxic WT8 cells (Figure 6B).
Figure 6.
HIF-α effects on c-Myc activity correlate with differential c-Myc interactions with Sp1, Mizl and Max. A. Co-precipitation of Sp1, Mizl, and Max after c-Myc IP and Max after MadI IP in HCT116 and WT8 cells grown at 0.5% O2 for 4 and 20 hrs. B. Co-precipitation of c-Myc after Sp1, Mizl or Max IP in HCT116 and WT8 cells grown under hypoxia for 20 hrs. C. HIF-1α and HIF-2α co-precipitation after Sp1, Mizl, and Max, or isotype control IP in HCT116 and WT8 cells treated DFX for 4 hrs. D. Vector control (V1) and HIF-1α overexpressing cell lines were cultured at 0.5% O2 for 20 hrs. and Max IP was performed and analyzed for co-precipitated HIF-1α, HIF-2α and c-Myc. E. Vector control (V1) and HIF-2α overexpressing cell lines were cultured at 0.5% O2 for 20 hrs. and Max IP was performed and analyzed for co-precipitated HIF-2α and c-Myc.
To identify a transcription-independent mechanism by which HIF-1α and HIF-2α might alter c-Myc/Max binding, we investigated the interaction of HIF-1α and HIF-2α with Sp1, Miz1 and Max. In HCTI16 cells, HIF-1α co-precipitated with Sp1 and Max, whereas in WT8 cells HIF-2α co-precipitated only with Max. Interestingly, we noted a difference in the stoichiometry of these interactions: the HIF-1α/Max complex accounted for approximately 0.25% of cellular HIF-1α, whereas the HIF-2α/Max complex accounted for 2% of cellular HIF-2α (Figure 6C). Control immunoblots were performed for specificity of the Max IP, showing no co-precipitation with the abundant transcription factor Rb (data not shown). Next, using WT8 cell lines overexpressing HIF-1α (1.1, 1.2, and 1.3) and HIF-2α (2.1, 2.2, and 2.3), we tested for effects of HIF-α levels on c-Myc/Max binding. When the HIF-1α overexpressing cell lines and a vector control (V1) were incubated at 0.5% O% for 20 hours, increasing levels of HIF-1α/Max binding correlated with decreasing c-Myc/Max binding and HIF-2α/Max binding, despite similar levels of c-Myc and Max in whole cell lysates. As noted before, there is a decrease in HIF-2α expression with increasing levels of HIF-1α (Raval et al., 2005). However, the decrease in HIF-2α/Max binding is greater than the difference in expression (Figure 6D). On the other hand, when HIF-2α overexpressing cell lines were incubated under hypoxia, a dose-dependent increase in c-Myc/Max binding was observed, correlating with the increase in HIF-2α/Max binding (Figure 6E). There was no difference in c-Myc/Max binding in any of these lines under normoxia (Supplemental Figure 6B,C). These data demonstrate the formation of complexes containing HIF-1α and Max or HIF-2α and Max. Both HIF-1α inhibition of c-Myc/Max complex formation, and HIF-2α promotion of c-Myc/Max complex formation correlated with the expression level of each α-subunit and its interaction with Max, suggesting a direct effect on c-Myc complex composition.
HIF-2α promotes cell cycle progression in NIH3T3 cells
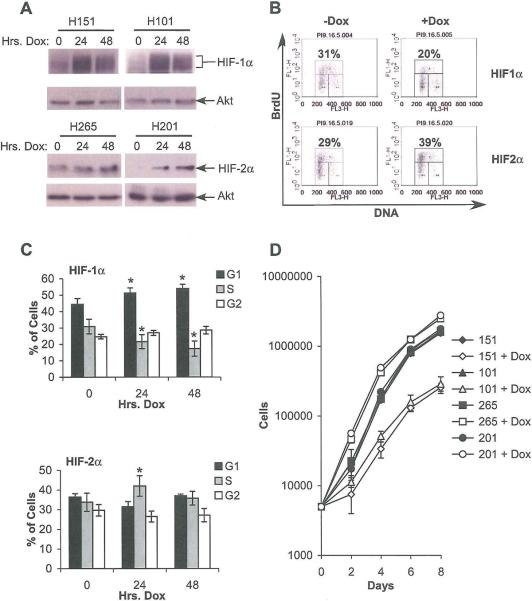
Having observed a differential effect of HIF-1α and HIF-2α on c-Myc targets in multiple cell types, we next confirmed their impact on cell cycle progression in NIH3T3 cells, which exhibit well-characterized cell cycle responses. For this purpose, we isolated NIH3T3 cell clones with doxycycline-responsive expression of DPM-HIF-1α-Myctag and DPM-HIF-2α-Myctag, allowing the separation of the HIF-α subunits from any HIF-independent hypoxic effects on cell cycle progression. Two clones expressing HIF-1α (151 and 101) and two expressing HIF-2α (265 and 201) were used for further analysis. The inducible HIF-1α was expressed at a level similar to that induced by 0.5% O2. While there is no endogenous HIF-2α in NIH3T3 cells, doxycycline induction of HIF-2α produced similar increases in VEGF mRNA to either hypoxia or HIF-1α induction (data not shown). Figure 7A shows the stable induction