Abstract
Glioblastoma multiforme (GBM) is a particularly aggressive brain tumor and remains a clinically devastating disease. Despite innovative therapies for the treatment of GBM, there has been no significant increase in patient survival over the past decade. Enzymes that control epigenetic alterations are of considerable interest as targets for cancer therapy because of their critical roles in cellular processes that lead to oncogenesis. Several inhibitors of histone deacetylases (HDACs) have been developed and tested in GBM with moderate success. We found that treatment of GBM cells with HDAC inhibitors caused the accumulation of histone methylation, a modification removed by the lysine specific demethylase 1 (LSD1). This led us to examine the effects of simultaneously inhibiting HDACs and LSD1 as a potential combination therapy. We evaluated induction of apoptosis in GBM cell lines after combined inhibition of LSD1 and HDACs. LSD1 was inhibited by targeted short hairpin RNA or pharmacological means and inhibition of HDACs was achieved by treatment with either vorinostat or PCI-24781. Caspase-dependent apoptosis was significantly increased (>2-fold) in LSD1-knockdown GBM cells treated with HDAC inhibitors. Moreover, pharmacologically inhibiting LSD1 with the monoamine oxidase inhibitor tranylcypromine, in combination with HDAC inhibitors, led to synergistic apoptotic cell death in GBM cells; this did not occur in normal human astrocytes. Taken together, these results indicate that LSD1 and HDACs cooperate to regulate key pathways of cell death in GBM cell lines but not in normal counterparts, and they validate the combined use of LSD1 and HDAC inhibitors as a therapeutic approach for GBM.
Keywords: combination therapy, epigenetics, glioblastoma, HDAC inhibitors
Glioblastoma multiforme (GBM) is the most aggressive and lethal type of brain tumor, with a median survival duration of 9–12 months.1 Current treatment options for GBM include surgery, radiation therapy, and standard chemotherapy. However, in the past few decades, there has been little effect of these options, or newly developed chemotherapeutic agents, on extending the survival of patients diagnosed with GBM.2 Therefore, the need to develop novel therapeutic approaches for the treatment of these tumors is of great importance. GBMs are characterized by heterogeneity and dysregulation of several signaling pathways, and studies have demonstrated that this occurs by both genetic and epigenetic mechanisms.3 Recently, the enzymes involved in the regulation of epigenetic mechanisms have become attractive targets for the treatment of cancer because of their ability to alter gene expression profiles to promote tumor cell death.
One of the most well-studied epigenetic modifications is histone acetylation, a process regulated by histone acetyltransferases (HATs) and histone deacetylases (HDACs). Dysregulation of HDACs in a variety of diseases, such as cancer and neurodegeneration, sparked the pharmacologic development of novel HDAC inhibitors (HDACis) for use in the treatment of these diseases. The first US Food and Drug Administration (FDA)-approved HDACi, vorinostat, was marketed in 2006 for the treatment of cutaneous T-cell lymphoma. Because of its success, dozens of other HDACis have been developed, and many of them are currently being tested in various stages of clinical trials. There are several different categories of HDACis that are structurally dissimilar and target different classes of HDACs, which include hydroxamates, cyclic peptides, aliphatic acids, and benzamides.4–7 Treatment of cancer cells with HDACis causes cell growth arrest and cell death; however, the exact mechanisms by which this occurs have not been completely elucidated.4,8,9 Although the primary mechanism of action of HDACis leads to the accumulation of acetylated histone and nonhistone proteins, they also can induce cell-cycle arrest, differentiation, apoptotic cell death, and autophagic pathways, as well as regulate reactive oxygen species (ROS) production, which facilitates cell death.9 Although the use of HDACis as monotherapy in the clinic has been validated in cutaneous T-cell lymphoma, they are less effective against solid tumors.7,10 In fact, the full potential of HDACis is more readily seen when used in combination with other chemotherapeutic agents, frequently producing either synergistic or additive effects.11–14 This has also been shown for GBM.15 In clinical trials, several HDACis have been tested, including vorinostat, the first FDA-approved HDACi. Response was minimal when HDACis were given as a single agent,16 and combination studies are currently underway. New hydroxamic acid-based HDACis are also in clinical development, including PCI-24781, a pan-HDACi that inhibits HDACs in the nanomolar range.17
HDACis not only influence acetylation of histones but also affect other histone modifications, such as methylation on lysine 4 of histone H3 (H3K4). Dimethylation of this residue generally leads to transcriptional activation, whereas loss of this mark leads to gene repression.18 The removal of H3K4 dimethylation occurs by the lysine-specific demethylase 1 (LSD1) enzyme.18 LSD1 (also known as BHC110, AOF2, or KDM1A) was the first lysine demethylase discovered, and it shares homology with the amine oxidase family of enzymes.18,19 Studies have demonstrated that several inhibitors of the mono- and polyamine oxidases (MAOi and PAOi, respectively) are effective at inhibiting LSD1 in vitro,20–25 and treatment of cells with these agents leads to changes in histone methylation and re-expression of aberrantly silenced genes in vivo.23,26,27 More recently, novel oligoamine analogs have been developed that are also effective in inhibiting LSD1 in vitro and in vivo.27 Neuroblastoma and colorectal cancer xenograft models treated with LSD1 inhibitors significantly inhibited tumor growth,27,28 suggesting that inhibitors of LSD1 may have therapeutic value.
Interestingly, HDAC1/2 and LSD1 exist in common protein complexes, and the activity of LSD1 is influenced by the acetylation status of neighboring histone residues.22 These data provide the rationale for targeting LSD1 in combination with HDACis for the treatment of cancer. Although studies have demonstrated that LSD1 inhibitors are effective as monotherapies for the treatment of neuroblastoma and colorectal cancer, there are no reports evaluating their effects on GBM, and only 2 studies combined LSD1 inhibitors with other agents.27 Here, we evaluated the effect of inhibition of LSD1, by genetic and pharmacological means, in combination with HDACis on GBM cell lines and immortalized human astrocytes. We discovered that the combination was selective for GBM cell lines and led to enhanced cell death through both caspase-dependent and caspase-independent mechanisms.
