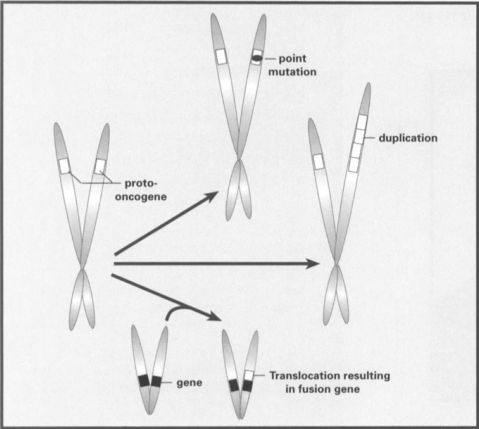
More than two decades ago, it was discovered that most cancer was the result of mutations occurring in normal genes in a single cell in the patient's body. This discovery of ‘oncogenes’ led to a massive effort to understand the genetic basis behind neoplasia. The typical types of mutations associated with oncogenes are outlined in Figure 1. These mutations generally result in a mutation that either changes the regulation of the protein coded by the oncogene or changes the level of expression of that protein in one or more cell types in the body, These mutations generally lead to an increase of that protein's normal activity, resulting in stimulated cell growth.
Figure 1.
Typical mutations transforming a normal proto-oncogene into an oncogene. The most common types of mutations associated with oncogenes involve point mutations that alter the control of the oncoprotein made from the gene (as in the ras family of oncogenes), duplication of oncogenes resulting in higher expression of the oncogene, or chromosomal translocations that fuse the oncogene with a different gene. The latter change either results in an oncoprotein of altered function or results in changes in expression pattern of the oncogene
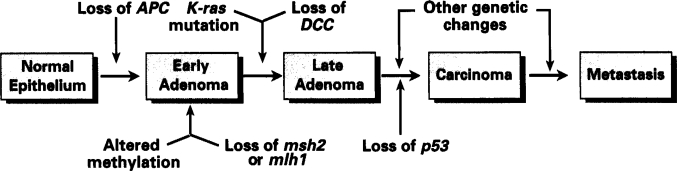
No single oncogene is capable of turning a normal cell into a transformed, neoplastic cell. It has been found that most cancers are the result of a series of mutations in a number of different genes. Figure 2 presents a simplified schematic of some of the specific genetic changes that are generally thought to be associated with colon cancer. A cell with only one or two genetic mutations in important genes may have a tendency to develop into a polyp or benign growth. If one of the cells within that polyp undergoes further mutation, it may have a tendency to become malignant and, as more genes in some of the cells accumulate mutations, malignant potential increases. Mutations in more genes are then required to make a tumor increase in metastatic potential.
Figure 2.
Simplified diagram of typical progression to colon cancer. The APC, DCC and p53 genes represent tumor suppressor genes that are often inactivated in this progression, resulting in poor growth control of the cells. K-ras represents one of several oncogenes that are commonly mutated in colon cancer progression, resulting in stimulation of tumor cell growth. Altered methylation of DNA results in changes in expression of genes associated with the methylation, and potentially contribute to some genomic instability. The msh2 and mlh1 genes are associated with DNA mismatch repair, and their mutation leads to rapid DNA mutation
The probability of many key mutations occurring in a single cell should normally be quite low. However, as discussed below, certain specific mutations lead to an increase in the mutation rate in some pre-cancerous cells. These increased mutation rates greatly increase the probability that more mutations will occur in other genes, which seems to be a critical and common step that leads to cancer progression.
