Abstract
Glioblastoma multiforme (GBM) is the most common type of primary brain tumor and a highly malignant and heterogeneous cancer. Current conventional therapies fail to eradicate or curb GBM cell growth. Hence, exploring the cellular and molecular basis of GBM cell growth is vital to develop novel therapeutic approaches. Neuroglia (NG)-2 is a transmembrane proteoglycan expressed by NG2+ progenitors and is strongly linked to cell proliferation in the normal brain. By using NG2 as a biomarker we identify a GBM cell population (GBM NG2+ cells) with robust proliferative, clonogenic, and tumorigenic capacity. We show that a significant proportion (mean 83%) of cells proliferating in the tumor mass express NG2 and that over 50% of GBM NG2+ cells are proliferating. Compared with the GBM NG2− cells from the same tumor, the GBM of NG2+ cells overexpress genes associated with aggressive tumorigenicity, including overexpression of Mitosis and Cell Cycling Module genes (e.g., MELK, CDC, MCM, E2F), which have been previously shown to correlate with poor survival in GBM. We also show that the coexpression pattern of NG2 with other glial progenitor markers in GBM does not recapitulate that described in the normal brain. The expression of NG2 by such an aggressive and actively cycling GBM population combined with its location on the cell surface identifies this cell population as a potential therapeutic target in a subset of patients with GBM.
Keywords: glioblastoma, molecular signature, NG2, proliferation, progenitor
Glioblastoma multiforme (GBM) is the most common type of primary brain malignancy, with a median life expectancy in optimally managed patients of only 14 months and with less than 25% surviving 24 months.1,2 The current clinical management of patients diagnosed with GBM involves a combination of surgery, radiotherapy, and chemotherapy, with radiotherapy as the principal therapeutic modality since the late 1970s.3,4 Additional targeted chemotherapy is of only modest benefit,2,5 and the need for new treatments has been unmet clinically.
Neuroglia (NG)-2, also known as chondroitin sulphate proteoglycan 4 (CSPG4), is a membrane-bound cell-surface proteoglycan of 450 kDa with a core protein of 250 kDa. It is a phylogenetically conserved protein with important roles in cell proliferation, migration, and angiogenesis.6 In the normal adult brain, NG2 is expressed by 5% of total neural cells, has a wide anatomical distribution, and identifies more than 70% of cycling progenitors.7–11 NG2+ progenitors have the ability to proliferate, self-renew, and produce different types of neural cells under normal12–14 and pathologic conditions.15–18 Collectively, these characteristics have led to a reevaluation of the lineage potential and function of what have previously been considered unipotent oligodendrocyte precursors.19–21
At the cellular level, normal NG2+ progenitors can be identified by the coexpression of NG2 with the transcription factor Olig2, platelet derived growth factor receptor alpha (PDGFRα; a tyrosine kinase receptor), and the ganglioside A2B5.9,22–30 Recent data demonstrate that NG2 expression identifies the vast majority of the proliferating cells in the murine and human normal adult brain.7,9,31
In GBM, the expression of these markers (NG2, Olig2, and PDGFRα) has also been reported.32,33 However, a detailed description of the proliferative and tumorigenic potential of NG2-expressing cells in GBM (GBM NG2+ cells) and correlation with their parent tumors has yet to be described. Consequently validation of the GBM NG2+ cells as a potential therapeutic target has yet to be established. Furthermore none of these reports have characterized quantitatively the coexpression pattern of markers associated with NG2+ progenitors (Olig2 and PDGFRα) by the GBM NG2+ cells. It is not known if the normal coexpression pattern of these markers reported in the normal CNS is recapitulated in GBM. It is also not known whether GBM samples have a common and distinct pattern of coexpression of these markers.
Therefore, we compared the proliferative, clonogenic, and tumorigenic capacities of the GBM NG2+ and GBM NG2− cells derived from the same GBM sample. We also explored the molecular genetic signature of the GBM NG2+ cells. Finally we screened the prevalence and interrogated the phenotypic identity of the GBM NG2+ cells in GBM.