Materials and Methods
Cell Culture and Reagents
LN-18, U87, SNB-19, and U251 brain tumor cell lines were obtained from American Type Culture Collection and maintained in DMEM/F12 medium supplemented with 10% fetal bovine serum, 2 mM L-glutamine, 100 U/mL penicillin, and 100 μg/mL streptomycin. Immortalized human astrocytes (NHA/E6/E7/Tert) have been described elsewhere,29 human neural stem cells were obtained from Lonza, and cancer stem cells derived from patients with GBM were obtained with appropriate informed consent after approval from The University of Texas MD Anderson Institutional Review Board. All cell lines were grown at 37°C in a humidified atmosphere containing 5% CO2. Vorinostat (suberoylanilide hydroxamic acid) was purchased from Cayman Chemical, PCI-24781 was kindly provided by Pharmacyclics, and tranylcypromine was purchased from Sigma-Aldrich. Antibodies used for this study were as follows: total histone H3 (Millipore), acetylated histone H3 (Millipore), dimethyl-lysine 4 of histone H3 (Abcam), and LSD1 (Abcam).
Acid Extraction of Histones
Nuclei were isolated by lysis of cells in phosphate-buffered saline (PBS) containing 0.5% Triton X-100 (v/v), 2 mM phenylmethylsulfonyl fluoride (PMSF), 0.02% (w/v) NaN3 for 10 min on ice. After centrifugation, the nuclei were washed with lysis buffer, and histones were extracted in 0.2 N HCl overnight at 4°C. Samples were centrifuged to pellet debris, and the concentration of histones in the supernatant was determined by Bradford assay (Sigma-Aldrich).
Transfection of shRNA into GBM cells
Control or vector containing sequence specific for LSD1 (GGCGAAGGTAGAGTACAGAGA) was described elsewhere.30 The shRNA constructs were transfected into LN-18 or NHA/E6/E7/Tert cells using the Lonza Group nucleofector technology in accordance with the manufacturer's instructions. Cells (1 × 106) were resuspended in Buffer V and nucleofected using program T-20.
DNA Fragmentation
Apoptosis was assessed by measuring DNA fragmentation using propidium iodide (PI; Sigma-Aldrich) staining followed by analysis by flow cytometry. Treated cells were harvested by trypsinization, washed with PBS, and fixed with 70% ethanol overnight. The next day, cells were washed with PBS and then resuspended in PI staining solution (PBS containing 25 μg/mL propidium iodide and 100 μg/mL RNase A) for 1 h before analysis by flow cytometry (FACS Calibur; Becton Dickinson) on the FL-3 channel. The percentage of cells in the subdiploid population was quantified using CellQuest software (BD Biosciences).
Western Blotting
Seventy-two hours after nucleofection, cells were harvested, and total cell lysates prepared by lysis with Triton X-100 lysis buffer (PBS, 25 mM Tris pH 7.5, 150 mM NaCl, 1% Triton X-100) containing protease inhibitors for 1 h at 4°C followed by centrifugation. Protein concentrations were determined by Bradford assay (Sigma-Aldrich), and 50 μg of protein was separated by sodium dodecyl sulfate-polyacrylamide gel electrophoresis (SDS-PAGE), transferred to nitrocellulose, and immunoblotted with specific antibodies as indicated. Immunoreactive bands were detected by chemiluminescence (GE Healthcare).
Caspase 3/7 Assay
Activity of caspase 3/7 was assessed as previously described.31 In brief, 1 × 106 cells were treated as indicated, lysed in PBS by freeze/thaw cycles on dry ice, clarified by centrifugation at 14k RPM, 15 min, 4°C; protein concentration was determine by Bradford assay. Cell extracts were adjusted to 1 mg/mL with PBS, and 50 μL was plated in duplicate wells of a 96-well plate and 150 μL of DEVD buffer containing 50 μM DEVD-AMC (Biomol International) was added. Samples were incubated for 3 h at room temperature, and fluorescence was read by spectrofluorimetry at 460 nm (ex: 355 nm). The fluorescence generated by the cleavage of the peptide and release of fluorogenic AMC correlates with the amount of caspase activity. Caspase activity was inhibited using the general caspase inhibitor z-VAD-fmk (25 μM; Calbiochem) for 30 min before addition of HDACis or tranylcypromine and added every 24 h throughout the duration of the experiment.
Statistical Analysis
Unless otherwise indicated, values in tables and figures are expressed as the mean ± standard error of the mean of at least triplicate determinations. Statistical comparisons were made using GraphPad Prism software, version 4.0 (GraphPad Software) by the Student t test. A probability value of <.05 was considered to be statistically significant. Synergism was calculated by determining the combination index by the method of Chou and Talalay36 using CalcuSyn software (Biosoft). Combination index values <0.8 indicate a synergistic combination, values of 0.8–1.0 are additive, and values >1.0 are antagonistic.
Results
HDACs Influence the Levels of Histone Methylation in Glioblastoma Cells
To lay the foundation for combined targeting of HDACs and LSD1, we sought to establish the relationship between acetylation and methylation in U87 (p53 wild-type) and LN-18 (p53 mutant) cells. The GBM cell lines were treated for 6 h with doses of vorinostat that have been previously described as effective in glioma cell lines,32 as well as other solid tumor cell lines,33,34 and the levels of histone H3 acetylation and methylation were evaluated by Western blot. We also treated the GBM cell lines with the HDACi PCI-24781. These 2 HDACis were selected to compare vorinostat, the current FDA-approved clinical inhibitor, with a novel hydroxamic acid-based HDACi, PCI-24781, which has greater affinity for HDACs, particularly HDAC1.17
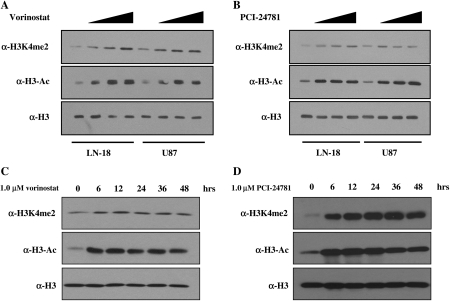
Treatment with vorinostat induced a dose-dependent accumulation of histone H3 acetylation in both LN-18 and U87 cell lines (Fig. 1A). We also observed a dose-dependent increase in di-methylation of lysine 4 of histone H3 (H3K4me2; Fig. 1A), suggesting that there is cross-talk between the enzyme activities in these cells. Similarly, treatment of cells with the novel hydroxamic acid-based HDACi PCI-24781 also caused the accumulation of histone H3 acetylation and H3K4me2 (Fig. 1B). To evaluate the dynamics of histone acetylation and methylation, we performed a time course in which LN-18 and U87 cells were treated with 1.0 μM of vorinostat or PCI-24781 and in which histone modifications were monitored by Western blot (Fig. 1C and D). Histone acetylation and methylation reached a maximum by 6 h and persisted for at least 48 h (Fig. 1C and D). These data strengthen the rationale for simultaneously targeting LSD1 and HDACs.
