Abstract
Despite its universal importance for controlling gene expression, messenger RNA degradation was initially thought to occur by disparate mechanisms in eukaryotes and bacteria. This conclusion was based on differences in the structures used by such organisms to protect mRNA termini and in the ribonucleases and modifying enzymes originally implicated in message decay. Subsequent discoveries have identified a number of striking parallels between the cellular factors and molecular events that govern mRNA degradation in those two kingdoms of life. Nevertheless, some key distinctions remain, the most fundamental of which may be related to the dissimilar mechanisms by which eukaryotes and bacteria control translation initiation.
Protein synthesis is one of the most important biochemical processes in all living cells. It is also among the costliest, requiring substantial investments of energy and resources. Therefore, to maximize their competitive advantage in an ever-changing environment, all organisms must precisely regulate this process so as to produce exactly the proteins that are needed in just the right amounts. Achieving that end requires an ability to degrade mRNA so that patterns of protein synthesis can be altered rapidly. In so doing, cells recycle ribonucleotides for incorporation into new RNA molecules.
mRNA degradation directly affects protein synthesis through its impact on the concentration of mRNA available for translation. Its influence on the expression of individual genes reflects the diverse lifetimes of mRNAs, whose half-lives can differ by as much as two orders of magnitude in the same cell. For example, in rapidly dividing bacterial cells, mRNA half-lives typically range from a fraction of a minute to as long as an hour, whereas in the cells of higher eukaryotes, which divide less frequently, those half-lives range from several minutes to more than a day. The lifetimes of mRNAs often are not invariant but are instead modulated in response to the changing needs of cells for the proteins those messages encode.
Because of the many real and presumed differences between bacterial and eukaryotic mRNAs and the enzymes available to degrade them, it was initially thought that the mechanisms by which messages are degraded in these two kingdoms of life were quite different as well. One by one, those distinctions have fallen by the wayside as a result of new discoveries that have revealed unexpected mechanistic parallels. These parallels, and the distinctions that remain, are the subject of this review. After summarizing earlier views that mRNA decay is generally governed by endonucleolytic events in bacteria and exonucleolytic events in eukaryotes, more recent evidence for the importance of 3’- and 5’-terminal degradative phenomena in bacterial cells and of internal cleavage in eukaryotic cells will be described. The influence of quality-control mechanisms and noncoding RNAs on bacterial and eukaryotic mRNA degradation will also be compared. Finally, possible explanations for some fundamental disparities between mRNA decay in bacteria and eukaryotes will be addressed. mRNA turnover in archaea will not be reviewed here because much less is known about it.
BREAKDOWN: FIRST IMPRESSIONS
The models initially conceived to explain mRNA decay were strongly influenced by differences in the structure of bacterial and eukaryotic mRNAs and by early studies of mRNA degradation in two model organisms: Escherichia coli and Saccharomyces cerevisiae.
Shapes of Things: mRNA Structural Dissimilarities
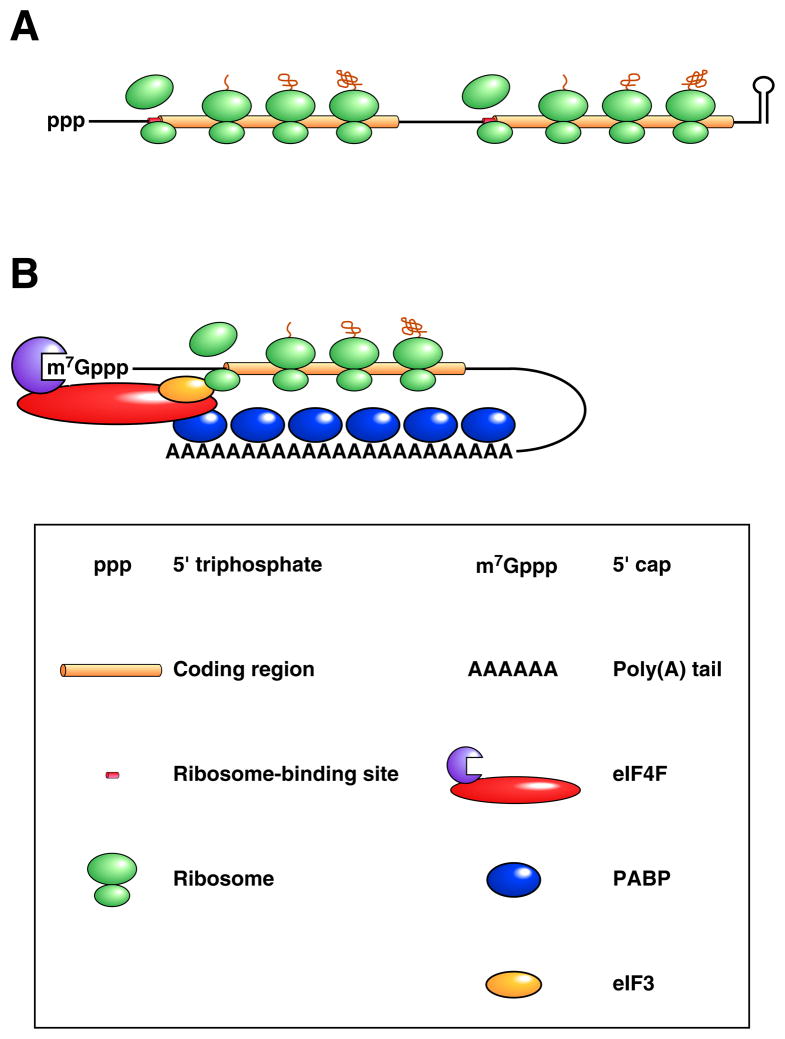
The structure and organization of bacterial and eukaryotic mRNAs differ in a number of significant ways (Figure 1). For example, while eukaryotic messages are capped at the 5’ end with a methylated guanosine connected via a 5’-5’ triphosphate linkage (m7GpppN…), their bacterial counterparts begin with a simple 5’-terminal triphosphate (pppN…). Furthermore, eukaryotic mRNAs typically end with a long, 3’-terminal poly(A) tail that is added post-transcriptionally, whereas few if any additional nucleotides are found at the 3’ terminus of bacterial messages, which instead typically end with a stem-loop structure. The protein complexes that assemble on the 5’-terminal cap and 3’-terminal poly(A) tail of eukaryotic mRNAs (eukaryotic translation initiation factor 4F [eIF4F] and poly(A)-binding protein [PABP], respectively) can interact with one another1, thereby causing messages to assume a noncovalent closed-loop conformation that is not thought to be characteristic of bacterial transcripts.
Figure 1. Differences in the structure and translation of bacterial and eukaryotic mRNAs.
(A) Bacterial mRNA. A translated dicistronic transcript that begins with a triphosphate and ends with a stem-loop is depicted. Ribosome binding to the beginning of each translational unit is aided by base pairing between the 3’ end of 16S ribosomal RNA (a component of the small ribosomal subunit) and a Shine-Dalgarno element adjacent to the initiation codon (collectively termed the ribosome binding site). (B) Eukaryotic mRNA. A message undergoing cap-dependent translation is shown. Ribosome binding to the translation initiation codon is guided by the affinity of the small ribosomal subunit for eukaryotic initiation factor 3 (eIF3), a protein multimer recruited to mRNA by the cap-binding complex eIF4F, whose affinity for mRNA is enhanced by its ability to bind PABP associated with the 3’ poly(A) tail.
The mechanisms by which ribosomes are recruited to bacterial and eukaryotic mRNAs are also quite different. In bacteria, ribosome binding is generally mediated by a complementary sequence element (the Shine-Dalgarno element) located just upstream of the initiation codon 2 (Figure 1A). The internal location of the signals that govern translation initiation makes it possible for a single polycistronic bacterial transcript to contain multiple translational units, each with its own ribosome-binding site and protein product. By contrast, eukaryotic ribosomes ordinarily are recruited to mRNA via the affinity of the small ribosomal subunit for the multiprotein complex (comprising eIF4F and eIF3) that assembles on the cap, leading to translation initiation at a nearby AUG codon 3, 4 (Figure 1B). As a result, most eukaryotic mRNAs encode only one protein. Although ribosomes are sometimes recruited directly to the initiation codon of eukaryotic messages by a cap-independent process involving an internal ribosome entry site (IRES), this alternative mechanism of translation initiation is uncommon 5, 6.
Go Your Own Way: Supposed Pathway Differences
Early studies suggested that the principal pathways for mRNA decay in bacteria and eukaryotes were quite different due to disparities in RNA structure, degradative enzymes, and the location of elements controlling mRNA stability.
The conventional model for mRNA degradation in bacterial cells (Figure 2A) was based entirely on studies in E. coli and strongly influenced by the kinds of ribonucleases that are present in that organism. E. coli contains a number of endonucleases and 3’ exonucleases (Table 1) but appears to lack any 5’ exonuclease capable of degrading RNA from the 5’ terminus. Because exonucleolytic digestion of E. coli mRNA from the 3’ end is impeded by the stem-loop structure typically present there, it was concluded that degradation of bacterial messages must begin with endonucleolytic cleavage at one or more internal sites to produce a pair of short-lived decay intermediates 7, 8. Lacking a protective 3’ stem-loop, the 5’ fragment thereby generated would be susceptible to 3’ exonuclease attack, while the 3’ fragment was assumed to undergo additional cycles of endonuclease cleavage and 3’ exonuclease digestion. Subsequent studies revealed that the endonuclease most important for mRNA turnover in E. coli is RNase E 9–14, a low-specificity ribonuclease that cleaves RNA in single-stranded regions that are AU-rich 15. Further investigation indicated that the rate at which RNase E degrades mRNA in E. coli is frequently determined by characteristics of the 5’ untranslated region (UTR), such as base pairing at the 5’ terminus and efficient ribosome binding, both of which have a protective effect 16–19. Four E. coli 3’ exonucleases – polynucleotide phosphorylase (PNPase), RNase II, RNase R, and oligoribonuclease – were also implicated in mRNA degradation as scavengers of RNA fragments lacking protection at the 3’ end 20–22. Interestingly, RNase E and PNPase associate with one another as subunits of the RNA degradosome, a multiprotein complex important for RNA processing and degradation that also contains an RNA helicase (RhlB) and a glycolytic enzyme (enolase) 23.
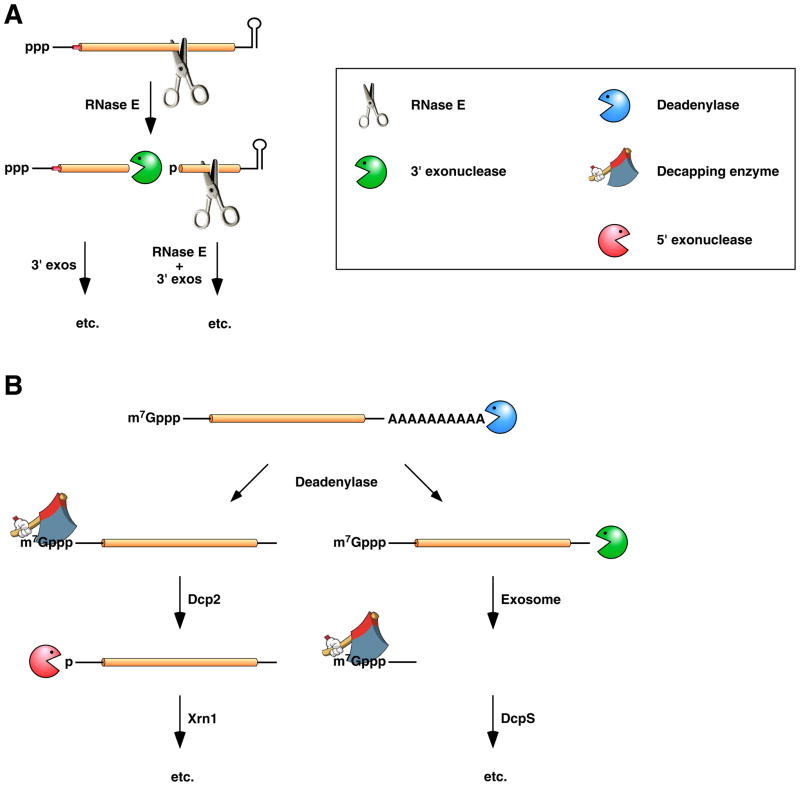
Figure 2. Conventional pathways for mRNA degradation in E. coli and in eukaryotic cells.