Gatekeeper Genes
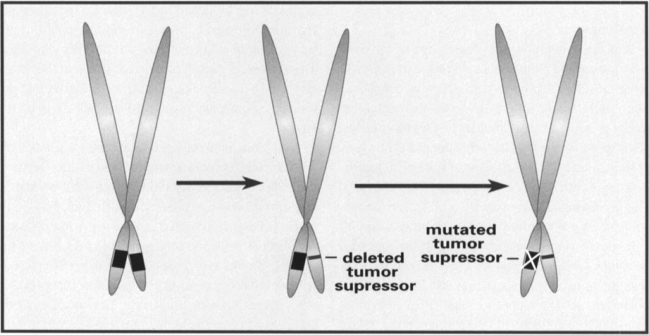
After the discovery that normal genes may become oncogenes, the next step in our understanding of cancer genetics involved the discovery of ‘gatekeeper’ genes that are responsible for controlling, or inhibiting, cell growth. These gatekeeper genes are generally of the type referred to as tumor suppressor genes. When both copies of a tumor suppressor gene are damaged in a cell, the inhibitory gene product can no longer be made, and the cells have one less control of their growth. This helps lead to the uncontrolled growth associated with tumors. The basic process of tumor suppressor gene inactivation is shown in Figure 3. These mutations generally lead to the loss of function by the tumor suppressor gene, resulting in increased cell growth.
Figure 3.
Loss of a tumor suppressor. The tumor suppressor gene is represented as a black box on the short arms of the chromosomes. The first step in loss of the tumor suppressor gene is often deletion of all or part of one copy of the gene. Families with strong predispositions to cancer often have one copy of a tumor suppressor gene deleted. There are no direct negative effects of loss of only one copy of the tumor suppressor gene. However, eventually something occurs to silence the second copy of the tumor suppressor gene, through either mutation or methylation. At this point, the protein coded by the tumor suppressor gene is no longer available to help suppress cell growth
A number of germ-line mutations in tumor suppressor genes have been found in families that have very high genetic risk for cancers. The prototype for this type of tumor suppressor is the retinoblastoma gene, Rb. A single copy of the gene can make enough tumor suppressor gene product to control cell growth. Someone born with a mutation in one of the two copies of Rb present in every cell will be completely healthy until a mutation occurs in the second copy of the Rb gene in a cell. That cell will then begin to proliferate and may form a tumor. This particular tumor suppressor gene seems to play a major role in the retina; therefore, defects lead to retinoblastoma.
Retinoblastoma can also occur in patients from families without familial Rb mutations. However, the probability of both copies of the Rb gene becoming defective in the same cell is quite small, making the sporadic occurrence of retinoblastoma relatively rare.
A number of other familial predispositions to cancer are associated with germ-line mutations in tumor suppressor genes. These include mutations in the APC gene, associated with familial adenomatous polyposis, which leads to very high incidence of colon cancer. The BRCA1 and BRCA2 genes have also been highly publicized for their role in familial predisposition to breast cancer.
Caretaker Genes
Why does mutation of a single copy of a tumor suppressor gene make an individual so prone to cancer if many genes must be mutated to reach the fully neoplastic state? Many of the tumor suppressor genes also play some role, directly or indirectly, as ‘caretakers.’ Caretaker genes are genes responsible for keeping other genes healthy (i.e. suppressing mutation). A good example of a tumor suppressor gene with some caretaker capability is the p53 gene. This gene is mutated in over 50% of all human tumors and is therefore a major contributor to the progression of cancer. Besides playing a direct growth regulatory role, p53 plays a secondary role in helping the genome recover from damaging mutations. It plays a critical role in a cell cycle ‘checkpoint’ prior to DNA replication, that either allows the cell to pause and repair damaged DNA, or signals the cell to destroy itself through a pathway called ‘apoptosis’ if it is too badly damaged. Thus, defects in p53 gene lead to a lessening of the cell's ability to care for damage to its own DNA. This then leads to higher mutation rates, either spontaneously or due to exposure to mutagens, leading to rapid accumulation of defects in other genes.
The true ‘caretaker’ genes, however, are generally more directly involved with the repair of the DNA and create much higher mutation levels than those normally associated with p53 mutations. In addition, they can cause high mutation rates even in cells that are not exposed to mutagens. Two of the best-studied caretaker genes are mlh1 and msh2, which are genes involved in mismatch repair of DNA bases that have been misincorporated during DNA replication. Thus, mutations in these genes greatly increase the rate of point mutations in genes. With this higher rate of mutation, it is then a question of when enough mutations will occur to cause tumor development, rather than if a tumor might develop.