Fig. 1.
Histone deacetylase inhibitors affect histone modifications removed by LSD1. LN-18 and U87 glioblastoma multiforme cells were treated with increasing doses (1.0–5.0 μM) of (A) vorinostat or (B) PCI-24781 for 6 h. To evaluate the dynamics of histone accumulation, LN-18 cells were treated with 1.0 μM (C) vorinostat or (D) PCI-24781 and harvested at the time points shown. Histone proteins were acid extracted, and changes in histone modifications were detected by Western blot using antibodies specific for acetylated H3 (α-H3-Ac), dimethyl-H3K4 (α-H3K4me2), and total H3 (α-H3). The Western blots shown are representative of 3 independent experiments.
LSD1 is Overexpressed in Glioblastoma
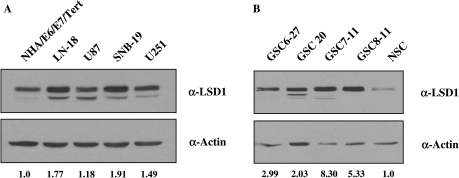
To determine whether LSD1 is a possible molecular target in GBM, we analyzed LSD1 protein expression by Western blot in a variety of established GBM cell lines and compared expression with that of immortalized human astrocytes (NHA/E6/E7/Tert). All GBM cell lines examined expressed more LSD1 than the immortalized astrocytes, with LN-18 and SNB-19 showing the greatest amount of overexpression (1.77- and 1.91-fold, respectively) (Fig. 2A). We then compared LSD1 protein expression in normal neural stem cells (NSCs) with that in cancer stem cells derived from patients with GBM (GSC). In all 4 of the samples tested, LSD1 protein was overexpressed as much as 8-fold in cancer stem cells obtained from GBM patients, compared with normal neural stem cells (Fig. 2B). These data demonstrated that LSD1 is more highly overexpressed at the protein level in GBM relative to immortalized human astrocytes or normal neural stem cells, suggesting that LSD1 may be a suitable molecular target for therapy.
Fig. 2.
LSD1 is overexpressed in glioblastoma multiforme (GBM) cell lines and patient specimens. Expression of LSD1 protein was assessed in total cell lysates by Western blot using an antibody for LSD1 (Abcam) in (A) GBM cell lines, compared with immortalized human astrocytes (NHA/E6/E7/Tert) and (B) cancer stem cells obtained from patients with GBM (GSC), compared with normal neural stem cells (NSCs). Actin was used as a loading control, and densitometry was performed using ImageJ software.
Loss of LSD1 Sensitizes Glioblastoma Cells to Treatment with HDACis
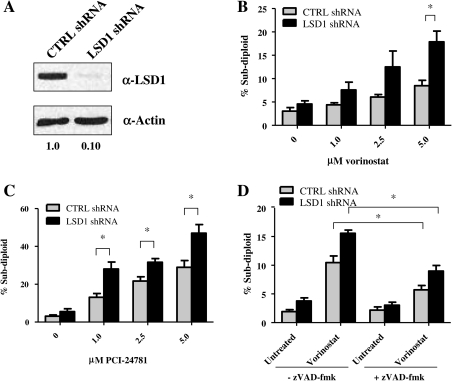
Having established higher LSD1 expression in GBM cell lines and patient-derived samples, we investigated the functional consequences of inhibiting LSD1 in combination with HDACis. To modulate levels of LSD1 protein, we transfected LSD1-directed short hairpin RNA (shRNA) constructs into LN-18 cells by electroporation and monitored knockdown by Western blot at 72 h after transfection. As shown in Fig. 3A, LSD1 protein levels were reduced by 90% in cells transfected with shRNA specific to LSD1, compared with control nonspecific shRNA.
Fig. 3.
Loss of LSD1 sensitizes glioblastoma multiforme cells to histone deacetylase inhibitors. (A) LN-18 cells were transfected (Lonza) with control (CTRL) or LSD1 shRNA, and knockdown was monitored at 72 h by Western blot (50 μg). For all Western blots, actin was used as a loading control, and densitometry was performed using ImageJ software. LN-18 cells transfected with control (CTRL; gray bar) or LSD1 (black bar) shRNA were treated with increasing amounts (1.0–5.0 μM) of (B) vorinostat or (C) PCI-24781 for 48 h, and the level of DNA fragmentation was assessed by PI staining and quantitation by flow cytometry. (D) Control (CTRL; gray bar) or LSD1 (black bar) shRNA transfected LN-18 cells were pretreated with the general caspase inhibitor zVAD-fmk (25 μM) for 30 min before treatment with 5.0 μM of vorinostat. Additional zVAD-fmk was added after 24 h, and DNA fragmentation was measured by PI staining 48 h after vorinostat addition. *P < .01. **P < .001. n = 3. Data are mean ± standard error of the mean.
Because of the functional interplay between the HDACs and LSD1 and the increased expression of LSD1 in GBM cell lines and cancer stem cells derived from patients with GBM, we asked whether loss of LSD1 would enhance apoptosis in GBM cells treated with HDACis. To evaluate whether loss of LSD1-sensitized cells to HDACis, we monitored DNA fragmentation as a measure of apoptosis in LSD1 knockdown cells treated with HDACis. We treated the LSD1 knockdown and control cells with increasing doses of the pan-HDACis vorinostat and PCI-24781 (1.0–5.0 μM) for 48 h and assessed DNA fragmentation by quantifying cells with sub-diploid DNA content by flow cytometry. Our data demonstrated that knockdown of LSD1 alone does not significantly induce apoptosis (Fig. 3B and C), indicating that targeting LSD1 alone is not sufficient to cause significant cell death. However, DNA fragmentation is significantly enhanced (P < .01) when LSD1 knockdown cells are treated in combination with a 5.0-μM dose of vorinostat (Fig. 3B). Similarly, all doses of PCI-24781 enhanced DNA fragmentation in LSD1 knockdown cells, with the 5.0-μM dose exhibiting the greatest effect (Fig. 3C). These data strongly suggest that loss of LSD1 in combination with 2 different hydroxamic acid-based HDACis has a combined effect on the induction of apoptosis in GBM cell lines.