of each transgene after 48 hrs. doxycycline treatment, with some baseline HIF-2α expression in clone 265. The effect of doxycycline treatment on cell cycle progression was assayed by BrdU incorporation. Representative FACS plots are shown (Figure 7B). Results from 4 independent experiments were averaged and a statistically significant decrease in the percentage of HIF-1α induced cells in S phase and increase in the percentage in G1 phase (p < 0.05) after 24 or 48 hrs. of doxycycline treatment was observed (Fig 7C, upper panel). Interestingly, a statistically significant increase in percentage in S phase (p < 0.05) was noted after 24 but not 48 hrs. in the HIF-2α inducible lines (Figure 7C, lower panel). When proliferation rates were measured, a significant decrease was observed in HIF-1α inducible cells following doxycycline treatment and a significant increase was observed in HIF-2α inducible cells, consistent with their cell cycle profiles (Figure 7D). Although HIF-2α enhanced S-phase entry for 1 day only, this was sufficient to cause increased cell numbers for the course of the experiment. We concluded that HIF-α subunit effects on cell cycle progression in tumor cell lines can be generalized to multiple cell types, including those with intact cell cycle regulation, and do not require HIF-independent hypoxic effects.
Figure 7.
Doxycycline-regulated NIH3T3 cells expressing normoxically stable HIF-1α or HIF-2α show differential effects on cell cycle progression. A. Western blots showing HIF-α subunit expression following 24 or 48 hrs. treatment with doxycycline. B. Representative FACS plot at 0 and 2 days doxycycline from a HIF-1α (151) and HIF-2α (201) expressing clone. C. Cell cycle progression in doxycycline-regulated 3T3s (clones 151 and 201) measured by BrdU incorporation. Results from 3 experiments are shown, error bars ±1 SEM, * p < 0.05. D. Proliferation measured by serial cell counts; data from one representative experiment. Error ±1 SD.
HIF-2α enhances RasV12G/c-Myc transformation of primary MEFs
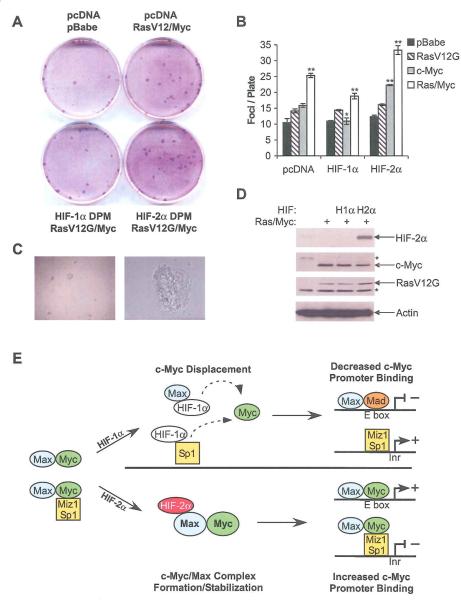
Given that HIF-2α promotes RCC tumor development (Mandriota et al., 2002), we tested HIF-2α's ability to promote transformation of primary MEFs. For this purpose, passage 4 MEFs from day 13.5 embryos were transfected with DPM-HIF-1α-FLAG or DPM-HIF-2α-FLAG, as well as RasV12G, c-Myc, or both. Cells were allowed to grow to confruency, and assessed for focus formation by Wright-Giemsa staining 30 days later. As demonstrated in a representative assay (Figure 8A), co-transfection of HIF-2α with RasV12G and c-Myc resulted in a 32% increase in focus formation compared to RasV12G and c-Myc alone, whereas co-transfection of HIF-1α resulted in a 25% decrease. The complete analysis of 3 experiments (Figure 8B) reveals a statistically significant difference between RasV12G/Myc with an empty vector, DPM-HIF-1α -FLAG and DPM-HIF-2α-FLAG (p < 0.01 in all cases). Interestingly, DPM-HIF-2α-FLAG co-transfection with c-Myc alone resulted in a 41% increase (p < 0.05), while DPM-HIF-1α-FLAG resulted in a 32% decrease (p < 0.05). Foci were picked from these plates and cell lines generated, all of which formed colonies in soft agar (Figure 8C). These clones were also evaluated for expression of the transfected proteins. Overexpressed c-Myc and RasV12G were detected in all established cell lines relative to mock transfected controls, with c-Myc overexpression leading to repression of endogenous c-Myc (endogenous protein is indicated with an asterisk). Interestingly, it appears that cell lines obtained from plates transfected with DPM-HIF-1α-FLAG failed to maintain expression of this transgene, but those transfected with DPM-HIF-2α-FLAG did (Figure 8D and data not shown). In aggregate, these data support a role for HIF-2α in promoting transformation, and suggest that HIF-1α can inhibit it.