(A) mRNA decay in E. coli. In this pathway, serial internal cleavage by RNase E generates degradation intermediates whose lack of base pairing at the 3’ end renders them susceptible to attack by the 3’ exonucleases polynucleotide phosphorylase, RNase II, RNase R, and (for very short RNA fragments) oligoribonuclease. By contrast, the intact transcript resists exonucleolytic degradation because it is protected by a 3’-terminal stem-loop, which hinders such attack. (B) mRNA decay in eukaryotic cells. In this pathway, poly(A) tail removal by a deadenylase (Ccr4-Not, Pan2-Pan3, or PARN) yields a deadenylated intermediate susceptible both to decapping by Dcp2 and to 3’-exonucleolytic degradation by exosomes. The decapped RNA generated by Dcp2 is then degraded by the 5’-exonuclease Xrn1, whereas the 5’-terminal RNA fragment that results from extensive exosome digestion undergoes cap removal by an alternative decapping enzyme (DcpS) specific for oligonucleotides 147. These pathways were deduced from early studies of mRNA degradation in E. coli, S. cerevisiae, and mammalian cells. Ribosomes, PABP, and translation factors have been omitted from this figure for the sake of simplicity.
Table 1.
Enzymes of broad importance for cytoplasmic mRNA decay
| Enzyme | Specificity/Function | |
|---|---|---|
| Endonucleases | ||
| Bacteria: | RNase E*, RNase G* | Single-stranded RNA |
| RNase III | Double-stranded RNA | |
| RNase J | Single-stranded RNA | |
| RNase Y | Single-stranded RNA | |
| Cmr complex | mRNA-CRISPR RNA duplexes | |
| Eukaryotes: | Ago | mRNA-siRNA or mRNA-miRNA duplexes that are fully paired |
| SMG6 | PTC-containing mRNAs | |
| 3’ Exonucleases | ||
| Bacteria: | Polynucleotide phosphorylase | Single-stranded 3’ end |
| RNase R | Single-stranded 3’ end | |
| RNase II | Single-stranded 3’ end | |
| Oligoribonuclease | RNA oligonucleotides | |
| Eukaryotes: | Exosome | 3’ end not protected by PABP |
| 5’ Exonucleases | ||
| Bacteria: | RNase J | Monophosphorylated 5’ end |
| Eukaryotes: | Xrn1 | Monophosphorylated 5’ end |
| 5’-end modification | ||
| Bacteria: | RppH | Pyrophosphate removal |
| Eukaryotes: | Dcp2 | Decapping of RNA polynucleotides |
| DcpS | Decapping of RNA oligonucleotides | |
| 3’-end modification | ||
| Bacteria: | Poly(A) polymerase (PcnB) | Polyadenylation |
| Polynucleotide phosphorylase | Heteropolymeric tail addition | |
| Eukaryotes: | Ccr4-Not | Deadenylation |
| Pan2-Pan3 | Deadenylation | |
| PARN | Deadenylation | |
| Cid1#, Zcchc11# | Oligouridylation | |
Homologous enzymes.
The standard model for mRNA degradation in eukaryotes (Figure 2B) was based primarily on studies in S. cerevisiae and mammalian cells, where a variety of degradative enzymes have been identified, including both 3’ and 5’ exonucleases, endonucleases, deadenylases, and decapping enzymes (Table 1). Although mRNA decay in those organisms is sometimes observed to begin with endonucleolytic cleavage or decapping, the most common mechanism of eukaryotic mRNA turnover appears to involve deadenylation as a first step 24, 25, an event whose rate is typically governed by discrete elements within the 3’ UTR or coding region 24, 26. Loss of the 3’ poly(A) tail and the PABP bound there triggers mRNA degradation by either of two mechanisms. On the one hand, by disrupting the mRNA closed loop, deadenylation facilitates 5’ cap removal by the decapping enzyme Dcp2, yielding a 5’-monophosphorylated decay intermediate that is then rapidly degraded by the monophosphate-dependent 5’ exoribonuclease Xrn1 27, 28. In addition, poly(A) tail removal renders messages more susceptible to 3’-terminal attack by the exosome, a multisubunit 3’ exonuclease that is present in both the cytoplasm and the nucleus and that can readily degrade RNA whose 3’ end is not protected by PABP 29, 30.
MAYBE I’M AMAZED: UNEXPECTED PARALLELS
In spite of its superficial appeal, the conventional model for bacterial mRNA degradation had a number of worrisome shortcomings. For one thing, it could not explain how stem-loop structures, either at the 3’ end or upstream of an RNase E cleavage site, would ever be degraded or why the 3’ product of initial endonucleolytic cleavage is typically so much more labile that its intact precursor 8, 31. In addition, the model could not account for the stabilizing influence of stem-loop structures at the 5’ terminus of bacterial mRNAs 16, 17, 32–35 or the ability of a stalled ribosome to selectively prolong the lifetime of the downstream mRNA segment in Bacillus subtilis 36. Finally, it offered no explanation for the striking absence of an RNase E sequence homolog in many bacterial species. These inadequacies of the standard model, as well as interest in how the stabilities of bacterial and eukaryotic messages are regulated, prompted additional investigations that revealed some unexpected similarities between mRNA degradation in those two kingdoms of life.
The End: 3’-Terminal Degradative Events
Despite the widespread belief that 3’ polyadenylation was a characteristic unique to eukaryotic messages, E. coli had long been known to contain a poly(A) polymerase 37, 38 and polyadenylated RNA 39, 40. However, it was not until many years later that the important role of polyadenylation in bacterial RNA decay was recognized 41, 42. It is now clear that the transient addition of poly(A) tails to bacterial RNAs is crucial for the 3’ exonucleolytic degradation of stem-loop structures in decay intermediates 43–45. Although exonucleases such as PNPase and RNase R are hindered when they encounter a significant stem-loop, they are nevertheless capable of inefficiently degrading such structures, but only if a single-stranded RNA segment is present downstream for them to start with 22, 43. (By contrast, RNase II seems unable to degrade structured RNA under any circumstances 46, 47.) Ordinarily the poly(A) tails added to bacterial RNAs are barely detectable because they are promptly digested by 3’ exonucleases 44, 48. However, their repeated addition provides those enzymes multiple opportunities to degrade through structured regions, and eventually they succeed, sometimes with the aid of an RNA helicase (such as RhlB, which assists PNPase) 22, 43, 46 (Figure 3). Bacteria that lack a poly(A) polymerase can use PNPase operating in reverse (i. e., synthetically rather than degradatively) to add heteropolymeric 3’-terminal tails that serve a similar purpose 49. Interestingly, this pathway for 3’-exonucleolytic degradation appears to be conserved in organelles such as chloroplasts 50, which are thought to be evolutionary descendents of bacteria.
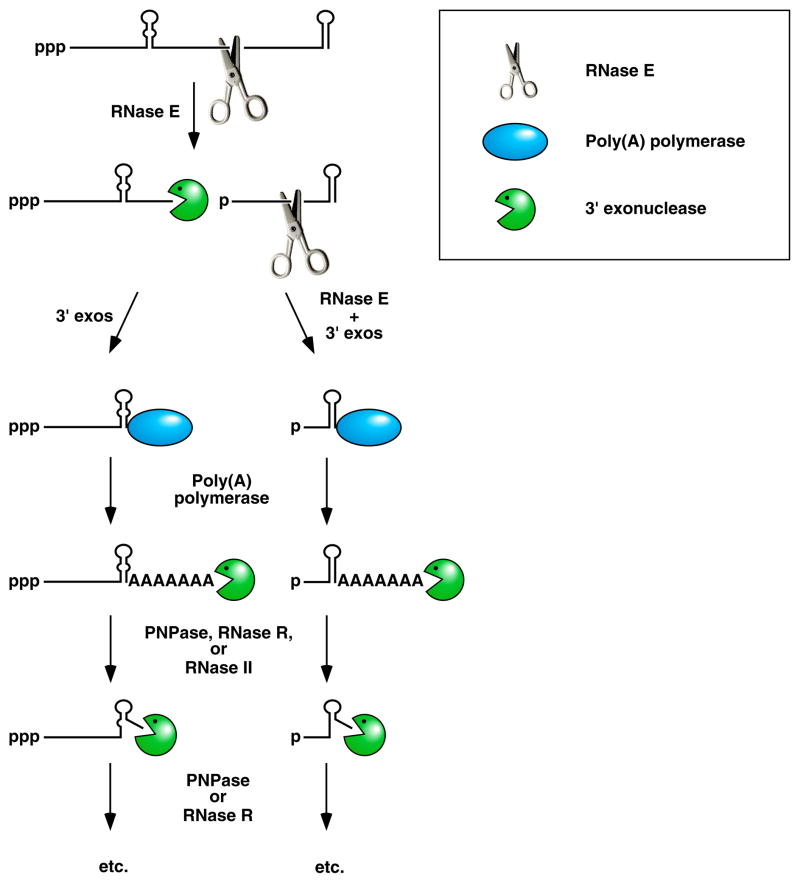
Figure 3. Facilitation of the 3’-exonucleolytic degradation of bacterial mRNA decay intermediates by polyadenylation.
Endonucleolytic cleavage of mRNA by RNase E generates multiple fragments, one of which ends with the original 3’-terminal stem-loop. The others undergo 3’-exonucleolytic attack by PNPase, RNase R, and/or RNase II until an upstream stem-loop is encountered, which interrupts further degradation due to the preference of those ribonucleases for 3’ ends that are unpaired. The resulting decay intermediates are then polyadenylated by poly(A) polymerase, thereby enabling the exonucleases to re-engage. The repeated addition of single-stranded poly(A) tails to the 3’ ends of these intermediates provides multiple opportunities for PNPase and RNase R to overcome structural impediments to exonucleolytic degradation, and eventually they succeed. The ability of PNPase to digest base-paired RNA is enhanced by its association with the RNA helicase RhlB, whereas RNase R requires no such assistance. By contrast, RNase II can degrade poly(A) and other kinds of unstructured RNA but not structured RNA. Ribosomes and coding regions have been omitted from this figure for the sake of simplicity.
The destabilizing influence of poly(A) on mRNA decay intermediates in bacteria may seem at odds with its stabilizing effect on messages in the cytoplasm of eukaryotic cells, where it must be removed by a specialized deadenylase (Ccr4-Not, Pan2-Pan3, or PARN (which is present only in vertebrates and insects) 51 (Table 1)) as a prelude to mRNA decay. However, recent data indicate that in eukaryotic nuclei a pair of noncanonical poly(A) polymerases, the TRAMP subunits Trf4 and Trf5, may have a function similar to that of bacterial poly(A) polymerase, in that they facilitate 3’ exonucleolytic degradation of defective RNAs by exosomes 52, 53. Interestingly, poly(A) addition by Trf4 or Trf5 appears to be necessary for the TRAMP complex to accelerate the decay of some but not all of its RNA targets 53–55.
The nuclear exosome and bacterial PNPase not only share a propensity to degrade polyadenylated RNA but also bear a striking structural resemblance to one another despite superficial differences in their subunit composition. The three identical subunits of PNPase each comprise two RNase PH-like domains and two RNA-binding domains (one each from the KH and S1 families) that together form a homotrimeric ring surrounding a central channel 56. An apparent product of divergent evolution, the core of the eukaryotic exosome contains an analogous set of protein domains organized into nine different polypeptides – six distinct RNase PH-like subunits and three distinct subunits containing KH and S1 domains – that assemble to form a similar toroidal structure 29, 57. However, unlike the bacterial enzyme, which contains one catalytically active RNase PH domain per subunit (three per trimer), none of the corresponding subunits of the core exosome in yeast or humans retains activity. Instead, those exosomes appear to rely for their exonucleolytic activity on either of two associated proteins, Rrp6 or Rrp44 (also known as Dis3) 57, 58. Moreover, unlike PNPase, which degrades RNA phosphorolytically to produce nucleoside diphosphates as reaction products, both Rrp6 and Rrp44/Dis3 are hydrolytic ribonucleases that yield nucleoside monophosphate products. Despite these catalytic differences, exosomes and PNPase both depend on assistance from an RNA helicase (Ski2 or Mtr4 in S. cerevisiae and RhlB in E. coli) for their efficient function 46, 59, 60.
Both Sides Now: 5’-Terminal Degradative Events
A key distinction between mRNA degradation in E. coli and eukaryotic cells is the apparent absence of a 5’-to-3’ exoribonuclease in the former, where mRNA degradation was thought to begin with endonucleolytic cleavage. Therefore, the discovery that 5’-terminal stem-loop structures can protect triphosphorylated primary transcripts from RNase E-mediated degradation in E. coli came as quite a surprise, as no mechanism was known that could account for that effect 16, 17, 32, 33.