Activation of the caspase enzymes is a central element in the induction of apoptosis. To confirm that the cell death observed in LSD1 knockdown cells treated with HDACi is proceeding through an apoptotic pathway, we evaluated DNA fragmentation in the presence of the general caspase inhibitor z-VAD-fmk. Control and LSD1 knockdown LN-18 cells were preincubated with z-VAD-fmk for 30 min prior to addition of vorinostat, and DNA fragmentation was measured after 48 h. As depicted in Fig. 3D, DNA fragmentation induced by vorinostat is significantly reduced (P < .01)—but not completely blocked—by the addition of z-VAD-fmk in both control and LSD1 knockdown LN-18 cells. These data suggest that targeting LSD1 in combination with HDAC inhibitors may function through both caspase-dependent as well as caspase-independent mechanisms. In fact, several studies demonstrate that vorinostat induces autophagy in a variety of cell types.35
Combination of Vorinostat and Tranylcypromine, a Monoamine Oxidase Inhibitor (MAOi), Results in Synergistic Cell Death of Glioblastoma Cells
Our results demonstrate that targeting both LSD1 and HDACs may be a viable therapeutic option to induce cell death. Because of the similarity between LSD1 and monoamine oxidases, several reports have established that MAOis, such as tranylcypromine, and polyamine oxidases also inhibit LSD1.20–25 In addition, studies evaluating monoamine oxidase inhibitors as monotherapies in mouse xenograft models led to reduced tumor size,27,28 suggesting that LSD1 inhibitors are effective chemotherapeutic agents that when combined with HDACi, cooperate to enhance cell death.
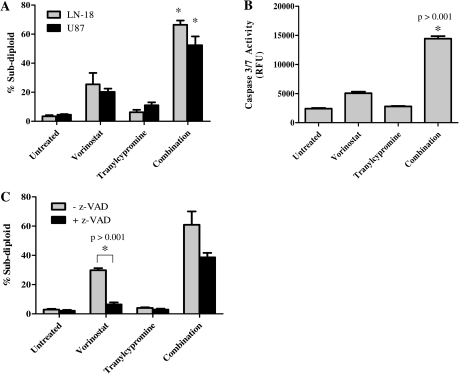
To evaluate this hypothesis, we treated GBM cell lines with different doses of vorinostat (1.0, 2.5, 5.0 μM) and tranylcypromine (0.1, 0.5, 1.0 mM) for 72 h and evaluated DNA fragmentation by PI staining. As depicted in Fig. 4A, tranylcypromine alone did not cause a significant increase in DNA fragmentation in any of the GBM cell lines. However, the combination of tranylcypromine and vorinostat led to a dramatic increase in DNA fragmentation in LN-18 and U87 cells, compared with either tranylcypromine or vorinostat alone (Fig. 4A). These results indicate that use of MAOis that target LSD1 in combination with HDACi promote the synergistic induction of DNA fragmentation.
Fig. 4.
Inhibition of LSD1 in glioblastoma multiforme cells leads to enhanced cell death in response to histone deacetylase inhibitors. (A) LN-18 and U87 GBM cells were treated concurrently with vorinostat (1.0, 2.5, or 5.0 μM) and increasing doses of the monoamine oxidase inhibitor tranylcypromine (0.1, 0.5, of 1.0 mM) for 72 h, followed by analysis of sub-diploid cells by PI staining. The concentrations depicted are 5.0 μM vorinostat, 1.0 mM tranylcypromine, and the combination. *P < .05. n = 3. Data are mean ± standard error of the mean (SEM). (B) LN-18 cells were treated with 5.0 μM of vorinostat, 1.0 mM of tranylcypromine, or the combination for 24 h. Caspase 3 activity was determined by adding the fluorogenic caspase 3/7 DEVD-AMC substrate and measuring the amount of fluorescence in each sample. (C) LN-18 cells were pretreated with the general caspase inhibtor zVAD-fmk (25 μM) for 30 min before addition of 5.0 μM of vorinostat, 1.0 mM tranylcypromine, or the combination. Additional zVAD-fmk was added every 24 h and DNA fragmentation was measured by PI staining 72 h after drug treatment. *P < .001. n = 3. Data are mean ± SEM.
To determine whether the increase in cell death observed during treatment with vorinostat and tranylcypromine corresponds to enhanced apoptosis, as was the case for genetic knockdown of LSD1, we measured caspase 3/7 activity in these cells. As shown in Fig. 4B, vorinostat alone induced a 2-fold increase in caspase 3 activity within 24 h. However, the combination of vorinostat and tranylcypromine led to a marked (6-fold) increase in caspase 3 activity. Moreover, pretreatment of LN-18 cells with the general caspase inhibitor z-VAD-fmk significantly reduced DNA fragmentation (P < .001) induced by vorinostat, and the data show evidence of trends towards a decrease in DNA fragmentation and protection from apoptosis (Fig. 4C). These data indicate that the cell death observed on LSD1 inhibition, chemically or genetically, in combination with HDACis at least partially occurs by activating apoptosis but may involve additional cell death pathways.
Immortalized Human Astrocytes are not Sensitive to the Combined Treatment of Vorinostat and Tranylcypromine
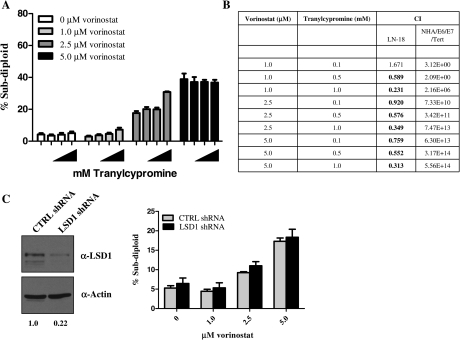
To assess the GBM-specific selectivity of simultaneous inhibition of HDACs and LSD1, we treated immortalized human astrocytes (NHA/E6/E7/Tert) with different doses of vorinostat (1.0, 2.5, and 5.0 μM) and tranylcypromine (0.1, 0.5, and 1.0 μM) for 72 h and evaluated DNA fragmentation by PI staining. Similar to the LN-18 and U87 cells, vorinostat caused an increase in DNA fragmentation in the immortalized astrocytes (Fig. 5A). Interestingly, simultaneous treatment of immortalized astrocytes with vorinostat and tranylcypromine failed to enhance DNA fragmentation (Fig. 5A). To determine whether tranylcypromine and vorinostat exhibit synergistic effects when used in combination, we calculated the combination index values36 for each combination as shown in Fig. 5B. Eight out of nine combination treatments in GBM cells lines resulted in synergistic effects with 5.0 μM of vorinostat and 1.0 mM of tranylcypromine displaying the strongest effect (combination index, 0.313). However, treatment of immortalized human astrocytes with the same doses of vorinostat and tranylcypromine did not cause synergistic effects (Fig. 5B). Moreover, we obtained similar results by knocking down LSD1 in the immortalized human astrocytes followed by treatment with vorinostat (Fig. 5C). We detected no increase in DNA fragmentation in the knockdown cells versus the control during treatment with vorinostat. These data provide evidence to suggest that the combination treatment is selective for GBM tumor cells versus normal cells.