Materials and Methods
Cell Derivation and Tumor Formation Assay
Tissue samples were obtained in accordance with local ethical guidelines. Cell derivation and the tumor formation assay have been described previously.33 Briefly, anonymized tissue was mechanically minced and cells were seeded in defined serum-free (SF) media and allowed to form primary aggregates. These spheroid aggregates were collected and plated, without dissociation, onto extracellular matrix (ECM)–coated flasks (ECM 1:10 dilution, Sigma) and allowed to form a primary monolayer. As the primary monolayer approached confluence, cells were passaged to generate the subsequent monolayers. Cells were cultured in 10 mL SF medium (phenol red free Neurobasal A; Invitrogen) with 20 mM l-glutamine and 1% volume/volume (v/v) penicillin/streptomycin/fungizone (PSF) solution with 20 ng/mL human epidermal growth factor (Sigma), 20 ng/mL basic fibroblast growth factor (R&D systems), 20 ng/mL heparin (Sigma), 2% v/v B27 SF supplement (Invitrogen), and 1% N2 SF supplement (Invitrogen).
Differentiation medium consisted of phenol-free n-butyl acrylate (NBA) with 20 mM l-glutamine, 1% v/v PSF solution, 2% v/v B27, and 1% v/v N2 and was supplemented with 10% v/v fetal calf serum (FCS; Gibco). Differentiation medium contained no mitogens.
Basic medium consisted of phenol-free NBA with 20 mM l-glutamine, 1% v/v PSF solution, 2% v/v B27, and 1% v/v N2. Basic media did not contain mitogens or FCS.
Conditioned media were made by mixing fresh SF medium with medium conditioned with the identical cell line growing for about 1 week. The ratio was 50% SF: 50% conditioned medium. All cultures were incubated at 37.5°C in 5% CO2.
Cell Proliferation Assay
The CellTiter 96 Aqueous One Solution Cell Proliferation Assay kit (Promega) was used. Cells were seeded (3000 cells per well) into 96-well plates precoated with ECM and incubated in 200 µL of defined SF media at 37°C. Cells were allowed to grow for 3 different days before assay was started. Solution of 5 µL of MTS (3-(4,5-dimethylthiazol-2-yl)-5-(3-carboxymethoxyphenyl)-2-(4-sulfophenyl)-2H-tetrazolium) was added into each well. After incubation for 1–2 h, by which the tetrazolium was reduced in the mitochondria to formazan, optical absorbance values at 490 nm from each well were measured using a plate reader (ELx 800; Bio-Tek Instruments).
Values measured from wells containing no cells were considered negative controls. All absorbance values were adjusted to the control by subtracting the absorbance values obtained from the control wells. All the adjusted absorbance values were also normalized to that of the corresponding GBM NG2+ cultures.
Limiting Dilution Analysis
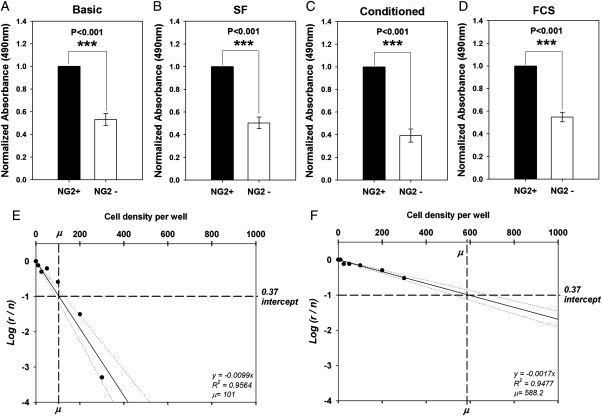
The assay was performed as described by Lefkovits and Waldmann (1999). Briefly, single cell suspensions from 2 different GBM cell lines were prepared. Cells were seeded by fluorescent-activated cell sorting (FACS) in decreasing densities of 300, 200, 100, 50, 25, and 10 cells per well into 96-well plates. Cells were allowed to grow for 1 week under defined SF culture conditions. Nonresponding wells, defined as those wells that did not contain colonies, were counted and divided by the overall number of wells. The values were plotted on a logarithmic axis against the cell densities (on a linear axis). The average number of colony-forming cells per well for each density was calculated and Poisson distribution was presented as a semilog plot pointing to the 37% intercept that corresponds to the cell density containing 1 colony-forming cell. Accordingly, linear regression was performed, as depicted in Figure 1.
Fig. 1.