A clue as to a possible explanation for that phenomenon came later when it was determined that RNase E has a strong preference for RNA substrates bearing a single phosphate at the 5’ end and will cut such RNAs at internal sites more than an order of magnitude faster than it will cut their triphosphorylated counterparts 61. The influence of 5’ phosphorylation on endonucleolytic cleavage by RNase E is a consequence of a discrete enzyme pocket where monophosphorylated 5’ ends can bind and promote downstream cleavage 62. This property was immediately accepted as the reason why the monophosphorylated 3’ products of internal cleavage are typically so much more labile than their intact triphosphorylated precursors. However, it could not account for the stabilizing influence of 5’-terminal stem-loop structures on primary transcripts, whose triphosphorylated 5’ ends are incapable of binding to RNase E.
This conundrum eventually led to the discovery of an important alternative decay pathway in which internal cleavage by RNase E is triggered by a prior event at the 5’ end: the conversion of the 5’-terminal triphosphate to a monophosphate by RppH, an E. coli RNA pyrophosphohydrolase that preferentially acts on 5’ termini that are single-stranded 63, 64 (Figure 4A). Interestingly, pyrophosphate removal from bacterial transcripts and the decapping of eukaryotic mRNAs not only bear a striking structural resemblance to one another (triphosphate cleavage to generate a monosphosphorylated product) but also are catalyzed by evolutionarily related enzymes (RppH and Dcp2, respectively (Table 1)) that are both members of the Nudix hydrolase family 28, 64. Moreover, the functional consequences of the two events are similar, as each involves removing a protective group to render RNA more susceptible to digestion by a 5’-monophosphate-dependent ribonuclease (the endonuclease RNase E or the 5’ exonuclease Xrn1) 27, 61, 63, 65.
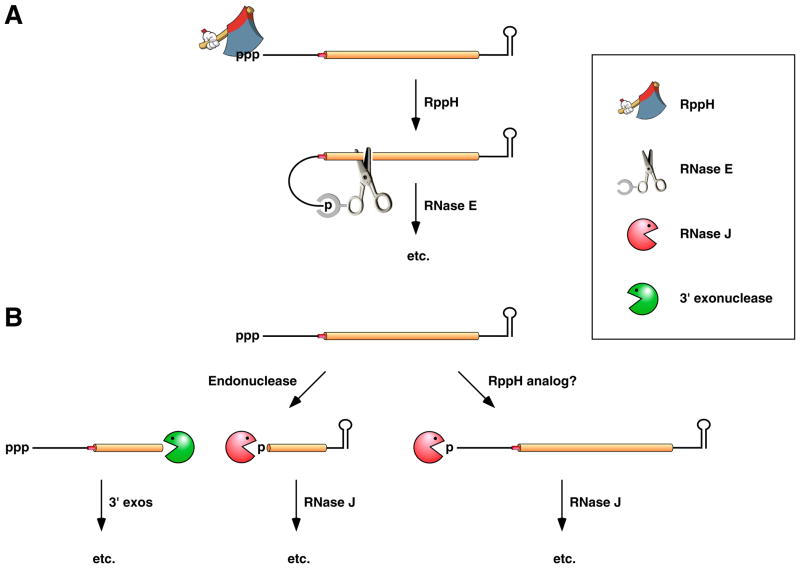
Figure 4. Pathways for 5’-end-dependent mRNA degradation in bacteria.
(A) 5’-end-dependent mRNA decay in bacteria that contain the endonuclease RNase E or a homolog thereof. Pyrophosphate removal by RppH generates a 5’-terminal monophosphate that binds to a discrete pocket on the surface of RNase E, thereby facilitating mRNA cleavage at a downstream location by the active site of that enzyme. In E. coli, RNase E cleavage of primary transcripts can also occur by an alternative, 5’-end-independent mechanism that does not require prior pyrophosphate removal (Figure 2A) 19, 33, 148. (B) 5’-end-dependent mRNA decay in bacteria that contain the 5’ exonuclease RNase J. Internal cleavage by an endonuclease generates a monophosphorylated intermediate susceptible to 5’-to-3’ digestion by RNase J, whose exonucleolytic activity is impeded by a 5’ triphosphate. Alternatively, it is possible that 5’-exonucleolytic digestion by RNase J may be triggered by pyrophosphate removal from primary transcripts by an as yet unidentified RppH analog. Ribosomes have been omitted from this figure for the sake of simplicity.
Remarkably, despite its central role in mRNA decay in E. coli, RNase E is absent from a number of bacterial species, including many firmicutes such as Bacillus subtilis and Staphylococcus aureus and even some proteobacteria such as Helicobacter pylori 66. Moreover, in contrast to what is observed in E. coli, a ribosome stalled on a B. subtilis transcript can protect the entire downstream RNA segment (but not the upstream segment) from degradation, suggesting a 5’-to-3’ directionality for message degradation in that organism 36. These mysteries were solved when it was discovered that, almost invariably, bacteria lacking RNase E instead contain the 5’-to-3’ exonuclease RNase J and/or the endonuclease RNase Y, two enzymes that are absent from E. coli (Table 1) 66–69. In B. subtilis, RNase J and RNase Y appear to play important roles in mRNA turnover 66, 70. Moreover, like Xrn1 and RNase E, these two bacterial ribonucleases preferentially degrade RNA substrates that bear only one phosphate at the 5’ end 66, 68. This property suggests that digestion of primary transcripts by RNase J or RNase Y may well be preceded by a deprotection step that generates a monophosphorylated intermediate. Interestingly, RNase J itself is capable of catalyzing that prior step, as it can act not only as an exonuclease but also, less efficiently, as a 5’-end-independent endonuclease 71, cleaving RNA at internal sites to generate 3’ fragments that it can then degrade exonucleolytically (Figure 4B). Alternatively, it has been hypothesized that exonucleolytic attack by RNase J may in many instances be triggered by pyrophosphate removal from primary transcripts to generate a monophosphorylated 5’ end 63. If so, the latter pathway would closely resemble the degradation mechanism that often ensues after deadenylation in eukaryotic cells: decapping followed by 5’ exonuclease digestion.
It has recently been reported that decapping and subsequent degradation of eukaryotic mRNAs can also be stimulated by the addition of an oligo(U) tract at the 3’ end 72, 73. No analogous oligo(U)-dependent decay pathway has been described in bacteria; nor do bacteria contain a homolog of the eukaryotic poly(U) polymerases Cid1 and Zcchc11 73–75.
Heartbreaker: Internal Cleavage of mRNA
In bacteria, endonucleases have long been thought to play a major role in mRNA degradation, particularly RNase E and its homolog RNase G (both of which cleave RNA in single-stranded regions 15, 76), RNase III (specific for double-stranded RNA 77), and more recently RNase Y and RNase J (both specific for single-stranded RNA 66, 67). By contrast, the important contribution of endonucleases to mRNA decay in eukaryotic cells was long overlooked as deprotection of the 5’ and 3’ ends grabbed the lion’s share of attention.
One of the earliest documented examples of endonucleolytic initiation of eukaryotic mRNA decay was the regulated degradation of transferrin receptor mRNA, whose vulnerability to site-specific cleavage within the 3’ untranslated region is controlled by iron 78. Although the enzyme responsible for that cleavage remains unknown, the recent implication of a number of metazoan endonucleases in mRNA turnover has revived interest in internal cleavage as a pathway for initiating mRNA decay in eukaryotes. Foremost among these endonucleases are the Argonaute (Ago) proteins important for RNA interference, many of which cleave messages at sites targeted by perfectly complementary small interfering RNAs (siRNAs) and microRNAs (miRNAs) 79, 80. Internal cleavage has also been implicated in nonsense-mediated decay and no-go decay, quality-control pathways for rapidly degrading translationally defective transcripts 81, 82 (see below). For example, in metazoans degradation via the former pathway is often initiated by SMG6, an endonuclease with a catalytically active PIN domain 83, 84. Lacking protection at one end or the other, the resulting RNA fragments are susceptible to swift exonucleolytic digestion. Interestingly, in addition to its RNase II-like 3’ exonuclease domain 85, the exosome-associated protein Rrp44/Dis3 has a PIN domain that is capable of cleaving RNA endonucleolytically 86, 87. Thus, not only do the exosome core and PNPase resemble one another structurally, but also each can form a multimeric complex with an endonuclease (Rrp44/Dis3 or RNase E, respectively). Other eukaryotic endonucleases that have been implicated in mRNA degradation include Zc3h12a, a PIN-domain ribonuclease induced by stimulation of Toll-like receptors 88, Swt1, a PIN-domain ribonuclease important for nuclear mRNP surveillance 89, RNase L, an enzyme involved in the host antiviral response 90, IRE-1, a mediator of the unfolded protein response 91, 92, and PMR1, a ribonuclease thought to contribute to hormone-dependent changes in mRNA stability 93. Nevertheless, the contribution of endonucleases to mRNA turnover appears to be more limited in eukaryotes than in bacteria.
You’re No Good: Quality Control via mRNA Degradation
Both bacteria and eukaryotes have evolved quality-control pathways for rapidly degrading messages that are unfit for protein synthesis due to defects in translation. In this manner, cells are able to minimize the synthesis of abnormal proteins, many of which may be toxic, while freeing ribosomes for more productive uses.
An example of such a translational defect is a premature termination codon (PTC, also known as a nonsense codon), which can arise by a number of mechanisms, including genetic mutation, transcription or translation initiation at a cryptic site, and aberrant or incomplete splicing. In both bacterial and eukaryotic cells, messages that contain a PTC are generally degraded much faster than their wild-type counterparts 94, 95. However, the mechanisms that govern nonsense-mediated mRNA decay (NMD) in these two kingdoms of life appear to be quite different from one another.
The specificity of NMD is dependent on the ability of cells to differentiate between PTCs and normal termination codons. To make this distinction, eukaryotic organisms rely on their capacity to recognize that the 3’ UTR downstream of a PTC is abnormal. The distinguishing characteristics of such a misconfigured 3’ UTR are not fully understood, but they appear to include an unusually long distance between the stop codon and the poly(A) tail and/or the presence there of one or more exon junctions, which are uncommon in natural 3’ UTRs 96–99. By contrast, although little is known about PTC recognition in bacteria, the highly variable number of translational units and the rarity of introns in bacterial mRNAs would seem to make distance measurements and splice sites downstream of stop codons unreliable gauges of normalcy.
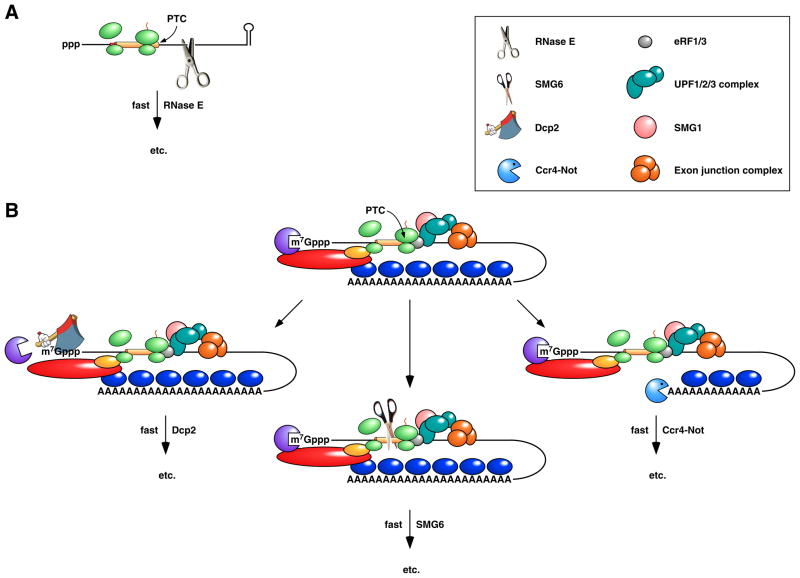
The mechanism of NMD in eukaryotes and bacteria is also different. In E. coli, it is thought to begin with 5’-end-independent RNase E cleavage at internal sites exposed by the premature release of ribosomes 19, 33 (Figure 5A), whereas a variety of triggering mechanisms have been reported in eukaryotes, including decapping, deadenylation, and internal cleavage near the site of premature translation termination 81, 100–102 (Figure 5B).
Figure 5. Pathways for the rapid degradation of bacterial and eukaryotic mRNAs that contain a premature termination codon.