Fig. 5.
The combined inhibition of histone deacetylases and LSD1 is specific for glioblastoma multiforme cells. (A) Immortalized normal human astrocytes (NHA/E6/E7/Tert) were treated concurrently with increasing doses of vorinostat (1.0, 2.5, or 5.0 μM) and tranylcypromine (0.1, 0.5, or 1.0 mM) for 72 h followed by analysis of sub-diploid cells by PI staining. (B) To determine synergy, the combination index values (CIs) were calculated by isobologram anaylsis using the CalcuSyn software. A CI value <0.9 indicates synergy, values of 0.9–1.1 indicate additive effects, and values >1.1 are considered antagonistic. The table defines the CI values for LN-18 cells treated with the dose combinations shown in (A). (C) Immortalized human astrocytes were nucleofected with control or LSD1-specific shRNA, and the degree of knockdown was monitored by Western blot. (D) Control (CTRL shRNA; gray bar) and LSD1 knockdown (LSD1 shRNA; black bar) cells were treated with increasing doses of vorinostat (1.0–5.0 μM) for 48 h and the level of DNA fragmentation was assessed by PI staining and quantitation by flow cytometry. n = 3. Data are mean ± standard error of the mean.
Discussion
Targeting enzymes involved in epigenetic regulation of gene expression has become increasingly popular as a cancer therapy. The success of the HDACi vorinostat, the first HDACi to have been approved by the FDA for treatment of cutaneous T-cell lymphoma, prompted the development of other small molecule HDACis for clinical use. Although HDACis show promise in many cancer types, they are not as effective against solid tumors as monotherapies.7,10 Therefore, the identification of agents that sensitize tumor cells to HDACis is highly clinically relevant. We evaluated the ability of 2 broad-spectrum HDACi, vorinostat and PCI-24781, to cause changes in histone acetylation and methylation in GBM cell lines. We found that both of these HDACis led to increased levels of histone H3 acetylation and H3K4 methylation, a modification removed by the LSD1 demethylase. On the basis of these results, we sought to determine whether targeting LSD1 and HDACs simultaneously caused enhanced GBM tumor cell death. We found that shRNA knockdown of LSD1 in GBM cells led to sensitization to the broad-spectrum HDACis vorinostat and PCI-24781. The cell death observed by the combination treatment was partially blocked (∼50%) by pretreatment with the general caspase inhibitor zVAD-fmk, suggesting that cell death occurs partially through the apoptotic pathway, but other caspase-independent pathways of cell death are also likely to contribute to our observations. Supporting these data are several reports of vorinostat inducing autophagy in a variety of cell types.35 We also obtained similar results when HDACis were combined with chemical inhibition of LSD1 with the monoamine oxidase inhibitor, tranylcypromine. Importantly, we demonstrated that combined inhibition of LSD1 and HDACs is selective for GBM tumor cell lines versus immortalized human astrocytes, an observation that may be due to elevated levels of LSD1 in GBM cell lines and patient samples (Fig. 2). Overall, these data offer a novel therapeutic option for the treatment of GBM.
It has been demonstrated that each HDACi has a different ability to inhibit various HDAC enzymes, some more specifically than others. The targets of each HDAC enzyme, both histone and nonhistone proteins, and their contribution to GBM is still unclear. In addition, most HDACis target the active site of the HDAC and function through moieties that chelate zinc. Therefore, other proteins that require zinc for their function may also be affected by very high doses of HDACis. However, efforts to improve HDACis have yielded compounds with chemical modifications that provide some selectivity against other zinc-containing enzymes. Our studies were performed with doses that have previously proven to be effective in inducing tumor cell death in vitro and in vivo32–34 and are clinically relevant. The maximum plasma concentration for vorinostat in patients is 2.5 μM for oral administration of 400 mg per day and 9 μM for 300 mg/m2 per day for intravenous administration,37 which is well within the range of doses used in the current study.
Histone tails are modified by a variety of posttranslational modifications, including phosphorylation, acetylation, methylation, and ubiquitination. An emerging hypothesis is that these modifications do not act alone but instead influence one another.38,39 Our data indicate that, in GBM cell lines, histone acetylation influences the demethylation of H3K4 (Fig. 1). Consistent with our results, inhibition of HDACs in HEK293 cells also influences methylation on nucleosomes in vivo and acetylation of nucleosomes decreases demethylation by LSD1 in vitro.22 One mechanism proposed to explain the interplay between histone acetylation and methylation is the physical association of HDAC1 and LSD1 by which each enzyme influences the activity of the other.22 More recently, Huang et al40 provided an alternative mechanism and demonstrated that treatment of prostate cancer cells with HDACi leads to the suppression of histone demethylase enzymes, including LSD1, through transcriptional repression of Sp1. These data indicate that there is a biological link between acetylation and H3K4 methylation and suggest targeting LSD1 as a possible therapeutic strategy. Similar to HDACs, LSD1 has also been reported to target proteins other than histones, which may play a role in the enhanced apoptosis observed on combined LSD1 and HDAC inhibition. For example, p53 is a substrate of LSD1.41,42 Demethylation of lysine 370 of p53 inhibits association of the p53-binding protein 1, leading to the down-regulation of p53-regulated genes, such as the pro-apoptotic p21 protein.41 In addition to regulating methylation of p53, LSD1 is also recruited by p53 to the α-fetoprotein gene during hepatic development, in which loss of p53 leads to decreased occupancy of LSD1, an increase in H3K4 methylation, and a loss of gene repression.42 To determine the contribution of p53 to the synergy observed with HDACis and tranylcypromine, we used GBM cell lines that contain wild-type p53 (U87) and mutant p53 (LN-18) in which there is a mutation in the DNA binding domain. We found no difference in the ability of tranylcypromine and HDACis to cause synergistic cell death in these 2 cell lines, suggesting that the cell death caused by these agents is p53-independent, although additional studies are required in isogenic cell lines to prove this concept. Additional studies aimed at determining the nonhistone targets of LSD1 will potentially reveal alternative pathways that contribute to the synergistic cell death observed in our studies.