Figure 8.
HIF-2α promotes focus formation by primary MEFs. A. Representative Wright-Giemsa stained plates from MEFs transfected with empty pcDNA3.1 and pBABE vectors, RasV12G, c-Myc, DPM-HIF-1α and DPM-HIF-2α. B. Colony counts from all transfection combinations are shown. Results from 3 experiments, error bars ±1 SEM, * p < 0.05, ** p < 0.01. C. Colony formation in soft agar by cell lines derived from foci. 20× and 1OO× magnifications are shown. D. Immunoblot analysis of HIF-2α, c-Myc and Ras overexpression in representative clones obtained from foci picked from plates transfected c-Myc, RasV12G, and pCDNA3.1, DPM-HIF-1α and DPM-HIF-2α. * indicates endogenous protein. E. Model for HIF-α regulation of c-Myc activity. We propose that HIF-1α specifically disrupts c-Myc/Max and c-Myc/Sp1 complexes, allowing more Mad/Max interaction and DNA binding. On the other hand, we hypothesize that HIF-2α stabilizes c-Myc/Max complexes, in turn promoting c-Myc DNA binding at both E boxes and Inrs.
Discussion
In this study, we found that HIF-2α promotes c-Myc transcriptional activity and cell cycle progression in renal carcinoma cells, NTH3T3 cells, HEK293 cells, and embryonic epithelial cells. Furthermore, HIF-2α promotes MEF transformation in concert with RasV12G and c-Myc. This is a novel pathway, and we believe it contributes substantially to the proliferation of normal and transformed cells under hypoxia, as well as the transformation of cells that have lost the VHL tumor suppressor. It is also the first description of HIF-1α and HIF-2α effects that directly oppose each other by altering a common pathway.
Previous data have demonstrated that HIF-1α modulates c-Myc function in hypoxic cells. In those studies, HIF-1α was shown to inhibit c-Myc transcriptional activity by inhibiting DNA binding, specifically through competition for Sp1 (Koshiji et al., 2004; Koshiji et al., 2005; To et al., 2006). We have repeated these findings, and also observed HIF-1α/Max binding, which may block c-Myc/Max interaction. While in previous studies, HIF-1α also bound DNA in the same location as c-Myc, we observed HIF-α binding at the p21 Inr but not the five other c-Myc target promoters tested, suggesting that displacement is not required to modulate c-Myc. HIF-2α also appears to regulate c-Myc by modulating c-Myc/Sp1, c-Myc/Miz1 and c-Myc/Max interactions. When HIF-2α is present, more c-Myc/Max, c-Myc/Miz and c-Myc/Sp1 complexes and fewer Mad/Max complexes are detected compared to normoxia. HIF-2α co-precipitated with Max, suggesting that it also interacts with these proteins, either through direct binding or as part of a larger complex. Based on these data and those previously reported (Koshiji et al., 2005; To et al., 2006), we propose the following model (Figure 8E): We believe that the presence of HIF-1α directly blocks c-Myc interaction with its DNA binding partners. HIF-2α, on the other hand, might promote c-Myc interaction with Max, and thus Sp1 and Miz1, by recruiting it directly to these complexes, or by stabilizing these complexes once they are formed. This might occur by selectively blocking Max interaction with Mad1, which competes with c-Myc for available cellular Max. Understanding the precise effects of HIF-1α/Max, HIF-1α/Sp1, and HIF-2α/Max interactions will require further analysis of the multimeric complexes formed.
Multiple HIF-2α targets besides c-Myc promote proliferation and transformation, and are necessary for tumor growth. HIF-2α directly activates the expression of Oct4, Cyclin D1, and TGFα, each of which can promote transformation in MEFs or NIH3T3 cells (Alt et al., 2000; Gidekel et al., 2003; Twardzik et al., 1982). HIF-2α activation of these targets is not responsible for transformation in our study as Oct4 and TGF-α were not detected in transformed MEF cell lines or NIH3T3 cells described above, and similar mRNA and protein levels were observed for Cyclin D1 regardless of HIF-α expression (data not shown). However, the activation of other HIF-2α targets may collaborate in c-Myc driven proliferation and transformation by blocking its effects on apoptosis.