(A) Rapid degradation of a premature termination codon (PTC)-containing mRNA in E. coli. Premature translation termination and the resulting loss of ribosome protection downstream of the PTC expose the mRNA to internal cleavage by RNase E. (B) Nonsense-mediated decay (NMD) in metazoans. Premature translation termination results in an unusually long 3’ UTR, often in conjunction with an exon junction downstream of the PTC. The assembly of the PTC surveillance proteins UPF1, UPF2, and UPF3 at the site of translation termination is guided by the presence there of termination factors eRF1 and eRF3 and can be enhanced by the interaction of UPF3 with an exon junction complex, a heteromultimer deposited on exon junctions during splicing, transported with mRNA to the cytoplasm, and displaced by translating ribosomes only if bound in the coding region. Phosphorylation of UPF1 by the serine/threonine kinase SMG1 triggers nonsense-mediated decay via any of three pathways: deadenylation-independent decapping by Dcp2 (left), endonucleolytic cleavage by SMG6 (centre), or poly(A) tail removal by Ccr4-Not (right). Deadenylation-independent decapping or poly(A) removal leads to degradation via the pathways depicted in Figure 2B. Endonucleolytic cleavage leads to 5’-exonucleolytic degradation of the 3’ fragment by Xrn1 and degradation of the 5’ fragment via the pathways depicted in Figure 2B.
A number of proteins required for NMD in eukaryotes have been identified, including three UPF proteins and, in metazoans, several SMG proteins 95. Functions have been assigned to some of these proteins, such as the UPFs, which form a surveillance complex important for PTC recognition 99, 103–106, and the endonuclease SMG6, which can cleave defective mRNAs near the site of premature translation termination 83, 84. By contrast, no ribonuclease or ancillary protein with a specialized role in NMD has been identified in bacteria, which lack homologs of the UPF and SMG proteins. Instead, the recognition and rapid turnover of PTC-containing transcripts in bacteria appears to be accomplished by the ordinary cellular apparatus for RNA degradation.
Another category of defective mRNAs are those that cannot release ribosomes due to the lack of an in-frame translation termination codon. Messages of this kind can arise by aberrant cleavage and polyadenylation within the coding region (eukaryotes) or degradation of the 3’-terminal portion of a transcript (eukaryotes or bacteria). In both types of organisms, the resulting “non-stop” mRNAs are rapidly degraded by 3’ exonucleases 107, 108; however, the mechanism by which the RNA 3’ end becomes exposed to exonuclease attack is quite different in each case. Bacteria utilize a process called “trans-translation” to free that end of the message from the ribosome trapped there 109. This mechanism of ribosome release depends on tmRNA, a specialized RNA that has properties of both a tRNA and an mRNA. An aminoacylated tmRNA molecule binds to the empty A-site of the stalled ribosome and serves first as an acceptor for peptidyl transfer and then as a template for renewed translation, which ends when the ribosome encounters a termination codon in the tmRNA and dissociates. Exonuclease digestion of the message then ensues. Lacking a homolog of tmRNA, eukaryotes rely instead on the exosome-associated protein Ski7 to stimulate the exonucleolytic degradation of non-stop mRNAs 107. The mechanism by which Ski7 accomplishes this feat is not yet clear.
A third type of surveillance mechanism, “no-go” decay, degrades mRNAs on which ribosomes have become stalled during translation. In both S. cerevisiae and E. coli, such ribosomal pausing can often induce endonucleolytic cleavage at a nearby site 82, 110-112. However, in neither case has the ribonuclease responsible for cleavage been identified, prompting speculation that ribosomes themselves might possess an intrinsic, but as yet unproven, endonuclease activity 110–113.
Every Little Thing: Destabilization by Noncoding RNAs
Short noncoding RNAs that control gene expression by base pairing with complementary sites in mRNA were first discovered in bacteria 114. Subsequently they were found to play a crucial regulatory role in eukaryotic organisms as well 115, 116. In both kingdoms of life, noncoding RNAs influence the translation and longevity of mRNAs to which they bind. However, despite similar regulatory outcomes, the mechanisms by which those outcomes are achieved are rather different.
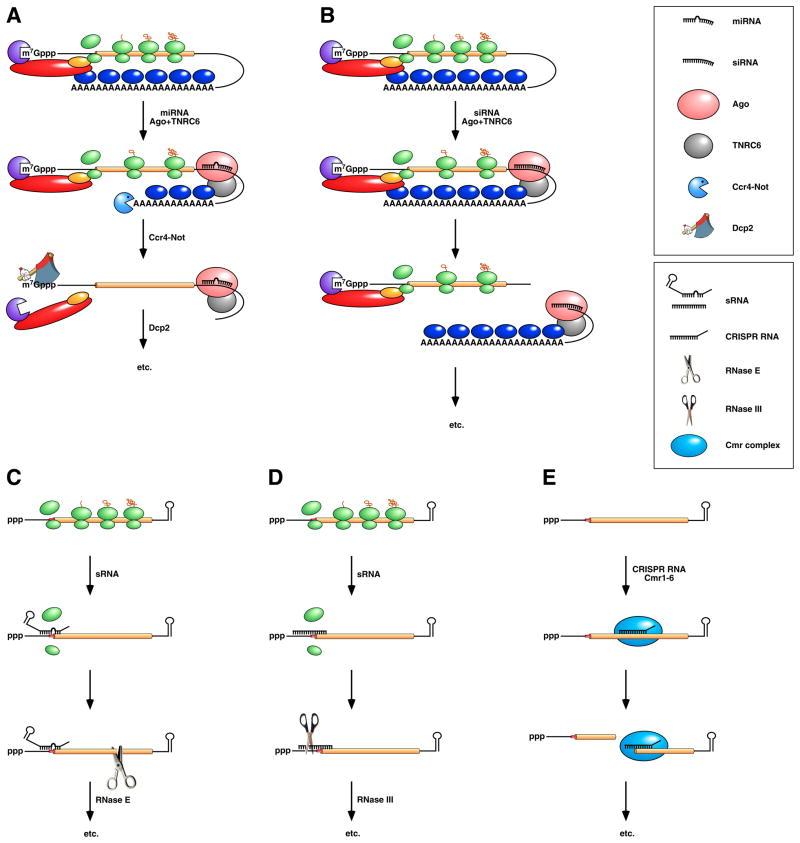
Metazoan organisms produce two major kinds of noncoding RNAs that act post-transcriptionally to control gene expression: miRNAs and siRNAs. These ~22-nucleotide RNAs differ somewhat in their biogenesis 117 but not in their regulatory potential, which depends on the degree of complementarity of their mRNA targets. When miRNAs or siRNAs base pair with a message to which they are partially complementary, they typically impair its translation while also hastening poly(A) removal and thereby destabilizing the message 115, 116, 118, 119 (Figure 6A). These two effects seem to be largely independent of one another and to contribute additively to downregulation 118, 119. Base pairing with perfectly complementary elements also results in faster decay and diminished translation; however, in this case decay is triggered by endonucleolytic cleavage at the site where the noncoding RNA binds rather than by deadenylation 120–122 (Figure 6B). Each of these effects results not from an intrinsic activity of the bound miRNA or siRNA but rather from the properties of two associated proteins that accompany it to its mRNA target: Ago, an endonuclease specific for fully base paired RNA duplexes 79, 80, and TNRC6 (also known as GW182), which mediates translational repression and deadenylation by mechanisms that are poorly understood 123–125.
Figure 6. Post-transcriptional downregulation by noncoding RNAs in eukaryotes and bacteria.
(Top) Repression by miRNAs in vertebrates and insects. (A) A miRNA associated with Ago and TNRC6 (also known as GW182) binds to the 3’ UTR of a message to which it is partially complementary, impeding (though not abolishing) translation and accelerating poly(A) tail removal by the deadenylase Ccr4-Not. The deadenylated mRNA is then decapped by Dcp2 and degraded exonucleolytically by Xrn1 and possibly also by exosomes. (B) An siRNA associated with Ago and TNRC6/GW182 binds to a message to which it is fully complementary, either within the 3’ UTR or somewhere upstream, and directs endonucleolytic cleavage there by Ago. Endonucleolytic cleavage leads to 5’-exonucleolytic degradation of the 3’ fragment by Xrn1 and degradation of the 5’ fragment via the pathways depicted in Figure 2B. Note that miRNAs and siRNAs have the same regulatory potential, their mode of action being determined by the degree of complementarity of their mRNA targets and the activity of the Ago and TNRC6/GW182 proteins with which they associate. (Bottom) Repression by sRNAs and CRISPR RNAs in bacteria. (C) sRNA binding to a partially complementary mRNA impairs translation initiation, often by occluding the ribosome binding site. No longer protected by ribosomes, the message becomes vulnerable to attack by RNase E. (D) Antisense sRNA binding to a fully complementary mRNA can create a long, perfectly paired duplex susceptible to cleavage by RNase III. (E) A Cmr-associated CRISPR RNA directs endonucleolytic cleavage of a complementary message by one of the six Cmr proteins. Ribosomes have been omitted from this panel because the effect of CRISPR RNAs on translation has not been investigated.
Unlike miRNAs and siRNAs, which are processed from long precursor transcripts, small noncoding RNAs (sRNAs) in bacteria typically are primary transcripts of diverse lengths (tens to hundreds of nucleotides). Chaperoned to their complementary mRNA targets by the Sm-like RNA-binding protein Hfq, bacterial sRNAs usually repress translation, often by competing directly with ribosomes for binding to sites of translation initiation (Figure 6C) but sometimes by binding at a distance and inhibiting translation by mechanisms that are not well understood 126–129. However, they occasionally have the opposite effect, antagonizing translational repression by disrupting intramolecular base pairing that would otherwise occlude the ribosome binding site 130–132. When they inhibit translation, bacterial sRNAs also destabilize their mRNA targets, apparently as a secondary consequence of diminished ribosomal protection 133, 134 (Figure 6C). Nevertheless, sRNAs sometimes destabilize mRNA as a primary effect that is not linked to translation, facilitating cleavage by either RNase III (when the two RNAs form a long, well paired duplex) (Figure 6D) or RNase E (when they do not) 135–137. In contrast to eukaryotic miRNAs and siRNAs, which determine specificity but rely entirely on specialized protein cofactors (Ago and TNRC6/GW182) to effect gene regulation, bacterial sRNAs bound to their targets can function as both specificity determinants and effectors, downregulating gene expression by acting either alone or in conjunction with components of the cell’s generalized machinery for RNA degradation. Moreover, whereas the target specificity of eukaryotic miRNAs is defined primarily by the sequence of nucleotides near the 5’ end owing to the manner in which those RNAs are bound by Ago 138–140, the location of the RNA segment that determines the specificity of bacterial sRNAs can vary.
Recent studies have identified a distinct class of short (~30–70 nucleotide) noncoding RNAs that may be able to downregulate gene expression by a mechanism that more closely resembles siRNA-directed RNA cleavage in eukaryotes. These regulatory RNAs are processed from long transcripts of CRISPR loci. In the archaeon Pyrococcus furiosus, CRISPR RNAs guide the hexameric Cmr complexes with which they associate to bind complementary RNAs and cleave them at a specific site within the base-paired region 141 (Figure 6E). By this and other means, CRISPR RNAs are thought to help their host resist invasion by viruses and plasmids. Eubacterial species that produce both CRISPR RNAs and Cmr proteins (e. g., Bacillus halodurans and Thermus thermophilus but not E. coli 142) are expected to share this defense mechanism. Notwithstanding the functional similarities of Cmr and Ago proteins, their sequences and cleavage site specificities are distinct 141.
WE JUST DISAGREE: KEY DIFFERENCES
Despite the growing number of parallels now evident between the mechanisms of mRNA degradation in bacteria and eukaryotes, a number of notable distinctions remain, some of which may be causally interrelated. Perhaps the most fundamental is the relative importance of internal versus terminal degradative events. Low-specificity endonucleases play a major role in bacterial mRNA decay. By contrast, in eukaryotes, where mRNA degradation is dominated by 3’- and 5’-terminal events (deadenylation, decapping, and exonuclease digestion), the contribution of endonucleases appears to be much more limited, and those that do participate seem to act with greater specificity than their bacterial counterparts. This difference appears to have had a number of important consequences. Foremost among these is that steric protection by ribosomes is generally crucial for the longevity of bacterial messages but relatively unimportant for eukaryotic mRNA stability. Thus, whereas inefficient translation initiation need not doom eukaryotic messages to rapid degradation 118, 143, a poor ribosome binding site almost always hastens bacterial mRNA decay, presumably by increasing the spacing between translating ribosomes and thereby exposing potential endonuclease cleavage sites in or near the protein-coding region 144. This difference might also explain why eukaryotes had to evolve a specialized machinery to recognize and degrade mRNAs that contain premature termination codons, while bacteria seem to have managed to achieve that end by employing the same proteins used to degrade ordinary messages. Furthermore, it may help to clarify why the 3’ UTRs of eukaryotic messages, which contain binding sites for proteins and noncoding RNAs that regulate translation, cellular localization, and decay, can be hundreds or even thousands of nucleotides long without adversely affecting mRNA stability, whereas intercistronic and 3’ untranslated regions in bacterial transcripts are generally much shorter.