Because of the homology of LSD1 with the amine oxidase family of enzymes, several mono- and polyamine oxidase inhibitors have been evaluated as inhibitors of LSD1.19,23,24 Inhibition of LSD1 in vivo leads to the reexpression of aberrantly silenced genes.23,26,27 These inhibitors have been used in mouse xenograft models with success,27,28 providing evidence for the use of these agents in the treatment of cancer. Our data demonstrate that the MAOi tranylcypromine enhances the cytotoxic effect of HDACis. Although tranylcypromine is a good candidate for use in combination studies for the treatment of GBM, because it is already clinically used for the treatment of depression, it is often associated with unfavorable side effects due to interactions with certain foods and medications.43 Moreover, the specificity of tranylcypromine for MAOs is much higher than that for LSD1,44 which may induce unwanted adverse effects when used for cancer chemotherapy. Several groups have developed new compounds that are more specific LSD1 inhibitors and are being evaluated preclinically.27,44–46 Our studies suggest that these new compounds could be evaluated, alone and in combination with HDACis, in GBM.
In conclusion, our work demonstrates that inhibition of LSD1 in combination with HDACis results in synergistic cell death. These data provide proof of principle experiments that indicate that combined inhibition of HDACs and LSD1 leads to enhanced GBM cell death. Future studies are focused on evaluating the combination of vorinostat and tranylcypromine in the intracranial glioblastoma mouse model described by Lal et al47 and understanding the mechanism(s) by which HDAC and LSD1 inhibition enhances tumor cell death in vitro and in vivo. One possible explanation is that HDACs and LSD1 cooperate to regulate the expression of genes involved in apoptosis. In fact, treatment of LSD1-knockdown cells with vorinostat yields changes in several genes known to be involved in apoptosis, including Bid, p53, and p73; several tumor necrosis factor receptors; and caspases (unpublished data). Overall, these studies are designed to make substantial contributions to the understanding of GBM biology, which may provide molecular explanations for resistance and identify novel targets for therapy.
Conflict of interest statement. None declared.
Funding
We gratefully acknowledge funding from the Center for Cancer Epigenetics at The University of Texas MD Anderson Cancer Center (Pilot Project Grant to J.C.), National Institutes of Health (R01CA115811 to J.C.), support by a Developmental Research grant from the SPORE in Brain Cancer (5 P50 CA127001-03 from NCI), The University of Texas MD Anderson Cancer Center (to J.C.), and a Cancer Prevention and Research Institute of Texas Research Training Award (RP101502 to C.A.M.). The content is solely the responsibility of the authors and does not necessarily represent the official views of the National Cancer Institute or the National Institutes of Health.
References
- 1.Maher EA, Furnari FB, Bachoo RM, et al. Malignant glioma: genetics and biology of a grave matter. Genes Dev. 2001;15(11):1311–1333. doi: 10.1101/gad.891601. doi:10.1101/gad.891601. [DOI] [PubMed] [Google Scholar]
- 2.Carpentier AF. Neuro-oncology: the growing role of chemotherapy in glioma. Lancet Neurol. 2005;4(1):4–5. doi: 10.1016/S1474-4422(04)00944-5. doi:10.1016/S1474-4422(04)00944-5. [DOI] [PubMed] [Google Scholar]
- 3.Nagarajan RP, Costello JF. Molecular epigenetics and genetics in neuro-oncology. Neurotherapeutics. 2009;6(3):436–446. doi: 10.1016/j.nurt.2009.04.002. doi:10.1016/j.nurt.2009.04.002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Bolden JE, Peart MJ, Johnstone RW. Anticancer activities of histone deacetylase inhibitors. Nat Rev Drug Discov. 2006;5(9):769–784. doi: 10.1038/nrd2133. doi:10.1038/nrd2133. [DOI] [PubMed] [Google Scholar]
- 5.Dokmanovic M, Marks PA. Prospects: histone deacetylase inhibitors. J Cell Biochem. 2005;96(2):293–304. doi: 10.1002/jcb.20532. doi:10.1002/jcb.20532. [DOI] [PubMed] [Google Scholar]
- 6.Marks PA, Dokmanovic M. Histone deacetylase inhibitors: discovery and development as anticancer agents. Expert Opin Investig Drugs. 2005;14(12):1497–1511. doi: 10.1517/13543784.14.12.1497. doi:10.1517/13543784.14.12.1497. [DOI] [PubMed] [Google Scholar]
- 7.Rasheed WK, Johnstone RW, Prince HM. Histone deacetylase inhibitors in cancer therapy. Expert Opin Investig Drugs. 2007;16(5):659–678. doi: 10.1517/13543784.16.5.659. doi:10.1517/13543784.16.5.659. [DOI] [PubMed] [Google Scholar]
- 8.Minucci S, Pelicci PG. Histone deacetylase inhibitors and the promise of epigenetic (and more) treatments for cancer. Nat Rev Cancer. 2006;6(1):38–51. doi: 10.1038/nrc1779. doi:10.1038/nrc1779. [DOI] [PubMed] [Google Scholar]
- 9.Xu WS, Parmigiani RB, Marks PA. Histone deacetylase inhibitors: molecular mechanisms of action. Oncogene. 2007;26(37):5541–5552. doi: 10.1038/sj.onc.1210620. doi:10.1038/sj.onc.1210620. [DOI] [PubMed] [Google Scholar]