Although HIF-2α is unique in its ability to promote proliferation and transformation, it shares HIF-1α's effects on angiogenesis, invasion, and metabolism, all of which contribute to tumor growth and progression. Many clinical studies have correlated the presence of either HIF-α subunit with poor patient outcomes. However, differential effects of the HIF-α submits may play a key role in cells that have more intact cell cycle control, as is the case for pre-neoplastic cells as well as many scientific model systems. Similarly, the amplitude of HIF-α activation may alter which pathways or effects are more dominant. For example, HIF-1α activation in Vhl−/− Ras-transformed MEFs inhibits subcutaneous allograft growth (Mack et al., 2005), while Hif-1α−/− Ras-transformed MEFs form smaller subcutaneous tumors than their WT counterparts (Ryan et al., 2000). Notably, the Vhl−/− MEFs exhibited enhanced vascularity but limited proliferation in vitro whereas the Hif-1α−/− MEFs exhibited limited VEGF expression. It appears that hypoxic HIF-1α activation is necessary for tumor angiogenesis, whereas continuous activation inhibits cell cycle progression. Modulation of hypoxic cell survival can also change tumor behavior, as observed in a recent study where HIF-1α or HIF-2α were overexpressed in glioma cells (Acker et al., 2005). Glioma is well known for extensive hypoxic domains with substantial cell death. This was promoted by HIF-α overexpression, leading to decreased tumor size due to apoptosis (Acker et al., 2005). Thus, each HIF-α may have the potential to be a tumor promoter or suppressor depending on the biology of a given tumor type and its stage of development.
These data have significant implications for tumor initiation and progression in multiple tumor types. In the context of RCC, several HIF-2α targets appear to be important in tumor initiation and progression, including VEGF, Oct4 and TGFα. Our data showing that HIF-2α promotes MEF transformation whereas HIF-1α inhibits it, support the evidence that relative increases in HIF-2α activity, as well as HIF-1α loss, correlate with the acquisition of dysplastic characteristics in VHL−/− renal lesions (Mandriota et al., 2002; Raval et al., 2005). As VEGF, Oct4, and Cyclin D1 are not relevant to MEF transformation in our studies, these data also suggest a unique role for HIF-2α mediated c-Myc activation in RCC initiation. Our findings are also relevant to other common malignancies. In recent studies of neuroblastoma and colorectal and non-small cell lung cancer, which express both α-subunits, the expression of HIF-2α was more conclusively associated with a poor prognosis than HIF-1α (Giatromanolaki et al., 2001; Holmquist-Mengelbier et al., 2006; Yoshimura et al., 2004). Given the data connecting c-Myc (or its family member n-Myc) to each tumor type, it is likely that HIF-2α mediated enhancement of c-Myc activity has a role in these, and other, human cancers.
Materials and Methods
Cell Culture
HCT116 (obtained from ATCC) and WT8 cells (kind gift of W.G. Kaelin) were cultured in DMEM with 10% FBS (Gemini Biosystems), 2 mM glutamine, 100 units/ml penicillin, 100 μg/ml streptomycin, and 0.1 mM MEM nonessential amino acids. MEFs were maintained in DMEM with 10% FBS (Hyclone), and the supplements above. ECs were derived and cultured as described (Mansfield et al., 2005). NIH3T3 cells (ATCC) were stably transfected with the reverse Tet-Activator according to the manufacturer's specifications (Clontech, Mountamview, CA), and then with pTRE-DPM-HIF-1α-Myctag and pTRE-DPM-HIF-2α-Myctag. Once clones were isolated, these lines (and the Tet-on HEK293 cells) were maintained in DMEM, 10% FBS (Clontech Tet-approved), 150 μg/ml Hygromycin B, and the supplements listed above. Doxycycline (Clontech) was used at 0.5 μg/ml in NIH3T3 cells and 2 μg/ml in HEK293 cells.
Hypoxia
Hypoxia (0.5% O2, 5% CO2, 94.5% N2) was achieved using an In Vivo2 hypoxic workstation (Ruskinn Technologies) or in a positive pressure chamber receiving gas from a custom mixed tank (Arrgas). DFX (Calbiochem) was used as a hypoxia mimetic at a final concentration of 200 μM.
Cell Cycle Analysis
For these experiments, cells were plated at a density such that they would be 50% confluent on the day of analysis. Treatment (hypoxia or doxycycline) was then initiated over the next several days, so that all cells were in culture for the same amount of time and at similar confluency when harvested. BrdU analysis was performed following the standard protocol (Becton Dickinson) after a 20 minute pulse with 10 μM BrdU. Cells were stained with Alexa-488 anti-BrdU (Invitrogen) and 0.1 M propidium iodide and analyzed in an LSR FACS machine (Becton Dickinson). For proliferation analysis, 104 cells were plated on 6 cm2 plates and 2 plates counted per time point in a hemocytometer over 8 days. Transformation assays are described in Supplementary Methods.