There are types of DNA instability, other than mismatch repair, that are even more common in cancer. The mechanisms of these types of instabilities are less well understood, but they involve deletion or duplication of major segments of chromosomes, chromosomal translocations, aneuploidy, and other chromosomal aberrations. These chromosomal instabilities contribute to cancer progression in a number of ways.
Deletions of segments of DNA can result in loss of tumor suppressor genes (Figure 3), which leads the tumor to grow more aggressively. Duplications of segments of chromosomes can cause increased copy numbers of protooncogenes (Figure 1), leading to higher levels of expression which lead these genes to stimulate tumor growth. Chromosomal translocations have also been found to contribute to tumorigenesis by causing the fusion of an oncogene with a different gene (Figure 1). The chromosomal translocation usually results in the fusion of the proto-oncogene with a different gene on the other chromosome. The expression pattern of the fusion gene in the tissues of the body is usually altered relative to the normal proto-oncogene expression. Again, this can result in specific tumor types.
In a number of genetic diseases, such as Bloom's syndrome and Fanconi's anemia, the primary defect seems to be chromosomal instability. Individuals with these syndromes are very susceptible to tumorigenesis as well. Thus, it is clear that a number of single genes can contribute to this type of instability. The Bloom's syndrome gene appears to be a helicase that is probably involved in some aspect of chromosome replication. A number of different genes may actually cause Fanconi's anemia, although their functions are unknown. Thus, a significant number of individual genes are likely to be able to contribute to the chromosomal instability of tumors if they are damaged.
Clinical Implications of Tumor DNA Instability
With the discovery of a number of common genetic defects associated with many tumors, it is now becoming possible to refine the classification of tumors on a molecular basis. Genetics is already used in some instances for diagnosis of tumors and predisposition to tumors. A number of molecular markers also serve as important prognostic indicators for some tumor types. One such marker is DNA instability. Numerous studies show that tumors with more unstable genomes are much more aggressive and more likely to recur following treatment. Such prognostic indicators may soon be useful tools in deciding how aggressively to treat a patient. In addition, it is becoming possible to direct individual therapies at tumors with specific genetic defects. One example involves trials with gene therapy vectors carrying p53 tumor suppressor genes. These gene therapy vectors are used to replace the defective gene in tumors that have defective p53. Defining the molecular defects associated with tumors and their genetic basis will allow us to tailor more specific treatments to the subsets of tumors with those specific defects. Any of these treatments, however, will not be long lasting unless they address any underlying genetic instabilities that have developed.
Suggested Further Reading
- 1.Lengauer C, Kinzler KW, Vogelstein B. Genetic instabilities in human cancers. Nature. 1998;396:643–649. doi: 10.1038/25292. [DOI] [PubMed] [Google Scholar]
- 2.Gupta RC, Lutz WK. Inherited and inducible chromosomal instability: a fragile bridge between genome integrity mechanisms and tumourigenesis. J Pathol. 1999;187:19–27. doi: 10.1002/(SICI)1096-9896(199901)187:1<19::AID-PATH233>3.0.CO;2-1. [DOI] [PubMed] [Google Scholar]
- 3.Stass SA, Mixson J. Oncogenes and tumor suppressor genes: therapeutic implications. Clin Cancer Res. 1997;3:2687–2695. [PubMed] [Google Scholar]
- 4.Bevilacqua RA, Nunes DN, Stroun M et al. The use of genetic instability as a clinical tool for cancer diagnosis. Semin Cancer Biol. 1998;8:447–453. doi: 10.1006/scbi.1998.0122. [DOI] [PubMed] [Google Scholar]
- 5.Lengauer C, Kinzler KW, Vogelstein B. Genetic instability in colorectal cancers. Nature. 1997;386:623–627. doi: 10.1038/386623a0. [DOI] [PubMed] [Google Scholar]