Glioblastoma multiforme (GBM) neuroglia (NG)-2+ cells show higher levels of cell proliferation compared with the GBM NG2− cells from the same tumor. Analysis of 8 GBM showed proliferative dominance by NG2+ cells (Supplementary Material, Table S1). Illustrative example from 1 cell line: (A) Normalized data from MTS assay shows higher signal in the GBM NG2+ cell cultures, indicating more robust cell growth (3 repeats per cell line). (B) Higher number of total cells in the GBM NG2+ cultures (3 repeats per cell line). (C) Ki67 staining of the fluorescent-activated cell–sorted GBM NG2+ and GBM NG2− cells (scale bars: 100 µm). (D) Quantification of the number of Ki67+ cells showing higher number in the GBM NG2+ cultures (3 repeats for each of 3 cell lines). (E) Quantification of the proportion of Ki67 cells showing higher proportion in the GBM NG2+ cultures. Note: Comparisons were made between NG2+ and NG2− populations within each tumor. Comparison between tumors was not made owing to heterogeneity between patients.
Animal Transplantation
All animals were housed and maintained in accordance with the UK Animal (Scientific Procedures) Act 1986 and the Cambridge University Commission for Animal Health. Tissue was implanted as single cell suspensions into subcutaneous tissue or forebrain of female CB17/ICR-Prkdcscid/Crl severe combined immunodeficient (SCID) mice aged 6–8 weeks.
For subcutaneous implantation, suspensions of 5 × 104 GBM cells were prepared in 50 μL of Hank's buffered salt solution (HBSS) and injected after being mixed with 100–200 μL of ECM into the dermal layer of the hind limbs.
For orthotopic intracerebral implantation, dissociated cells were implanted at a density of 5 × 104 cells/μL in a total volume of 1 μL HBSS over 5–10 min using a 1-μL microsyringe with a 200-μm needle in a minimally traumatic technique (SGE Europe) into anaesthetized animals (using isoflurane). The implants were made at the following coordinates relative to bregma: anteroposterior +2 mm, lateral (L) +1 mm, and dorsoventral (DV) +2 mm from the skull surface.
Immunochemistry
For the immunohistochemistry protocol, free-floating sections (30–40 μm thick) were washed with tris-balanced saline (TBS; pH 7.4) twice for 10 min. Antigen retrieval for nuclear markers was performed by 2 M HCl for 20 min at 37°C, then the sections were washed 3 times with TBS, permeabilized with 0.2% Triton-X, and incubated with blocking buffer for 2 h before the primary antibodies were added for overnight incubation at 4°C. Sections were then washed and incubated with secondary antibodies for 2 h at 4°C. After further washes, sections were incubated with Hoechst 1:10 000 for 15 min, washed with TBS for 10 min, and mounted on slides using FluorSave™ Reagent (Galbiochem).
For the immunocytochemistry protocol, cells were fixed with ice cold ethanol absolute and then washed with phosphate buffered saline (PBS; pH 7.4), incubated with 0.02% Triton-X for 15 min at room temperature, and incubated with blocking buffer for 30–60 min at room temperature. Block was removed and cells were then incubated overnight with primary antibodies at 4°C. After washing and incubating at room temperature with fluorophore conjugated secondary antibodies (goat anti-rabbit and goat anti-mouse), cells were washed again and incubated for 10 min with 1:10 000 dilution Hoechst to label cell nuclei, then were mounted onto borosilicate slides using VectaShield (Vector Labs) and coverslipped. All immunofluorescence was analyzed using either a Leica DM6000B fluorescent microscope or a Leica TCS (true confocal scanner) SPE (spectral) confocal microscope mounted onto an axiovert inverted stage.
Fluorescent-Activated Cell Sorting Analysis
Live cells were suspended in 500 µL FACS buffer (PBS containing 1% FCS and 1% bovine serum albumin) and incubated for 60 min with primary antibodies. Cells were washed before incubating with the appropriate secondary antibodies for 30 min. Finally, cells were washed twice before resuspension and FACS analysis.
For intracellular staining, cells were fixed by incubation with ice cold ethanol at −20°C for at least 20–30 min. Cells were then washed twice with PBS before being permeablized by 0.02% Triton-X in PBS for 5 min. Cells were washed again with PBS and seeded into 96-well plates, and the primary and appropriate secondary antibodies were added as described above.
The FL1 (for Alex488) and FL4 (for Alexa647) gates were adjusted by the autofluorescent activity of the unlabeled cells (negative control). All labeled cells with fluorescent activity above the values obtained from the negative control were considered positive.