QRT-PCR
Total RNA was extracted from cells with Trizol reagent following the manufacturer's instructions (Invitrogen). cDNA was produced from 2μg of RNA using Superscript II (Invitrogen) with random hexamer primers (Boehringer Mannheim). Primers against 18S (Applied Biosystems) were used for the endogenous control in ΔΔ CT analysis. Primer sets were generated against human and mouse VEGF, PGK, Oct4, p27, and Cyclin D2, as well as human p21 and E2F1 (sequences available on request). Analysis was performed in an Applied Biosystems 7900HT Sequence Detection System, with amplification quantified with SYBR green.
siRNA Analysis
Specific knockdown was achieved using siRNAs against HIF-1α (Hs_HIF1_2 and 3), HIF-2α (Hs_HIF1_2 and 4) and c-Myc (Hs_Myc_2 and 3) or a control siRNA (all from Qiagen). Transfection was performed using HiPerfect reagent (Qiagen) as directed. Six hrs. after transfection, media was changed and cells were placed under hypoxia for 20 hrs. (for expression analysis) or 48 hrs. (for cell cycle analysis). Cells were harvested for protein and RNA analysis as described above.
Immunoprecipitation and Western Blot Analysis
For immunoprecipitation assays, cells were lysed in 25 mM Tris pH 8.0, 100 mM NaCl, and 1% Triton X-100 containing Complete protease inhibitors (Roche) and 200 μM DFX. For all other Western blots, lysis was performed in RIPA. Subcellular fractionation was performed as previously described (Qu et al., 2004). Antibodies and protocol for Chromatin IP are listed in the supplementary text.
Supplementary Material
Significance.
Activation of the Hypoxia Inducible Factors (HIFs) is a key feature of solid tumor biology. Here we describe a fundamental distinction between the two HIF-α subunits: Tumor cells expressing HIF-2α exhibit increased proliferation by promoting c-Myc transcriptional activity, while HIF-1α inhibits cell cycle progression by opposing c-Myc. HIF-2α's unique enhancement of cell cycle progression, combined with the effects of either HIF-α on angiogenesis, invasion and metastasis, is likely to produce worse clinical outcomes than activation of HIF-1α alone. This is consistent with clinical data from RCC, neuroblastoma, colorectal and non-small cell lung cancer, and highlights the importance of HIF-2α as a specific target of anticancer therapy.
Acknowledgements
We are grateful for the helpful advice of many members of the Abramson Family Cancer Research Institute, in particular Craig Thompson, and also Steve McMahon and Gerd Blobel. Brian Keith and Bryan Barnhart provided critical evaluation of the manuscript. This work was supported by NIH Program Project Grant no. CA104838 and the Howard Hughes Medical Institute. J.D.G. was supported by a Medical Scientist Training Grant and a NRSA T32 grant for Hemostasis and Thrombosis. J.A.B. was supported by a Veterinary Scientist Training Grant. M.C.S. is an investigator of the Howard Hughes Medical Institute.
References
- Acker T, Diez-Juan A, Aragones J, Tjwa M, Brusselmans K, Moons L, Fukumura D, Moreno-Murciano MP, Herbert JM, Burger A, et al. Genetic evidence for a tumor suppressor role of HIF-2alpha. Cancer Cell. 2005;8:131–141. doi: 10.1016/j.ccr.2005.07.003. [DOI] [PubMed] [Google Scholar]
- Adhikary S, Eilers M. Transcriptional regulation and transformation by Myc proteins. Nat Rev Mol Cell Biol. 2005;6:635–645. doi: 10.1038/nrm1703. [DOI] [PubMed] [Google Scholar]
- Alt JR, Cleveland JL, Hannink M, Diehl JA. Phosphorylation-dependent regulation of cyclin D1 nuclear export and cyclin D1-dependent cellular transformation. Genes Dev. 2000;14:3102–3114. doi: 10.1101/gad.854900. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Baba M, Hirai S, Yamada-Okabe H, Hamada K, Tabuchi H, Kobayashi K, Kondo K, Yoshida M, Yamashita A, Kishida T, et al. Loss of von Hippel-Lindau protein causes cell density dependent deregulation of Cyclin D1 expression through hypoxia-inducible factor. Oncogene. 2003;22:2728–2738. doi: 10.1038/sj.onc.1206373. [DOI] [PubMed] [Google Scholar]