- 10.Lee MJ, Kim YS, Kummar S, Giaccone G, Trepel JB. Histone deacetylase inhibitors in cancer therapy. Curr Opin Oncol. 2008;20(6):639–649. doi: 10.1097/CCO.0b013e3283127095. doi:10.1097/CCO.0b013e3283127095. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Miller CP, Rudra S, Keating MJ, Wierda WG, Palladino M, Chandra J. Caspase-8 dependent histone acetylation by a novel proteasome inhibitor, NPI-0052: a mechanism for synergy in leukemia cells. Blood. 2009;113(18):4289–4299. doi: 10.1182/blood-2008-08-174797. doi:10.1182/blood-2008-08-174797. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Rahmani M, Reese E, Dai Y, et al. Cotreatment with suberanoylanilide hydroxamic acid and 17-allylamino 17-demethoxygeldanamycin synergistically induces apoptosis in Bcr-Abl+ cells sensitive and resistant to STI571 (imatinib mesylate) in association with down-regulation of Bcr-Abl, abrogation of signal transducer and activator of transcription 5 activity, and Bax conformational change. Mol Pharmacol. 2005;67(4):1166–1176. doi: 10.1124/mol.104.007831. doi:10.1124/mol.104.007831. [DOI] [PubMed] [Google Scholar]
- 13.Rahmani M, Reese E, Dai Y, et al. Coadministration of histone deacetylase inhibitors and perifosine synergistically induces apoptosis in human leukemia cells through Akt and ERK1/2 inactivation and the generation of ceramide and reactive oxygen species. Cancer Res. 2005;65(6):2422–2432. doi: 10.1158/0008-5472.CAN-04-2440. doi:10.1158/0008-5472.CAN-04-2440. [DOI] [PubMed] [Google Scholar]
- 14.Yu C, Rahmani M, Conrad D, Subler M, Dent P, Grant S. The proteasome inhibitor bortezomib interacts synergistically with histone deacetylase inhibitors to induce apoptosis in Bcr/Abl+ cells sensitive and resistant to STI571. Blood. 2003;102(10):3765–3774. doi: 10.1182/blood-2003-03-0737. doi:10.1182/blood-2003-03-0737. [DOI] [PubMed] [Google Scholar]
- 15.Yu C, Friday BB, Yang L, et al. Mitochondrial Bax translocation partially mediates synergistic cytotoxicity between histone deacetylase inhibitors and proteasome inhibitors in glioma cells. Neuro Oncol. 2008;10(3):309–319. doi: 10.1215/15228517-2007-063. doi:10.1215/15228517-2007-063. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Galanis E, Jaeckle KA, Maurer MJ, et al. Phase II trial of vorinostat in recurrent glioblastoma multiforme: a north central cancer treatment group study. J Clin Oncol. 2009;27(12):2052–2058. doi: 10.1200/JCO.2008.19.0694. doi:10.1200/JCO.2008.19.0694. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Buggy JJ, Cao ZA, Bass KE, et al. CRA-024781: a novel synthetic inhibitor of histone deacetylase enzymes with antitumor activity in vitro and in vivo. Mol Cancer Ther. 2006;5(5):1309–1317. doi: 10.1158/1535-7163.MCT-05-0442. doi:10.1158/1535-7163.MCT-05-0442. [DOI] [PubMed] [Google Scholar]
- 18.Shi Y, Lan F, Matson C, et al. Histone demethylation mediated by the nuclear amine oxidase homolog LSD1. Cell. 2004;119(7):941–953. doi: 10.1016/j.cell.2004.12.012. doi:10.1016/j.cell.2004.12.012. [DOI] [PubMed] [Google Scholar]
- 19.Metzger E, Wissmann M, Yin N, et al. LSD1 demethylates repressive histone marks to promote androgen-receptor-dependent transcription. Nature. 2005;437(7057):436–439. doi: 10.1038/nature04020. [DOI] [PubMed] [Google Scholar]
- 20.Culhane JC, Szewczuk LM, Liu X, Da G, Marmorstein R, Cole PA. A mechanism-based inactivator for histone demethylase LSD1. J Am Chem Soc. 2006;128(14):4536–4537. doi: 10.1021/ja0602748. doi:10.1021/ja0602748. [DOI] [PubMed] [Google Scholar]
- 21.Gooden DM, Schmidt DM, Pollock JA, Kabadi AM, McCafferty DG. Facile synthesis of substituted trans-2-arylcyclopropylamine inhibitors of the human histone demethylase LSD1 and monoamine oxidases A and B. Bioorg Med Chem Lett. 2008;18(10):3047–3051. doi: 10.1016/j.bmcl.2008.01.003. doi:10.1016/j.bmcl.2008.01.003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Lee MG, Wynder C, Bochar DA, Hakimi MA, Cooch N, Shiekhattar R. Functional interplay between histone demethylase and deacetylase enzymes. Mol Cell Biol. 2006;26(17):6395–6402. doi: 10.1128/MCB.00723-06. doi:10.1128/MCB.00723-06. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Lee MG, Wynder C, Schmidt DM, McCafferty DG, Shiekhattar R. Histone H3 lysine 4 demethylation is a target of nonselective antidepressive medications. Chem Biol. 2006;13(6):563–567. doi: 10.1016/j.chembiol.2006.05.004. doi:10.1016/j.chembiol.2006.05.004. [DOI] [PubMed] [Google Scholar]
- 24.Schmidt DM, McCafferty DG. trans-2-Phenylcyclopropylamine is a mechanism-based inactivator of the histone demethylase LSD1. Biochemistry. 2007;46(14):4408–4416. doi: 10.1021/bi0618621. doi:10.1021/bi0618621. [DOI] [PubMed] [Google Scholar]
- 25.Yang M, Culhane JC, Szewczuk LM, et al. Structural basis for the inhibition of the LSD1 histone demethylase by the antidepressant trans-2-phenylcyclopropylamine. Biochemistry. 2007;46(27):8058–8065. doi: 10.1021/bi700664y. doi:10.1021/bi700664y. [DOI] [PubMed] [Google Scholar]
- 26.Huang Y, Greene E, Murray Stewart T, et al. Inhibition of lysine-specific demethylase 1 by polyamine analogues results in reexpression of aberrantly silenced genes. Proc Natl Acad Sci USA. 2007;104(19):8023–8028. doi: 10.1073/pnas.0700720104. doi:10.1073/pnas.0700720104. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Huang Y, Vasilatos SN, Boric L, Shaw PG, Davidson NE. Inhibitors of histone demethylation and histone deacetylation cooperate in regulating gene expression and inhibiting growth in human breast cancer cells. Breast Cancer Res Treat. 2011 doi: 10.1007/s10549-011-1480-8. Mar 31. [Epub ahead of print] doi:10.1158/1078-0432.CCR-09-1293. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Schulte JH, Lim S, Schramm A, et al. Lysine-specific demethylase 1 is strongly expressed in poorly differentiated neuroblastoma: implications for therapy. Cancer Res. 2009;69(5):2065–2071. doi: 10.1158/0008-5472.CAN-08-1735. doi:10.1158/0008-5472.CAN-08-1735. [DOI] [PubMed] [Google Scholar]