FACS was performed using a Moflo Cell Sorter (Beckman Coulter) and Summit software. FACS analysis was performed using a FACSCalibur flow cytometer (Becton Dickinson). Data analysis and quantification were performed using Cell Quest software (Becton Dickinson) and FCS press (Ray Hicks).
Antibodies
Table 1 summarizes the primary and secondary antibodies used for immunohistochemistry and immunocytochemistry in this study. Antibody concentrations used for FACS and flow cytometry analysis were similar to those used for immunocytochemistry.
Table 1.
Primary and secondary antibodies used for immunohistochemistry and immunocytochemistry
| Primary Antibodies | Host | Source | IHC | ICC | Secondary Antibodies | IHC/ICC |
|---|---|---|---|---|---|---|
| A2B5 | M | Millipore, UK | – | 1:500 | G anti-M | 1:500 |
| HN | M | Millipore, UK | 1:500 | – | G anti-M | 1:500 |
| Ki67 | R | Abcam, UK | 1:100 | 1:100 | G anti-R | 1:500 |
| NG2 | M | Millipore, UK | 1:200 | 1:200 | G anti-R | 1:500 |
| Olig2 | R | Drs Stiles and Rowitch | 1:200 | 1:500 | G anti-R | 1:500 |
| Olig2 | R | Abcam, UK | 1:200 | 1:200 | G anti-R | 1:500 |
| PDGFRα | R | Dr Stallcup | 1:100 | 1:100 | G anti-R | 1:500 |
| PDGFRβ | R | Dr Stallcup | 1:100 | 1:100 | G anti-R | 1:500 |
| vWF | R | Dr Al-Lamki | 1:200 | – | G anti-R | 1:500 |
Abbreviations: IHC, immunohistochemistry; ICC, immunocytochemistry; G, goat; HN, human nuclei; M, mouse; R, rabbit; NG2, neuroglia 2; PDGFR, platelet derived growth factor receptor; vWF, von Willebrand factor. The source for all antibodies was Invitrogen.
DNA and RNA Purification and Comparative Genomic Hybridization
DNA and RNA were extracted using the Allprep DNA/RNA kit (Qiagen) according to the manufacturer's instructions. Final concentration and purity were assessed using the Nanodrop system.
Comparative genomic hybridization (CGH) of a whole genome array was constructed as described previously.34 For array CGH, a set of clones representing sequence at an approximately 1-Mb interval across the whole genome (1-Mb clone set) was obtained from the Wellcome Trust Sanger Institute. Of these clones, 3038 at an average interval of 0.97 Mb were used for analysis. Cloned DNA was extracted and then amplified with 3 degenerate oligonucleotide primers (DOPs). Products of DOP PCR were then amplified with a 5′ amine primer and spotted onto amine-binding slides. Test- and sex-mismatched reference DNA were labeled with Cy5- and Cy3-dCTP (a fluorescent dye + deoxycytidine triphosphate), respectively, using a Bio Prime labeling kit (Invitrogen) and hybridized to arrays that had been prehybridized to minimize nonspecific binding and binding to repetitive sequence. Arrays were scanned with an Axon GenePix 4100A device and signal-quantified with GenePix Pro 6.1 software (Molecular Devices). Spots with poor morphology or with low intensity compared with Drosophila controls were excluded. The test/reference signal ratios of the remaining spots were normalized against all autosomal clones within each subarray and the duplicates were averaged. The log2 value of the normalized ratio (log2 ratio) was plotted on the abscissa against clones on ordinate. All analysis was performed using Microsoft Excel.
Expression Microarray Platforms and Data Analysis
Screening of GBM samples for whole-genome expression was performed using Illumina platform HumanRef8_V3 arrays (Centre for Microarray Resources, Department of Pathology, University of Cambridge). The data are available online in the Gene Omnibus Database with accession number 15 846. The gene is considered present when the detection value in the raw data tables is greater than 0.99. Expression array of NG2+ and NG2− fractions was performed using Illumina platform HumanWG6_V3 (Cancer Research Institute). For data analysis, values were filtered according to the following criteria: average detection rate >6.5, log2 fold change of <−.5 or >.5 and adjusted false discovery rate (FDR) P< .05. Heat map, clustering, and general statistical analysis were produced by Dr R. Russell at the Cancer Research Institute. Gene ontology analysis was performed using GeneTrail enrichment tools (http://genetrail.bioinf.uni-sb.de/).