- Bouchard C, Thieke K, Maier A, Saffrich R, Hanley-Hyde J, Ansorge W, Reed S, Sicinski P, Bartek J, Eilers M. Direct induction of cyclin D2 by Myc contributes to cell cycle progression and sequestration of p27. Embo J. 1999;18:5321–5333. doi: 10.1093/emboj/18.19.5321. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Carmeliet P, Dor Y, Herbert JM, Fukumura D, Brusselmans K, Dewerchin M, Neeman M, Bono F, Abramovitch R, Maxwell P, et al. Role of HIF-1alpha in hypoxia-mediated apoptosis, cell proliferation and tumour angiogenesis. Nature. 1998;394:485–490. doi: 10.1038/28867. [DOI] [PubMed] [Google Scholar]
- Coller HA, Grandori C, Tamayo P, Colbert T, Lander ES, Eisenman RN, Golub TR. Expression analysis with oligonucleotide microarrays reveals that MYC regulates genes involved in growth, cell cycle, signaling, and adhesion. Proc Natl Acad Sci USA. 2000;97:3260–3265. doi: 10.1073/pnas.97.7.3260. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Compernolle V, Brusselmans K, Acker T, Hoet P, Tjwa M, Beck H, Plaisance S, Dor Y, Keshet E, Lupu F, et al. Loss of HIF-2alpha and inhibition of VEGF impair fetal lung maturation, whereas treatment with VEGF prevents fatal respiratory distress in premature mice. Nat Med. 2002;8:702–710. doi: 10.1038/nm721. [DOI] [PubMed] [Google Scholar]
- Covello KL, Kehler J, Yu H, Gordan JD, Arsham AM, Hu CJ, Labosky PA, Simon MC, Keith B. HIF-2alpha regulates Oct-4: effects of hypoxia on stem cell function, embryonic development, and tumor growth. Genes Dev. 2006;20:557–570. doi: 10.1101/gad.1399906. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Covello KL, Simon MC, Keith B. Targeted replacement of hypoxia-inducible factor-1alpha by a hypoxia-inducible factor-2alpha knock-in allete promotes tumor growth. Cancer Res. 2005;65:2277–2286. doi: 10.1158/0008-5472.CAN-04-3246. [DOI] [PubMed] [Google Scholar]
- Ema M, Taya S, Yokotani N, Sogawa K, Matsuda Y, Fujii-Kuriyama Y. A novel bHLH-PAS factor with close sequence similarity to hypoxia-inducible factor 1alpha regulates the VEGF expression and is potentially involved in lung and vascular development. Proc Natl Acad Sci U S A. 1997;94:4273–4278. doi: 10.1073/pnas.94.9.4273. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Erler JT, Bennewith KL, Nicolau M, Dornhofer N, Kong C, Le QT, Chi JT, Jeffrey SS, Giaccia AJ. Lysyl oxidaseis is essential for hypoxia-induced metastasis. Nature. 2006;440:1222–1226. doi: 10.1038/nature04695. [DOI] [PubMed] [Google Scholar]
- Fernandez PC, Frank SR, Wang L, Schroede M, Liu S, Greene J, Cocito A, Amati B. Genomic targets of the human c-Myc protein. Genes Dev. 2003;17:1115–1129. doi: 10.1101/gad.1067003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gardner LB, Li Q, Park MS, Flanagan WM, Semenza GL, Dang CV. Hypoxia inhibits G1/S transition through regulation of p27 expression. J Biol Chem. 2001;276:7919–7926. doi: 10.1074/jbc.M010189200. [DOI] [PubMed] [Google Scholar]
- Giatromanolaki A, Koukourakis MI, Sivridis E, Turley H, Talks K, Pezzella F, Gatter KC, Harris AL. Relation of hypoxia inducible factor 1 alpha and 2 alpha in operable non-small cell lung cancer to angiogenic/molecular profile of tumours and survival. Br J Cancer. 2001;85:881–890. doi: 10.1054/bjoc.2001.2018. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gidekel S, Pizov G, Bergman Y, Pikarsky E. Oct-3/4 is a dose-dependent oncogenic fate determinant. Cancer Cell. 2003;4:361–370. doi: 10.1016/s1535-6108(03)00270-8. [DOI] [PubMed] [Google Scholar]
- Goda N, Ryan HE, Khadivi B, McNutty W, Rickert RC, Johnson RS. Hypoxia-inducible factor 1 alpha is essential for cell cycle arrest during hypoxia. Mol Cell Biol. 2003;23:359–369. doi: 10.1128/MCB.23.1.359-369.2003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Green SL, Freiberg RA, Giaccia AJ. p21(Cip1) and p27(Kip1) regulate cell cycle reentry after hypoxic stress but are not necessary for hypoxia-induced arrest. Mol Cell Biol. 2001;21:1196–1206. doi: 10.1128/MCB.21.4.1196-1206.2001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gunaratnam L, Morley M, Franovic A, de Paulsen N, Mekhail K, Parolin DA, Nakamura E, Lorimer IA, Lee S. Hypoxia inducible factor activates the transforming growth factor-alpha/epidermal growth factor receptor growth stimulatory pathway in VHL(−/−) renal ceil carcinoma cells. J Biol Chem. 2003;278:44966–44974. doi: 10.1074/jbc.M305502200. [DOI] [PubMed] [Google Scholar]