Conversely, eukaryotes depend heavily on deadenylation to govern rates of cytoplasmic mRNA decay. That reliance has necessitated the protective influence of poly(A)-binding protein, without which rapid and uncontrollable degradation would ensue, and the existence of specialized deadenylases capable of degrading PABP-associated poly(A) tails in an orderly manner. By contrast to its stabilizing effect in the cytoplasm of eukaryotes, poly(A) appears to serve only a destabilizing function in bacteria despite its affinity for the bacterial RNA-binding protein Hfq, whose ability to impede the exonucleolytic destruction of poly(A) tails 145 is not sufficient for those tails to persist at significant steady-state lengths in vivo.
The evolutionary imperatives for these differences may be related to the distinct mechanisms by which eukaryotes and bacteria control translation initiation. In eukaryotic organisms, ribosome binding is ordinarily governed by a protein complex (eIF4F) that associates with both the 5’-terminal cap and the PABP on the 3’-terminal poly(A) tail. Deadenylation disrupts those interactions, thereby inhibiting translation 3, 4. Employing poly(A) tail loss also as the principal mechanism for triggering mRNA degradation allows this decrease in translational activity to be tightly coupled to the destruction of eukaryotic messages. By contrast, the reliance of bacteria on internal ribosome binding sites rather than terminal structures to control translation initiation has enabled them to coordinate the expression of genes by organizing them into co-transcribed polycistronic operons. Although removing pyrophosphate from the 5’ end or adding poly(A) to the 3’ end may trigger exonucleolytic mRNA degradation, a heavy reliance on endonucleolytic cleavage (sometimes triggered by pyrophosphate removal) makes it possible for bacteria to selectively degrade discrete segments of polycistronic transcripts, irrespective of the position of those segments. Consequently, a translational unit anywhere within such a transcript may be either longer or shorter lived than the others 146. Together with individualized signals for translational control, such segmental differences in mRNA stability are an important mechanism by which bacteria can differentially regulate the expression of genes within operons. This kind of degradative flexibility would be of little use in eukaryotes, where the mechanism of translation initiation prevents most mRNAs from encoding more than one polypeptide.
CHANGES: EVOLVING PERCEPTIONS
It is clear that what had once seemed a broad divide between the degradation mechanisms of mRNA in bacteria and eukaryotes has been steadily shrinking in recent years due to a growing awareness of the importance of 5’- and 3’-terminal events (such as pyrophosphate removal, 5’ exonuclease attack, and polyadenylation) in bacteria and internal cleavage (such as during RNA interference or NMD) in eukaryotes. This gap is likely to narrow further as new findings reveal previously unrecognized parallels. For example, the recent discovery of CRISPR RNA-guided RNA cleavage in archaea suggests that eubacteria that produce CRISPR RNAs and Cmr proteins will be found to have a similar ability to trigger mRNA degradation by a mechanism very reminiscent of siRNA-guided mRNA cleavage in eukaryotes. Nonetheless, although infrequent exceptions can be found to almost any rule, some important distinctions are likely to persist due to fundamental differences in how mRNAs are synthesized, compartmentalized, and translated in bacterial and eukaryotic cells. Thus, the pervasive importance of low-specificity endonucleases for bacterial mRNA decay seems unlikely to be replicated in eukaryotes. The valuable insights that can be derived from such comparisons ensure that studies of the mechanisms of mRNA degradation in these two kingdoms of life will continue to inform one another.
Acknowledgments
I am grateful to Ciarán Condon and Lionel Bénard for their helpful comments. The writing of this review was supported by research grants to J. G. B. from the National Institutes of Health (GM35769 and GM79477).
GLOSSARY
- Argonaute/Ago
A member of a family of proteins that contain PAZ and PIWI domains and help to mediate RNA interference by binding siRNAs and miRNAs and delivering them to complementary mRNAs
- A-site
The ribosomal site where aminoacylated tRNA binds
- CRISPR
A cluster of regularly interspaced short palindromic repeats in bacterial or archaeal DNA
- CRISPR RNA
A bacterial or archaeal regulatory RNA, ~30-60 nucleotides long, that is processed from the transcript of clustered regularly interspaced short palindromic repeats (CRISPRs) in chromosomal DNA
- Deadenylase
A 3’ exonuclease specific for degrading poly(A) tails
- eIF3
A multisubunit eukaryotic translation initiation factor that binds to cap-associated eIF4F and guides small ribosomal subunits to the translation initiation codon
- eIF4F
A multisubunit eukaryotic translation initiation factor that binds the mRNA cap, eIF3, and poly(A)-binding protein
- Endonuclease
An enzyme that cleaves RNA or DNA at an internal position
- eRF1 and eRF3
Eukaryotic release factors that mediate translation termination at UAA, UAG, and UGA codons
- Exon junction
A site in a spliced eukaryotic mRNA where an intron was excised and the two flanking exons were joined
- Exon junction complex
A protein complex deposited on exon junctions during splicing and transported with mRNA to the cytoplasm
- Exonuclease
An enzyme that degrades RNA or DNA by removing mononucleotides sequentially from the 5’ or 3’ end
- Exosome
A protein complex that, in eukaryotes, comprises a nine-subunit core and other associated polypeptides and that utilizes its 3’ exonuclease activity and to a lesser extent its endonuclease activity to function in RNA processing and decay in the nucleus and cytoplasm
- Hfq
A bacterial RNA-binding protein, homologous to eukaryotic Sm and Lsm proteins, that acts as a chaperone for many sRNAs and can also bind to mRNA and poly(A) tails
- KH domain
A member of a family of RNA-binding domains homologous to the RNA-binding domains of heterogeneous nuclear ribonucleoprotein K (hnRNP K)
- miRNA
A microRNA, ~22 nucleotides long, that binds to Ago and mediates translational repression and accelerated degradation of complementary mRNAs
- mRNP
A complex of mRNA and the proteins with which it associates
- NMD
Nonsense-mediated degradation of mRNAs that contain a premature termination codon
- Nudix hydrolase
A member of a family of hydrolytic enzymes that share a characteristic sequence motif and catalyze the hydrolysis of substrates containing a nucleoside diphosphate as a constituent unit
- PIN domain
A member of a family of homologous protein domains that have endoribonuclease activity and are present in both eukaryotes and bacteria
- PNPase
Polynucleotide phosphorylase, a phosphorolytic bacterial 3’ exoribonuclease that can also act synthetically to add heteropolymeric tails to RNA
- Poly(A)-binding protein
A protein that binds 3’ poly(A) tails and eIF4F, thereby facilitating translation initiation and protecting mRNA from attack by exosomes and decapping enzymes
- Polycistronic mRNA
An mRNA that contains multiple translational units, each encoding a distinct protein
- Premature termination codon (PTC)
An in-frame UAA, UAG, or UGA triplet that causes ribosomes to terminate translation upstream of the normal termination codon
- RNA degradosome
A bacterial protein complex comprising multiple enzymes important for RNA processing and degradation (e. g., RNase E, a 3’ exonuclease, and an RNA helicase)
- RNA interference
A eukaryotic regulatory process in which siRNAs or miRNAs repress gene expression by inhibiting the translation and accelerating the degradation of complementary mRNAs
- RNA pyrophosphohydrolase
An enzyme that can remove the γ and β phosphates from the 5’ end of a triphosphorylated transcript, converting it to a 5’-monophosphorylated RNA
- RNase PH
A phosphorolytic 3’ exoribonuclease important for the maturation of the 3’ ends of bacterial tRNAs
- Rrp6
A hydrolytic 3’ exoribonuclease associated with nuclear exosomes
- Rrp44/Dis3
A bifunctional exosome-associated ribonuclease that contains both a hydrolytic 3’ exonuclease domain and an endonucleolytic PIN domain
- S1 domain
A member of a family of RNA-binding domains homologous to the RNA-binding domains of ribosomal protein S1
- Shine-Dalgarno element
A bacterial mRNA element that guides translation initiation at a downstream start codon by base pairing with the 3’ end of 16S rRNA.
- siRNA
A small interfering RNA, ~22 nucleotides long, that binds to Ago and mediates translational repression and accelerated degradation of complementary mRNAs
- SMG1
A serine/threonine kinase that helps to trigger NMD in metazoans by phosphorylating UPF1
- Sm protein
A eukaryotic RNA-binding protein that assembles into a multimeric ring and binds RNA (e. g., spliceosomal snRNA) in single-stranded regions that typically are U-rich.
- sRNA
A noncoding bacterial RNA that influences the translation and/or degradation of complementary mRNAs to which it binds
- tmRNA
A bifunctional aminoacylated RNA that has properties of both a tRNA and an mRNA and mediates the release of ribosomes from bacterial mRNAs that lack an in-frame translation termination codon
- TNRC6/GW182
A member of a family of proteins that bind to Argonaute and help to mediate the effects of miRNAs and siRNAs on translation and deadenylation
- TRAMP
A eukaryotic polyadenylation complex, comprising Trf, Air, and Mtr4 proteins, that facilitates the 3’-exonucleolytic degradation of defective RNAs by nuclear exosomes
- UPF protein
One of three proteins (UPF1, UPF2, and UPF3) that are important for the recognition of premature termination codons during nonsense-mediated mRNA decay in eukaryotic cells
- UTR
An mRNA segment that is not translated by ribosomes and does not encode a protein
References
- 1.Tarun SZ, Jr, Sachs AB. Association of the yeast poly(A) tail binding protein with translation initiation factor eIF-4G. EMBO J. 1996;15:7168–7177. [PMC free article] [PubMed] [Google Scholar]
- 2.Laursen BS, Sorensen HP, Mortensen KK, Sperling-Petersen HU. Initiation of protein synthesis in bacteria. Microbiol Mol Biol Rev. 2005;69:101–123. doi: 10.1128/MMBR.69.1.101-123.2005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Sonenberg N, Hinnebusch AG. Regulation of translation initiation in eukaryotes: mechanisms and biological targets. Cell. 2009;136:731–745. doi: 10.1016/j.cell.2009.01.042. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Jackson RJ, Hellen CU, Pestova TV. The mechanism of eukaryotic translation initiation and principles of its regulation. Nat Rev Mol Cell Biol. 2010;11:113–127. doi: 10.1038/nrm2838. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Baird SD, Turcotte M, Korneluk RG, Holcik M. Searching for IRES. RNA. 2006;12:1755–1785. doi: 10.1261/rna.157806. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Filbin ME, Kieft JS. Toward a structural understanding of IRES RNA function. Curr Opin Struct Biol. 2009;19:267–276. doi: 10.1016/j.sbi.2009.03.005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Apirion D. Degradation of RNA in Escherichia coli. A hypothesis. Mol Gen Genet. 1973;122:313–322. doi: 10.1007/BF00269431. [DOI] [PubMed] [Google Scholar]
- 8.Belasco JG, Higgins CF. Mechanisms of mRNA decay in bacteria: a perspective. Gene. 1988;72:15–23. doi: 10.1016/0378-1119(88)90123-0. [DOI] [PubMed] [Google Scholar]