- 29.Sonoda Y, Ozawa T, Hirose Y, et al. Formation of intracranial tumors by genetically modified human astrocytes defines four pathways critical in the development of human anaplastic astrocytoma. Cancer Res. 2001;61(13):4956–4960. [PubMed] [Google Scholar]
- 30.Sui G, Soohoo C, Affar el B, Gay F, Shi Y, Forrester WC. A DNA vector-based RNAi technology to suppress gene expression in mammalian cells. Proc Natl Acad Sci USA. 2002;99(8):5515–5520. doi: 10.1073/pnas.082117599. doi:10.1073/pnas.082117599. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Miller CP, Ban K, Dujka ME, et al. NPI-0052, a novel proteasome inhibitor, induces caspase-8 and ROS-dependent apoptosis alone and in combination with HDAC inhibitors in leukemia cells. Blood. 2007;110(1):267–277. doi: 10.1182/blood-2006-03-013128. doi:10.1182/blood-2006-03-013128. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Ugur HC, Ramakrishna N, Bello L, et al. Continuous intracranial administration of suberoylanilide hydroxamic acid (SAHA) inhibits tumor growth in an orthotopic glioma model. J Neurooncol. 2007;83(3):267–275. doi: 10.1007/s11060-007-9337-z. doi:10.1007/s11060-007-9337-z. [DOI] [PubMed] [Google Scholar]
- 33.Munshi A, Tanaka T, Hobbs ML, Tucker SL, Richon VM, Meyn RE. Vorinostat, a histone deacetylase inhibitor, enhances the response of human tumor cells to ionizing radiation through prolongation of gamma-H2AX foci. Mol Cancer Ther. 2006;5(8):1967–1974. doi: 10.1158/1535-7163.MCT-06-0022. doi:10.1158/1535-7163.MCT-06-0022. [DOI] [PubMed] [Google Scholar]
- 34.Marks PA, Breslow R. Dimethyl sulfoxide to vorinostat: development of this histone deacetylase inhibitor as an anticancer drug. Nat Biotechnol. 2007;25(1):84–90. doi: 10.1038/nbt1272. doi:10.1038/nbt1272. [DOI] [PubMed] [Google Scholar]
- 35.Schrump DS. Cytotoxicity mediated by histone deacetylase inhibitors in cancer cells: mechanisms and potential clinical implications. Clin Cancer Res. 2009;15(12):3947–3957. doi: 10.1158/1078-0432.CCR-08-2787. doi:10.1158/1078-0432.CCR-08-2787. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Chou TC, Talalay P. Analysis of combined drug effects: a new look at a very old problem. Trends in Pharmacological Sciences. 1983;4:450–454. doi:10.1016/0165-6147(83)90490-X. [Google Scholar]
- 37.O'Connor OA, Heaney ML, Schwartz L, et al. Clinical experience with intravenous and oral formulations of the novel histone deacetylase inhibitor suberoylanilide hydroxamic acid in patients with advanced hematologic malignancies. J Clin Oncol. 2006;24(1):166–173. doi: 10.1200/JCO.2005.01.9679. doi:10.1200/JCO.2005.01.9679. [DOI] [PubMed] [Google Scholar]
- 38.Latham JA, Dent SY. Cross-regulation of histone modifications. Nat Struct Mol Biol. 2007;14(11):1017–1024. doi: 10.1038/nsmb1307. doi:10.1038/nsmb1307. [DOI] [PubMed] [Google Scholar]
- 39.Suganuma T, Workman JL. Crosstalk among histone modifications. Cell. 2008;135(4):604–607. doi: 10.1016/j.cell.2008.10.036. doi:10.1016/j.cell.2008.10.036. [DOI] [PubMed] [Google Scholar]
- 40.Huang PH, Chen CH, Chou CC, et al. Histone deacetylase inhibitors stimulate histone H3 lysine 4 methylation in part via transcriptional repression of histone H3 lysine 4 demethylases. Mol Pharmacol. 2011;79(1):197–206. doi: 10.1124/mol.110.067702. doi:10.1124/mol.110.067702. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Huang J, Sengupta R, Espejo AB, et al. p53 is regulated by the lysine demethylase LSD1. Nature. 2007;449(7158):105–108. doi: 10.1038/nature06092. doi:10.1038/nature06092. [DOI] [PubMed] [Google Scholar]
- 42.Tsai WW, Nguyen TT, Shi Y, Barton MC. p53-targeted LSD1 functions in repression of chromatin structure and transcription in vivo. Mol Cell Biol. 2008;28(17):5139–5146. doi: 10.1128/MCB.00287-08. doi:10.1128/MCB.00287-08. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Krishnan KR. Revisiting monoamine oxidase inhibitors. J Clin Psychiatry. 2007;68(suppl 8):35–41. [PubMed] [Google Scholar]
- 44.Binda C, Valente S, Romanenghi M, et al. Biochemical, structural, and biological evaluation of tranylcypromine derivatives as inhibitors of histone demethylases LSD1 and LSD2. J Am Chem Soc. 2010;132(19):6827–6833. doi: 10.1021/ja101557k. [DOI] [PubMed] [Google Scholar]
- 45.Culhane JC, Wang D, Yen PM, Cole PA. Comparative analysis of small molecules and histone substrate analogues as LSD1 lysine demethylase inhibitors. J Am Chem Soc. 2010;132(9):3164–3176. doi: 10.1021/ja909996p. doi:10.1021/ja909996p. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Ueda R, Suzuki T, Mino K, et al. Identification of cell-active lysine specific demethylase 1-selective inhibitors. J Am Chem Soc. 2009;131(48):17536–17537. doi: 10.1021/ja907055q. doi:10.1021/ja907055q. [DOI] [PubMed] [Google Scholar]
- 47.Lal S, Lacroix M, Tofilon P, Fuller GN, Sawaya R, Lang FF. An implantable guide-screw system for brain tumor studies in small animals. J Neurosurg. 2000;92(2):326–333. doi: 10.3171/jns.2000.92.2.0326. [DOI] [PubMed] [Google Scholar]