- Holmquist-Mengelbier L, Fredlund E, Lofstedt T, Noguera R, Navarro S, Nilsson H, Pietras A, Vallon-Christersson J, Borg A, Gradin K, et al. Recruitment of HIF-1alpha and HIF-2alpha to common target genes is differentially regulated in neuroblastoma: HIF-2alpha promotes an aggressive phenotype. Cancer Cell. 2006;10:413–423. doi: 10.1016/j.ccr.2006.08.026. [DOI] [PubMed] [Google Scholar]
- Hopfl G, Wenger RH, Ziegler U, Stalhnach T, Gardelle O, Achermann R, Wergin M, Kaser-Hotz B, Saunders HM, Williams KJ, et al. Rescue of hypoxia-inducible factor-1alpha-deficient tumor growth by wild-type cells is independent of vascular endothelial growth factor. Cancer Res. 2002;62:2962–2970. [PubMed] [Google Scholar]
- Hu CJ, Wang LY, Chodosh LA, Keith B, Simon MC. Differential roles of hypoxia-inducible factor 1alpha (HIF-1alpha) and HIF-2alpha in hypoxic gene regulation. Mol Cell Biol. 2003;23:9361–9374. doi: 10.1128/MCB.23.24.9361-9374.2003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Iyer NV, Kotch LE, Agani F, Leung SW, Laughner E, Wenger RH, Gassmann M, Gearhart JD, Lawler AM, Yu AY, Semenza GL. Cellular and developmental control of O2 homeostasis by hypoxia-inducible factor 1 alpha. Genes Dev. 1998;12:149–162. doi: 10.1101/gad.12.2.149. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kondo K, Kim WY, Lechpammer M, Kaelin WG., Jr. Inhibition of HIF2alpha is sufficient to suppress pVHL-defective tumor growth. PLoS Biol. 2003;1:E83. doi: 10.1371/journal.pbio.0000083. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kondo K, Klco J, Nakamura E, Lechpammer M, Kaelin WG., Jr. Inhibition of HIF is necessary for tumor suppression by the von Hippel-Lindau protein. Cancer Cell. 2002;1:237–246. doi: 10.1016/s1535-6108(02)00043-0. [DOI] [PubMed] [Google Scholar]
- Koshiji M, Kageyama Y, Pete EA, Horikawa I, Barrett JC, Huang LE. HIF-1alpha induces cell cycle arrest by functionally counteracting Myc. Embo J. 2004;23:1949–1956. doi: 10.1038/sj.emboj.7600196. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Koshiji M, To KK, Hammer S, Kumamoto K, Harris AL, Modrich P, Huang LE. HIF-1alpha induces genetic instability by transcriptionally downregulating MutSalpha expression. Mol Cell. 2005;17:793–803. doi: 10.1016/j.molcel.2005.02.015. [DOI] [PubMed] [Google Scholar]
- Mack FA, Patel JH, Biju MP, Flaase VH, Simon MC. Decreased growth of Vhl−/−fibrosarcomas is associated with elevated levels of cyclin kinase inhibitors p21 and p27. Mol Cell Biol. 2005;25:4565–4578. doi: 10.1128/MCB.25.11.4565-4578.2005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mandriota SJ, Turner KJ, Davies DR, Murray PG, Morgan NV, Sowter HM, Wykoff CC, Maher ER, Harris AL, Ratcliffe PJ, Maxwell PH. HIF activation identifies early lesions in VHL kidneys: evidence for site-specific tumor suppressor function in the nephron. Cancer Cell. 2002;1:459–468. doi: 10.1016/s1535-6108(02)00071-5. [DOI] [PubMed] [Google Scholar]
- Mansfield KD, Guzy RD, Pan Y, Young RM, Cash TP, Schumacker PT, Simon MC. Mitochondria] dysfunction resulting from loss of cytochrome c impairs cellular oxygen sensing and hypoxic HIF-alpha activation. Cell Metab. 2005;1:393–399. doi: 10.1016/j.cmet.2005.05.003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Maranchie JK, Vasselli JR, Riss J, Bonifacino JS, Linehan WM, Klausner RD. The contribution of VHL substrate binding and HIF1-alpha to the phenotype of VHL loss in renal cell carcinoma. Cancer Cell. 2002;1:247–255. doi: 10.1016/s1535-6108(02)00044-2. [DOI] [PubMed] [Google Scholar]
- Maxwell PH, Wiesener MS, Chang GW, Clifford SC, Vaux EC, Cockman ME, Wykoff CC, Pugh CW, Maher ER, Ratcliffe PJ. The tumour suppressor protein VHL targets hypoxia-inducible factors for oxygen-dependent proteolysis. Nature. 1999;399:271–275. doi: 10.1038/20459. [DOI] [PubMed] [Google Scholar]
- Nilsson JA, Cleveland JL. Myc pathways provoking cell suicide and cancer. Oncogene. 2003;22:9007–9021. doi: 10.1038/sj.onc.1207261. [DOI] [PubMed] [Google Scholar]