- 9.Apirion D. Isolation, genetic mapping, and some characterization of a mutation in Escherichia coli that affects the processing of ribonucleic acids. Genetics. 1978;90:659–671. doi: 10.1093/genetics/90.4.659. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Ono M, Kuwano M. A conditional lethal mutation in an Escherichia coli strain with a longer chemical lifetime of mRNA. J Mol Biol. 1979;129:343–357. doi: 10.1016/0022-2836(79)90500-x. [DOI] [PubMed] [Google Scholar]
- 11.Mudd EA, Krisch HM, Higgins CF. RNase E, an endoribonuclease, has a general role in the chemical decay of E. coli mRNA: evidence that rne and ams are the same genetic locus. Mol Microbiol. 1990;4:2127–2135. doi: 10.1111/j.1365-2958.1990.tb00574.x. [DOI] [PubMed] [Google Scholar]
- 12.Babitzke P, Kushner SR. The Ams (altered mRNA stability) protein and ribonuclease E are encoded by the same structural gene of Escherichia coli. Proc Natl Acad Sci USA. 1991;88:1–5. doi: 10.1073/pnas.88.1.1. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Melefors Ö, von Gabain A. Genetic studies of cleavage-initiated mRNA decay and processing of ribosomal 9S RNA show that the Escherichia coli ams and rne loci are the same. Mol Microbiol. 1991;5:857–864. doi: 10.1111/j.1365-2958.1991.tb00759.x. [DOI] [PubMed] [Google Scholar]
- 14.Taraseviciene L, Miczak A, Apirion D. The gene specifying RNase E (rne) and a gene affecting mRNA stability (ams) are the same gene. Mol Microbiol. 1991;5:851–855. doi: 10.1111/j.1365-2958.1991.tb00758.x. [DOI] [PubMed] [Google Scholar]
- 15.McDowall KJ, Lin-Chao S, Cohen SN. A+U content rather than a particular nucleotide order determines the specificity of RNase E cleavage. J Biol Chem. 1994;269:10790–10796. [PubMed] [Google Scholar]
- 16.Emory SA, Bouvet P, Belasco JG. A 5′-terminal stem-loop structure can stabilize mRNA in Escherichia coli. Genes Dev. 1992;6:135–148. doi: 10.1101/gad.6.1.135. [DOI] [PubMed] [Google Scholar]
- 17.Bouvet P, Belasco JG. Control of RNase E-mediated RNA degradation by 5′-terminal base pairing in E. coli. Nature. 1992;360:488–491. doi: 10.1038/360488a0. [DOI] [PubMed] [Google Scholar]
- 18.Hansen MJ, Chen LH, Fejzo MLS, Belasco JG. The ompA 5′ untranslated region impedes a major pathway for mRNA degradation in Escherichia coli. Mol Microbiol. 1994;12:707–716. doi: 10.1111/j.1365-2958.1994.tb01058.x. [DOI] [PubMed] [Google Scholar]
- 19.Arnold TE, Yu J, Belasco JG. mRNA stabilization by the ompA 5′ untranslated region: two protective elements hinder distinct pathways for mRNA degradation. RNA. 1998;4:319–330. [PMC free article] [PubMed] [Google Scholar]
- 20.Donovan WP, Kushner SR. Polynucleotide phosphorylase and ribonuclease II are required for cell viability and mRNA turnover in Escherichia coli K-12. Proc Natl Acad Sci USA. 1986;83:120–124. doi: 10.1073/pnas.83.1.120. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Ghosh S, Deutscher MP. Oligoribonuclease is an essential component of the mRNA decay pathway. Proc Natl Acad Sci USA. 1999;96:4372–4377. doi: 10.1073/pnas.96.8.4372. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Cheng ZF, Deutscher MP. An important role for RNase R in mRNA decay. Mol Cell. 2005;17:313–318. doi: 10.1016/j.molcel.2004.11.048. [DOI] [PubMed] [Google Scholar]
- 23.Carpousis AJ. The RNA degradosome of Escherichia coli: an mRNA-degrading machine assembled on RNase E. Annu Rev Microbiol. 2007;61:71–87. doi: 10.1146/annurev.micro.61.080706.093440. [DOI] [PubMed] [Google Scholar]
- 24.Shyu AB, Belasco JG, Greenberg ME. Two distinct destabilizing elements in the c-fos message trigger deadenylation as a first step in rapid mRNA decay. Genes Dev. 1991;5:221–231. doi: 10.1101/gad.5.2.221. [DOI] [PubMed] [Google Scholar]
- 25.Decker CJ, Parker R. A turnover pathway for both stable and unstable mRNAs in yeast: evidence for a requirement for deadenylation. Genes Dev. 1993;7:1632–1643. doi: 10.1101/gad.7.8.1632. [DOI] [PubMed] [Google Scholar]
- 26.Muhlrad D, Parker R. Mutations affecting stability and deadenylation of the yeast MFA2 transcript. Genes Dev. 1992;6:2100–2111. doi: 10.1101/gad.6.11.2100. [DOI] [PubMed] [Google Scholar]
- 27.Muhlrad D, Decker CJ, Parker R. Deadenylation of the unstable mRNA encoded by the yeast MFA2 gene leads to decapping followed by 5′-3′ digestion of the transcript. Genes Dev. 1994;8:855–866. doi: 10.1101/gad.8.7.855. This paper was the first to report that the deadenylation of eukaryotic mRNA triggers cap removal, thereby exposing the 5’ end to exonucleolytic attack by Xrn1. [DOI] [PubMed] [Google Scholar]
- 28.Dunckley T, Parker R. The DCP2 protein is required for mRNA decapping in Saccharomyces cerevisiae and contains a functional MutT motif. EMBO J. 1999;18:5411–5422. doi: 10.1093/emboj/18.19.5411. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Mitchell P, Petfalski E, Shevchenko A, Mann M, Tollervey D. The exosome: a conserved eukaryotic RNA processing complex containing multiple 3′–5′ exoribonucleases. Cell. 1997;91:457–466. doi: 10.1016/s0092-8674(00)80432-8. [DOI] [PubMed] [Google Scholar]
- 30.Jacobs Anderson JS, Parker RP. The 3′ to 5′ degradation of yeast mRNAs is a general mechanism for mRNA turnover that requires the SKI2 DEVH box protein and 3′ to 5′ exonucleases of the exosome complex. EMBO J. 1998;17:1497–1506. doi: 10.1093/emboj/17.5.1497. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.von Gabain A, Belasco JG, Schottel JL, Chang ACY, Cohen SN. Decay of mRNA in Escherichia coli: investigation of the fate of specific segments of transcripts. Proc Natl Acad Sci USA. 1983;80:653–657. doi: 10.1073/pnas.80.3.653. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Bricker AL, Belasco JG. Importance of a 5’ stem-loop for longevity of papA mRNA in Escherichia coli. J Bacteriol. 1999;181:3587–3590. doi: 10.1128/jb.181.11.3587-3590.1999. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Baker KE, Mackie GA. Ectopic RNase E sites promote bypass of 5′-end-dependent mRNA decay in Escherichia coli. Mol Microbiol. 2003;47:75–88. doi: 10.1046/j.1365-2958.2003.03292.x. [DOI] [PubMed] [Google Scholar]
- 34.Hambraeus G, Karhumaa K, Rutberg B. A 5′ stem-loop and ribosome binding but not translation are important for the stability of Bacillus subtilis aprE leader mRNA. Microbiology. 2002;148:1795–1803. doi: 10.1099/00221287-148-6-1795. [DOI] [PubMed] [Google Scholar]
- 35.Sharp JS, Bechhofer DH. Effect of 5′-proximal elements on decay of a model mRNA in Bacillus subtilis. Mol Microbiol. 2005;57:484–495. doi: 10.1111/j.1365-2958.2005.04683.x. [DOI] [PubMed] [Google Scholar]
- 36.Bechhofer DH, Zen KH. Mechanism of erythromycin-induced ermC mRNA stability in Bacillus subtilis. J Bacteriol. 1989;171:5803–5811. doi: 10.1128/jb.171.11.5803-5811.1989. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Gottesman ME, Canellakis ZN, Canellakis ES. Studies on the polymerization of adenylic acid by an enzyme of Escherichia coli. Biochim Biophys Acta. 1962;61:34–42. doi: 10.1016/0926-6550(62)90026-9. [DOI] [PubMed] [Google Scholar]
- 38.August JT, Ortiz PJ, Hurwitz J. Ribonucleic acid-dependent ribonucleotide incorporation. I. Purification and properties of the enzyme. J Biol Chem. 1962;237:3786–3793. [PubMed] [Google Scholar]
- 39.Nakazato H, Venkatesan S, Edmonds M. Polyadenylic acid sequences in E. coli messenger RNA. Nature. 1975;256:144–146. doi: 10.1038/256144a0. [DOI] [PubMed] [Google Scholar]
- 40.Srinivasan PR, Ramanarayanan M, Rabbani E. Presence of polyriboadenylate sequences in pulse-labeled RNA of Escherichia coli. Proc Natl Acad Sci USA. 1975;72:2910–2914. doi: 10.1073/pnas.72.8.2910. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Cao G, Sarkar N. Identification of the gene for an Escherichia coli poly(A) polymerase. Proc Natl Acad Sci USA. 1992;89:10380–10384. doi: 10.1073/pnas.89.21.10380. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Xu F, Lin-Chao S, Cohen SN. The Escherichia coli pcnB gene promotes adenylylation of antisense RNAI of ColE1-type plasmids in vivo and degradation of RNAI decay intermediates. Proc Natl Acad Sci USA. 1993;90:6756–6760. doi: 10.1073/pnas.90.14.6756. This report was the first to reveal the function of bacterial poly(A) tails by showing that they expedite the degradation of RNA decay intermediates in E. coli. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Xu F, Cohen SN. RNA degradation in Escherichia coli regulated by 3' adenylation and 5′ phosphorylation. Nature. 1995;374:180–183. doi: 10.1038/374180a0. [DOI] [PubMed] [Google Scholar]
- 44.Hajnsdorf E, Braun F, Haugel-Nielsen J, Régnier P. Polyadenylylation destabilizes the rpsO mRNA of Escherichia coli. Proc Natl Acad Sci USA. 1995;92:3973–3977. doi: 10.1073/pnas.92.9.3973. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Haugel-Nielsen J, Hajnsdorf E, Régnier P. The rpsO mRNA of Escherichia coli is polyadenylated at multiple sites resulting from endonucleolytic processing and exonucleolytic degradation. EMBO J. 1996;15:3144–3152. [PMC free article] [PubMed] [Google Scholar]
- 46.Coburn GA, Miao X, Briant DJ, Mackie GA. Reconstitution of a minimal RNA degradosome demonstrates functional coordination between a 3′ exonuclease and a DEAD-box RNA helicase. Genes Dev. 1999;13:2594–2603. doi: 10.1101/gad.13.19.2594. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Marujo PE, et al. RNase II removes the oligo(A) tails that destabilize the rpsO mRNA of Escherichia coli. RNA. 2000;6:1185–1193. doi: 10.1017/s135583820000073x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.O'Hara EB, et al. Polyadenylylation helps regulate mRNA decay in Escherichia coli. Proc Natl Acad Sci USA. 1995;92:1807–1811. doi: 10.1073/pnas.92.6.1807. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Mohanty BK, Kushner SR. Polynucleotide phosphorylase functions both as a 3′-5′ exonuclease and a poly(A) polymerase in Escherichia coli. Proc Natl Acad Sci USA. 2000;97:11966–11971. doi: 10.1073/pnas.220295997. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Schuster G, Stern D. RNA polyadenylation and decay in mitochondria and chloroplasts. Prog Mol Biol Transl Sci. 2009;85:393–422. doi: 10.1016/S0079-6603(08)00810-6. [DOI] [PubMed] [Google Scholar]
- 51.Meyer S, Temme C, Wahle E. Messenger RNA turnover in eukaryotes: pathways and enzymes. Crit Rev Biochem Mol Biol. 2004;39:197–216. doi: 10.1080/10409230490513991. [DOI] [PubMed] [Google Scholar]
- 52.LaCava J, et al. RNA degradation by the exosome is promoted by a nuclear polyadenylation complex. Cell. 2005;121:713–724. doi: 10.1016/j.cell.2005.04.029. [DOI] [PubMed] [Google Scholar]
- 53.Vaňáčová S, et al. A new yeast poly(A) polymerase complex involved in RNA quality control. PLoS Biol. 2005;3:e189. doi: 10.1371/journal.pbio.0030189. References 52 and 53 show that, as in bacteria, polyadenylation in yeast can stimulate 3’-exonucleolytic degradation of RNA in the nucleus. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Rougemaille M, et al. Dissecting mechanisms of nuclear mRNA surveillance in THO/sub2 complex mutants. EMBO J. 2007;26:2317–2326. doi: 10.1038/sj.emboj.7601669. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55.San Paolo S, et al. Distinct roles of non-canonical poly(A) polymerases in RNA metabolism. PLoS Genet. 2009;5:e1000555. doi: 10.1371/journal.pgen.1000555. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56.Nurmohamed S, Vaidialingam B, Callaghan AJ, Luisi BF. Crystal structure of Escherichia coli polynucleotide phosphorylase core bound to RNase E, RNA and manganese: implications for catalytic mechanism and RNA degradosome assembly. J Mol Biol. 2009;389:17–33. doi: 10.1016/j.jmb.2009.03.051. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57.Liu Q, Greimann JC, Lima CD. Reconstitution, activities, and structure of the eukaryotic RNA exosome. Cell. 2006;127:1223–1237. doi: 10.1016/j.cell.2006.10.037. [DOI] [PubMed] [Google Scholar]