- Peng J, Zhang L, Drysdale L, Fong GH. The transcription factor EPAS-1/hypoxia-inducible factor 2alpha plays an important role in vascular remodeling. Proc Natl Acad Sci U S A. 2000;97:8386–8391. doi: 10.1073/pnas.140087397. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pugh CW, Ratcliffe PJ. Regulation of angiogenesis by hypoxia: role of the HIF system. Nat Med. 2003;9:677–684. doi: 10.1038/nm0603-677. [DOI] [PubMed] [Google Scholar]
- Qu L, Huang S, Baltzis D, Rivas-Estilla AM, Pluquet O, Hatzoglou M, Koumenis C, Taya Y, Yoshimura A, Koromilas AE. Endoplasmic reticulum stress induces p53 cytoplasmic localization and prevents p53-dependent apoptosis by a pathway involving glycogen synthase kinase-3beta. Genes Dev. 2004;18:261–277. doi: 10.1101/gad.1165804. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Raval RR, Lau KW, Tran MG, Sowter HM, Mandriota SJ, Li JL, Pugh CW, Maxwell PH, Harris AL, Ratcliffe PJ. Contrasting properties of hypoxia-inducible factor 1 (HIF-1) and HIF-2 in von Hippel-Lindau-associated renal cell carcinoma. Mol Cell Biol. 2005;25:5675–5686. doi: 10.1128/MCB.25.13.5675-5686.2005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ryan HE, Lo J, Johnson RS. HIF-1 alpha is required for solid tumor formation and embryonic vascularization. Embo J. 1998;17:3005–3015. doi: 10.1093/emboj/17.11.3005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ryan HE, Poloni M, McNulty W, Elson D, Gassmann M, Arbeit JM, Johnson RS. Hypoxia-inducible factor-1 alpha is a positive factor in solid tumor growth. Cancer Res. 2000;60:4010–4015. [PubMed] [Google Scholar]
- Scortegagna M, Ding K, Oktay Y, Gaur A, Thurmond F, Yan LJ, Marck BT, Matsumoto AM, Shelton JM, Richardson JA, et al. Multiple organ pathology, metabolic abnormalities and impaired homeostasis of reactive oxygen species in Epas1−/− mice. Nat Genet. 2003;35:331–340. doi: 10.1038/ng1266. [DOI] [PubMed] [Google Scholar]
- Semenza GL. Targeting HIF-1 for cancer therapy. Nat Rev Cancer. 2003;3:721–732. doi: 10.1038/nrc1187. [DOI] [PubMed] [Google Scholar]
- Tian H, Hammer RE, Matsumoto AM, Russell DW, McKnight SL. The hypoxia-responsive transcription factor EPAS1 is essential for catecholamine homeostasis and protection against heart failure during embryonic development. Genes Dev. 1998;12:3320–3324. doi: 10.1101/gad.12.21.3320. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tian H, McKnight SL, Russell DW. Endothelial PAS domain protein 1 (EPAS1), a transcription factor selectively expressed in endothelial cells. Genes Dev. 1997;11:72–82. doi: 10.1101/gad.11.1.72. [DOI] [PubMed] [Google Scholar]
- To KK, Sedelnikova OA, Samons M, Bonner WM, Huang LE. The phosphorylation status of PAS-B distinguishes HIF-1alpha from HIF-2alpha in NBS1 repression. Embo J. 2006;25:4784–4794. doi: 10.1038/sj.emboj.7601369. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Twardzik DR, Todaro GJ, Marquardt H, Reynolds FH, Jr., Stephenson JR. Transformation induced by Abelson murine leukemia virus involves production of a polypeptide growth factor. Science. 1982;216:894–897. doi: 10.1126/science.6177040. [DOI] [PubMed] [Google Scholar]
- Wang V, Davis DA, Haque M, Huang LE, Yarchoan R. Differential gene up-regulation by hypoxia-inducible factor-1alpha and hypoxia-inducible factor-2alpha in HEK293T cells. Cancer Res. 2005;65:3299–3306. doi: 10.1158/0008-5472.CAN-04-4130. [DOI] [PubMed] [Google Scholar]
- Wiesener MS, Jurgensen JS, Rosenberger C, Scholze CK, Horstrup JH, Warnecke C, Mandriota S, Bechmann I, Frei UA, Pugh CW, et al. Widespread hypoxia-inducible expression of HIF-2alpha in distinct cell populations of different organs. Faseb J. 2003;77:271–273. doi: 10.1096/fj.02-0445fje. [DOI] [PubMed] [Google Scholar]
- Yoshimura H, Dhar DK, Kohno H, Kubota H, Fujii T, Ueda S, Kinugasa S, Tachibana M, Nagasue N. Prognostic impact of hypoxia-inducible factors 1alpha and 2alpha in colorectal cancer patients: correlation with tumor angiogenesis and cyclooxygenase-2 expression. Clin Cancer Res. 2004;10:8554–8560. doi: 10.1158/1078-0432.CCR-0946-03. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.