- 58.Dziembowski A, Lorentzen E, Conti E, Seraphin B. A single subunit, Dis3, is essentially responsible for yeast exosome core activity. Nat Struct Mol Biol. 2007;14:15–22. doi: 10.1038/nsmb1184. This paper demonstrates that yeast exosomes derive their 3’ exonuclease activity from one or more associated ribonucleases rather than from the exosome core. [DOI] [PubMed] [Google Scholar]
- 59.Anderson JS, Parker RP. The 3′ to 5′ degradation of yeast mRNAs is a general mechanism for mRNA turnover that requires the SKI2 DEVH box protein and 3′ to 5′ exonucleases of the exosome complex. EMBO J. 1998;17:1497–1506. doi: 10.1093/emboj/17.5.1497. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 60.de la Cruz J, Kressler D, Tollervey D, Linder P. Dob1p (Mtr4p) is a putative ATP-dependent RNA helicase required for the 3′ end formation of 5.8S rRNA in Saccharomyces cerevisiae. EMBO J. 1998;17:1128–1140. doi: 10.1093/emboj/17.4.1128. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 61.Mackie GA. Ribonuclease E is a 5′-end-dependent endonuclease. Nature. 1998;395:720–723. doi: 10.1038/27246. This is the original report showing that the rate of internal RNA cleavage by RNase E is strongly influenced by the phosphorylation state of the RNA 5’ end. [DOI] [PubMed] [Google Scholar]
- 62.Callaghan AJ, et al. Structure of Escherichia coli RNase E catalytic domain and implications for RNA turnover. Nature. 2005;437:1187–1191. doi: 10.1038/nature04084. [DOI] [PubMed] [Google Scholar]
- 63.Celesnik H, Deana A, Belasco JG. Initiation of RNA decay in Escherichia coli by 5′ pyrophosphate removal. Mol Cell. 2007;27:79–90. doi: 10.1016/j.molcel.2007.05.038. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 64.Deana A, Celesnik H, Belasco JG. The bacterial enzyme RppH triggers messenger RNA degradation by 5′ pyrophosphate removal. Nature. 2008;451:355–358. doi: 10.1038/nature06475. This paper identifies the RNA pyrophosphohydrolase that triggers 5’-end-dependent mRNA decay in E. coli, revealing it to be a homolog of the eukaryotic decapping enzyme Dcp2. [DOI] [PubMed] [Google Scholar]
- 65.Stevens A, Poole TL. 5′-exonuclease-2 of Saccharomyces cerevisiae. Purification and features of ribonuclease activity with comparison to 5′-exonuclease-1. J Biol Chem. 1995;270:16063–16069. doi: 10.1074/jbc.270.27.16063. [DOI] [PubMed] [Google Scholar]
- 66.Shahbabian K, Jamalli A, Zig L, Putzer H. RNase Y, a novel endoribonuclease, initiates riboswitch turnover in Bacillus subtilis. EMBO J. 2009;28:3523–3533. doi: 10.1038/emboj.2009.283. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 67.Even S, et al. Ribonucleases J1 and J2: two novel endoribonucleases in B. subtilis with functional homology to E. coli RNase E. Nucleic Acids Res. 2005;33:2141–2152. doi: 10.1093/nar/gki505. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 68.Mathy N, et al. 5′-to-3′ exoribonuclease activity in bacteria: role of RNase J1 in rRNA maturation and 5′ stability of mRNA. Cell. 2007;129:681–692. doi: 10.1016/j.cell.2007.02.051. This article demonstrates that, like eukaryotes, some bacterial species contain a 5’ exoribonuclease. [DOI] [PubMed] [Google Scholar]
- 69.Commichau FM, et al. Novel activities of glycolytic enzymes in Bacillus subtilis: interactions with essential proteins involved in mRNA processing. Mol Cell Proteomics. 2009;8:1350–1360. doi: 10.1074/mcp.M800546-MCP200. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 70.Mäder U, Zig L, Kretschmer J, Homuth G, Putzer H. mRNA processing by RNases J1 and J2 affects Bacillus subtilis gene expression on a global scale. Mol Microbiol. 2008;70:183–196. doi: 10.1111/j.1365-2958.2008.06400.x. [DOI] [PubMed] [Google Scholar]
- 71.de la Sierra-Gallay IL, Zig L, Jamalli A, Putzer H. Structural insights into the dual activity of RNase J. Nat Struct Mol Biol. 2008;15:206–212. doi: 10.1038/nsmb.1376. [DOI] [PubMed] [Google Scholar]
- 72.Mullen TE, Marzluff WF. Degradation of histone mRNA requires oligouridylation followed by decapping and simultaneous degradation of the mRNA both 5′ to 3′ and 3′ to 5′. Genes Dev. 2008;22 :50–65. doi: 10.1101/gad.1622708. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 73.Rissland OS, Norbury CJ. Decapping is preceded by 3′ uridylation in a novel pathway of bulk mRNA turnover. Nat Struct Mol Biol. 2009;16:616–623. doi: 10.1038/nsmb.1601. This paper shows that Cid1-mediated uridylation at the 3’ end can trigger 5’ decapping and degradation of mRNA in Schizosaccharomyces pombe. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 74.Hagan JP, Piskounova E, Gregory RI. Lin28 recruits the TUTase Zcchc11 to inhibit let-7 maturation in mouse embryonic stem cells. Nat Struct Mol Biol. 2009;16:1021–1025. doi: 10.1038/nsmb.1676. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 75.Jones MR, et al. Zcchc11-dependent uridylation of microRNA directs cytokine expression. Nat Cell Biol. 2009;11:1157–1163. doi: 10.1038/ncb1931. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 76.Tock MR, Walsh AP, Carroll G, McDowall KJ. The CafA protein required for the 5′-maturation of 16 S rRNA is a 5′-end-dependent ribonuclease that has context-dependent broad sequence specificity. J Biol Chem. 2000;275:8726–8732. doi: 10.1074/jbc.275.12.8726. [DOI] [PubMed] [Google Scholar]
- 77.Nicholson AW. Function, mechanism and regulation of bacterial ribonucleases. FEMS Microbiol Rev. 1999;23:371–390. doi: 10.1111/j.1574-6976.1999.tb00405.x. [DOI] [PubMed] [Google Scholar]
- 78.Binder R, et al. Evidence that the pathway of transferrin receptor mRNA degradation involves an endonucleolytic cleavage within the 3′ UTR and does not involve poly(A) tail shortening. EMBO J. 1994;13:1969–1980. doi: 10.1002/j.1460-2075.1994.tb06466.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 79.Liu J, et al. Argonaute2 is the catalytic engine of mammalian RNAi. Science. 2004;305:1437–1441. doi: 10.1126/science.1102513. [DOI] [PubMed] [Google Scholar]
- 80.Meister G, et al. Human Argonaute2 mediates RNA cleavage targeted by miRNAs and siRNAs. Mol Cell. 2004;15:185–197. doi: 10.1016/j.molcel.2004.07.007. References 79 and 80 identify Ago2 as the endonuclease that cleaves mRNAs targeted by fully complementary siRNAs during RNA interference in mammalian cells. [DOI] [PubMed] [Google Scholar]
- 81.Gatfield D, Izaurralde E. Nonsense-mediated messenger RNA decay is initiated by endonucleolytic cleavage in Drosophila. Nature. 2004;429:575–578. doi: 10.1038/nature02559. [DOI] [PubMed] [Google Scholar]
- 82.Doma MK, Parker R. Endonucleolytic cleavage of eukaryotic mRNAs with stalls in translation elongation. Nature. 2006;440:561–564. doi: 10.1038/nature04530. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 83.Huntzinger E, Kashima I, Fauser M, Sauliere J, Izaurralde E. SMG6 is the catalytic endonuclease that cleaves mRNAs containing nonsense codons in metazoan. RNA. 2008;14:2609–2617. doi: 10.1261/rna.1386208. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 84.Eberle AB, Lykke-Andersen S, Muhlemann O, Jensen TH. SMG6 promotes endonucleolytic cleavage of nonsense mRNA in human cells. Nat Struct Mol Biol. 2009;16:49–55. doi: 10.1038/nsmb.1530. References 83 and 84 identify SMG6 as the endonuclease that cleaves metazoan mRNAs containing a premature termination codon. [DOI] [PubMed] [Google Scholar]
- 85.Lorentzen E, Basquin J, Tomecki R, Dziembowski A, Conti E. Structure of the active subunit of the yeast exosome core, Rrp44: diverse modes of substrate recruitment in the RNase II nuclease family. Mol Cell. 2008;29:717–728. doi: 10.1016/j.molcel.2008.02.018. [DOI] [PubMed] [Google Scholar]
- 86.Schaeffer D, et al. The exosome contains domains with specific endoribonuclease, exoribonuclease and cytoplasmic mRNA decay activities. Nat Struct Mol Biol. 2009;16:56–62. doi: 10.1038/nsmb.1528. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 87.Lebreton A, Tomecki R, Dziembowski A, Seraphin B. Endonucleolytic RNA cleavage by a eukaryotic exosome. Nature. 2008;456:993–996. doi: 10.1038/nature07480. [DOI] [PubMed] [Google Scholar]
- 88.Matsushita K, et al. Zc3h12a is an RNase essential for controlling immune responses by regulating mRNA decay. Nature. 2009;458:1185–1190. doi: 10.1038/nature07924. [DOI] [PubMed] [Google Scholar]
- 89.Skružný M, et al. An endoribonuclease functionally linked to perinuclear mRNP quality control associates with the nuclear pore complexes. PLoS Biol. 2009;7:e8. doi: 10.1371/journal.pbio.1000008. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 90.Liang SL, Quirk D, Zhou A. RNase L: its biological roles and regulation. IUBMB Life. 2006;58:508–514. doi: 10.1080/15216540600838232. [DOI] [PubMed] [Google Scholar]
- 91.Hollien J, Weissman JS. Decay of endoplasmic reticulum-localized mRNAs during the unfolded protein response. Science. 2006;313:104–107. doi: 10.1126/science.1129631. [DOI] [PubMed] [Google Scholar]
- 92.Hollien J, et al. Regulated Ire1-dependent decay of messenger RNAs in mammalian cells. J Cell Biol. 2009;186:323–331. doi: 10.1083/jcb.200903014. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 93.Pastori RL, Moskaitis JE, Schoenberg DR. Estrogen-induced ribonuclease activity in Xenopus liver. Biochemistry. 1991;30:10490–10498. doi: 10.1021/bi00107a018. [DOI] [PubMed] [Google Scholar]
- 94.Nilsson G, Belasco JG, Cohen SN, von Gabain A. Effect of premature termination of translation on mRNA stability depends on the site of ribosome release. Proc Natl Acad Sci USA. 1987;84:4890–4894. doi: 10.1073/pnas.84.14.4890. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 95.Isken O, Maquat LE. Quality control of eukaryotic mRNA: safeguarding cells from abnormal mRNA function. Genes Dev. 2007;21:1833–1856. doi: 10.1101/gad.1566807. [DOI] [PubMed] [Google Scholar]
- 96.Zhang J, Sun X, Qian Y, Maquat LE. Intron function in the nonsense-mediated decay of β-globin mRNA: indications that pre-mRNA splicing in the nucleus can influence mRNA translation in the cytoplasm. RNA. 1998;4:801–815. doi: 10.1017/s1355838298971849. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 97.Amrani N, et al. A faux 3′-UTR promotes aberrant termination and triggers nonsense-mediated mRNA decay. Nature. 2004;432:112–118. doi: 10.1038/nature03060. [DOI] [PubMed] [Google Scholar]
- 98.Bühler M, Steiner S, Mohn F, Paillusson A, Mühlemann O. EJC-independent degradation of nonsense immunoglobulin-μ mRNA depends on 3′ UTR length. Nat Struct Mol Biol. 2006;13:462–464. doi: 10.1038/nsmb1081. [DOI] [PubMed] [Google Scholar]
- 99.Singh G, Rebbapragada I, Lykke-Andersen J. A competition between stimulators and antagonists of Upf complex recruitment governs human nonsense-mediated mRNA decay. PLoS Biol. 2008;6:e111. doi: 10.1371/journal.pbio.0060111. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 100.Muhlrad D, Parker R. Premature translational termination triggers mRNA decapping. Nature. 1994;370:578–581. doi: 10.1038/370578a0. [DOI] [PubMed] [Google Scholar]
- 101.Mitchell P, Tollervey D. An NMD pathway in yeast involving accelerated deadenylation and exosome-mediated 3′-5′ degradation. Mol Cell. 2003;11:1405–1413. doi: 10.1016/s1097-2765(03)00190-4. [DOI] [PubMed] [Google Scholar]
- 102.Chen CY, Shyu AB. Rapid deadenylation triggered by a nonsense codon precedes decay of the RNA body in a mammalian cytoplasmic nonsense-mediated decay pathway. Mol Cell Biol. 2003;23 :4805–4813. doi: 10.1128/MCB.23.14.4805-4813.2003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 103.Leeds P, Peltz SW, Jacobson A, Culbertson MR. The product of the yeast UPF1 gene is required for rapid turnover of mRNAs containing a premature translational termination codon. Genes Dev. 1991;5:2303–2314. doi: 10.1101/gad.5.12a.2303. [DOI] [PubMed] [Google Scholar]
- 104.Leeds P, Wood JM, Lee BS, Culbertson MR. Gene products that promote mRNA turnover in Saccharomyces cerevisiae. Mol Cell Biol. 1992;12:2165–2177. doi: 10.1128/mcb.12.5.2165. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 105.Cui Y, Hagan KW, Zhang S, Peltz SW. Identification and characterization of genes that are required for the accelerated degradation of mRNAs containing a premature translational termination codon. Genes Dev. 1995;9:423–436. doi: 10.1101/gad.9.4.423. [DOI] [PubMed] [Google Scholar]
- 106.He F, Brown AH, Jacobson A. Upf1p, Nmd2p, and Upf3p are interacting components of the yeast nonsense-mediated mRNA decay pathway. Mol Cell Biol. 1997;17:1580–1594. doi: 10.1128/mcb.17.3.1580. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 107.van Hoof A, Frischmeyer PA, Dietz HC, Parker R. Exosome-mediated recognition and degradation of mRNAs lacking a termination codon. Science. 2002;295:2262–2264. doi: 10.1126/science.1067272. [DOI] [PubMed] [Google Scholar]
- 108.Richards J, Mehta P, Karzai AW. RNase R degrades non-stop mRNAs selectively in an SmpB-tmRNA-dependent manner. Mol Microbiol. 2006;62:1700–1712. doi: 10.1111/j.1365-2958.2006.05472.x. [DOI] [PubMed] [Google Scholar]
- 109.Karzai AW, Roche ED, Sauer RT. The SsrA-SmpB system for protein tagging, directed degradation and ribosome rescue. Nat Struct Biol. 2000;7:449–455. doi: 10.1038/75843. [DOI] [PubMed] [Google Scholar]
- 110.Hayes CS, Sauer RT. Cleavage of the A site mRNA codon during ribosome pausing provides a mechanism for translational quality control. Mol Cell. 2003;12:903–911. doi: 10.1016/s1097-2765(03)00385-x. [DOI] [PubMed] [Google Scholar]
- 111.Sunohara T, Jojima K, Yamamoto Y, Inada T, Aiba H. Nascent-peptide-mediated ribosome stalling at a stop codon induces mRNA cleavage resulting in nonstop mRNA that is recognized by tmRNA. RNA. 2004;10:378–386. doi: 10.1261/rna.5169404. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 112.Sunohara T, Jojima K, Tagami H, Inada T, Aiba H. Ribosome stalling during translation elongation induces cleavage of mRNA being translated in Escherichia coli. J Biol Chem. 2004;279:15368–15375. doi: 10.1074/jbc.M312805200. [DOI] [PubMed] [Google Scholar]
- 113.Passos DO, et al. Analysis of Dom34 and its function in no-go decay. Mol Biol Cell. 2009;20:3025– 3032. doi: 10.1091/mbc.E09-01-0028. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 114.Mizuno T, Chou MY, Inouye M. A unique mechanism regulating gene expression: translational inhibition by a complementary RNA transcript (micRNA) Proc Natl Acad Sci USA. 1984;81:1966–1970. doi: 10.1073/pnas.81.7.1966. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 115.Lee RC, Feinbaum RL, Ambros V. The C. elegans heterochronic gene lin-4 encodes small RNAs with antisense complementarity to lin-14. Cell. 1993;75:843–854. doi: 10.1016/0092-8674(93)90529-y. [DOI] [PubMed] [Google Scholar]
- 116.Wightman B, Ha I, Ruvkun G. Posttranscriptional regulation of the heterochronic gene lin- 14 by lin-4 mediates temporal pattern formation in C. elegans. Cell. 1993;75:855–862. doi: 10.1016/0092-8674(93)90530-4. [DOI] [PubMed] [Google Scholar]
- 117.Kim VN, Han J, Siomi MC. Biogenesis of small RNAs in animals. Nat Rev Mol Cell Biol. 2009;10:126–139. doi: 10.1038/nrm2632. [DOI] [PubMed] [Google Scholar]
- 118.Wu L, Fan J, Belasco JG. MicroRNAs direct rapid deadenylation of mRNA. Proc Natl Acad Sci USA. 2006;103:4034–4039. doi: 10.1073/pnas.0510928103. This report demonstrates that miRNAs accelerate the degradation of partially complementary mRNAs by expediting their deadenylation. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 119.Eulalio A, et al. Deadenylation is a widespread effect of miRNA regulation. RNA. 2009;15:21–32. doi: 10.1261/rna.1399509. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 120.Zamore PD, Tuschl T, Sharp PA, Bartel DP. RNAi: double-stranded RNA directs the ATP-dependent cleavage of mRNA at 21 to 23 nucleotide intervals. Cell. 2000;101:25–33. doi: 10.1016/S0092-8674(00)80620-0. This paper was the first to determine the mechanism of RNA interference by showing that siRNAs direct endonucleolytic cleavage of mRNAs to which they are fully complementary. [DOI] [PubMed] [Google Scholar]
- 121.Elbashir SM, Lendeckel W, Tuschl T. RNA interference is mediated by 21- and 22- nucleotide RNAs. Genes Dev. 2001;15:188–200. doi: 10.1101/gad.862301. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 122.Wu L, Fan J, Belasco JG. Importance of translation and nonnucleolytic Ago proteins for on-target RNA interference. Curr Biol. 2008;18:1327–1332. doi: 10.1016/j.cub.2008.07.072. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 123.Chekulaeva M, Filipowicz W, Parker R. Multiple independent domains of dGW182 function in miRNA-mediated repression in Drosophila. RNA. 2009;15:794–803. doi: 10.1261/rna.1364909. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 124.Zipprich JT, Bhattacharyya S, Mathys H, Filipowicz W. Importance of the C-terminal domain of the human GW182 protein TNRC6C for translational repression. RNA. 2009;15:781–793. doi: 10.1261/rna.1448009. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 125.Lazzaretti D, Tournier I, Izaurralde E. The C-terminal domains of human TNRC6A, TNRC6B, and TNRC6C silence bound transcripts independently of Argonaute proteins. RNA. 2009;15:1059–1066. doi: 10.1261/rna.1606309. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 126.Waters LS, Storz G. Regulatory RNAs in bacteria. Cell. 2009;136:615–628. doi: 10.1016/j.cell.2009.01.043. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 127.Bouvier M, Sharma CM, Mika F, Nierhaus KH, Vogel J. Small RNA binding to 5′ mRNA coding region inhibits translational initiation. Mol Cell. 2008;32:827–837. doi: 10.1016/j.molcel.2008.10.027. [DOI] [PubMed] [Google Scholar]
- 128.Darfeuille F, Unoson C, Vogel J, Wagner EG. An antisense RNA inhibits translation by competing with standby ribosomes. Mol Cell. 2007;26:381–392. doi: 10.1016/j.molcel.2007.04.003. [DOI] [PubMed] [Google Scholar]
- 129.Sharma CM, Darfeuille F, Plantinga TH, Vogel J. A small RNA regulates multiple ABC transporter mRNAs by targeting C/A-rich elements inside and upstream of ribosome-binding sites. Genes Dev. 2007;21:2804–2817. doi: 10.1101/gad.447207. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 130.Majdalani N, Cunning C, Sledjeski D, Elliott T, Gottesman S. DsrA RNA regulates translation of RpoS message by an anti-antisense mechanism, independent of its action as an antisilencer of transcription. Proc Natl Acad Sci USA. 1998;95:12462–12467. doi: 10.1073/pnas.95.21.12462. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 131.Majdalani N, Hernandez D, Gottesman S. Regulation and mode of action of the second small RNA activator of RpoS translation, RprA. Mol Microbiol. 2002;46:813–826. doi: 10.1046/j.1365-2958.2002.03203.x. [DOI] [PubMed] [Google Scholar]
- 132.Fröhlich KS, Vogel J. Activation of gene expression by small RNA. Curr Opin Microbiol. 2009;12 :674–682. doi: 10.1016/j.mib.2009.09.009. [DOI] [PubMed] [Google Scholar]
- 133.Massé E, Escorcia FE, Gottesman S. Coupled degradation of a small regulatory RNA and its mRNA targets in Escherichia coli. Genes Dev. 2003;17:2374–2383. doi: 10.1101/gad.1127103. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 134.Udekwu KI, et al. Hfq-dependent regulation of OmpA synthesis is mediated by an antisense RNA. Genes Dev. 2005;19:2355–2366. doi: 10.1101/gad.354405. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 135.Case CC, Simons EL, Simons RW. The IS10 transposase mRNA is destabilized during antisense RNA control. EMBO J. 1990;9:1259–1266. doi: 10.1002/j.1460-2075.1990.tb08234.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 136.Huntzinger E, et al. Staphylococcus aureus RNAIII and the endoribonuclease III coordinately regulate spa gene expression. EMBO J. 2005;24:824–835. doi: 10.1038/sj.emboj.7600572. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 137.Pfeiffer V, Papenfort K, Lucchini S, Hinton JC, Vogel J. Coding sequence targeting by MicC RNA reveals bacterial mRNA silencing downstream of translational initiation. Nat Struct Mol Biol. 2009;16:840–846. doi: 10.1038/nsmb.1631. [DOI] [PubMed] [Google Scholar]
- 138.Doench JG, Sharp PA. Specificity of microRNA target selection in translational repression. Genes Dev. 2004;18:504–511. doi: 10.1101/gad.1184404. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 139.Bartel DP. MicroRNAs: genomics, biogenesis, mechanism, and function. Cell. 2004;116:281–297. doi: 10.1016/s0092-8674(04)00045-5. [DOI] [PubMed] [Google Scholar]
- 140.Wang Y, Sheng G, Juranek S, Tuschl T, Patel DJ. Structure of the guide-strand- containing argonaute silencing complex. Nature. 2008;456:209–213. doi: 10.1038/nature07315. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 141.Hale CR, et al. RNA-guided RNA cleavage by a CRISPR RNA-Cas protein complex. Cell. 2009;139:945–956. doi: 10.1016/j.cell.2009.07.040. This article reports in vitro evidence that CRISPR RNAs, like eukaryotic siRNAs, can guide the endonucleolytic cleavage of fully complementary RNAs in microorganisms that contain Cmr proteins. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 142.Haft DH, Selengut J, Mongodin EF, Nelson KE. A guild of 45 CRISPR-associated (Cas) protein families and multiple CRISPR/Cas subtypes exist in prokaryotic genomes. PLoS Comput Biol. 2005;1:e60. doi: 10.1371/journal.pcbi.0010060. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 143.Chen CY, Xu N, Shyu AB. mRNA decay mediated by two distinct AU-rich elements from c-fos and granulocyte-macrophage colony-stimulating factor transcripts: different deadenylation kinetics and uncoupling from translation. Mol Cell Biol. 1995;15:5777–5788. doi: 10.1128/mcb.15.10.5777. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 144.Deana A, Belasco JG. Lost in translation: the influence of ribosomes on bacterial mRNA decay. Genes Dev. 2005;19:2526–2533. doi: 10.1101/gad.1348805. [DOI] [PubMed] [Google Scholar]
- 145.Folichon M, et al. The poly(A) binding protein Hfq protects RNA from RNase E and exoribonucleolytic degradation. Nucleic Acids Res. 2003;31:7302–7310. doi: 10.1093/nar/gkg915. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 146.Selinger DW, Saxena RM, Cheung KJ, Church GM, Rosenow C. Global RNA half-life analysis in Escherichia coli reveals positional patterns of transcript degradation. Genome Res. 2003;13 :216–223. doi: 10.1101/gr.912603. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 147.Wang Z, Kiledjian M. Functional link between the mammalian exosome and mRNA decapping. Cell. 2001;107:751–762. doi: 10.1016/s0092-8674(01)00592-x. [DOI] [PubMed] [Google Scholar]
- 148.Mackie GA. Stabilization of circular rpsT mRNA demonstrates the 5′-end dependence of RNase E action in vivo. J Biol Chem. 2000;275:25069–25072. doi: 10.1074/jbc.C000363200. [DOI] [PubMed] [Google Scholar]