Statistical Analysis
Comparisons between 2 groups with one variable were made using a 1-way analysis of variance (ANOVA). This was preceded by testing the normality by the Kolmogorov–Smirnov test. If the normality test failed or the variances of the tested groups were not equal, then rank transformation of the nonparametric data into parametric data was performed before conducting parametric statistical tests. Alternatively, if the rank transformation failed, then a Kruskal–Wallis 1-way ANOVA on ranks was performed. For comparisons between more than 2 groups with one or more variables or treatments, a 1-way or 2-way ANOVA was performed as appropriate. These were preceded by testing the normality by a Kolmogorov–Smirnov test. If the normality test failed, then rank transformation of the nonparametric data into parametric data was performed before conducting parametric statistical tests. Alternatively, the nonparametric data were rearranged to conduct a Kruskal–Wallis 1-way ANOVA on ranks. Post hoc studies of the Holm–Sidak, Tukey, or Dunnett method were run for pairwise comparisons as appropriate.
Linear regression was made through the origin, and the coefficient of determination R2, the corresponding equation, and the 95% confidence intervals were provided.
All error bars generated and depicted in the figures represent standard error of means. An alpha level of less than .05 (P< .05) was used for statistical significance in all tests. All statistical analysis was performed using Microsoft Excel and Sigma Stat software and all diagrams were generated using Sigma Plot software (Systat).
Results
GBM NG2+ Cells Show Higher Levels of Cell Proliferation Compared With GBM NG2− Cells
We first addressed this issue using our GBM cell lines derived and maintained under defined SF conditions according to our protocol.33 We have previously demonstrated that our cell lines are an improved model system for GBM compared with traditional cell lines because of a close recapitulation of the molecular genetic characteristics of the parent tumor. We have also shown that our cell lines endogenously express NG2 (in addition to Olig2, PDGFRα, and A2B534; Supplementary Material, Fig. S2).
Using FACS, GBM cell lines were sorted based on their expression of NG2, and equal numbers of cells from the GBM NG2+ and GBM NG2− fractions were cultured under identical conditions. We examined cell growth in each fraction at day 3 in culture using the MTS assay.35 All tested cell lines (n = 8) showed that the GBM NG2+ cultures gave higher absorbance signals and optical density values compared with their GBM NG2− counterparts (Supplementary Material, Table S1, Fig. 1A; P< .001), suggesting a higher level of cell activity.
To make sure that this observation was due to the generation of more cells rather than to higher intrinsic metabolic activity, we quantified the number of total cells in the GBM NG2+ and GBM NG2− populations after 3 days in culture. When we compared populations from the same sample, we found that the GBM NG2+ cultures from each of 3 different GBM cell lines contained a higher number of cells (Fig. 1B; mean = 154; range = 102–188 cells per field) compared with the GBM NG2− cultures (Fig. 1B; mean = 69; range = 41–88 cells per field; P< .001).
Next, we stained the GBM NG2+ and GBM NG2− cultures for the proliferative marker Ki67 and quantified the Ki67+ cells in each culture (Fig. 1C). Again, data showed that the GBM NG2+ cultures had a higher number of Ki67+ cells (Fig. 1C and D; mean = 66; range = 47–78 Ki67+ cells per field) compared with the GBM NG2− cultures (Fig. 1C and D; mean = 13; range = 5–18 Ki67+ cells per field; P< .001).
Furthermore, the proportion of Ki67+ cells was higher in the GBM NG2+ population (Fig. 1E; mean = 44%; range = 39%–46% Ki67+ cells per field) compared with the GBM NG2− population (Fig. 1E; mean = 18%; range = 12%–23% Ki67+ cells per field; P< .001).
These data confirm a proliferative advantage in the GBM NG2+ cells over the GBM NG2− cells isolated from the same GBM cell lines.
The Proliferative Advantage of the GBM NG2+ Cells Is Cell Autonomous
Next we asked whether the high proliferative capacity of the GBM NG2+ cells was dependent on, or could be manipulated by, external cues.
We isolated the GBM NG2+ and GBM NG2− fractions from 3 GBM cell lines. Then we cultured both fractions under 4 different types of culture media as follows: basic media (NBA + B27 + N2), SF media, conditioned media from the identical cell line, and differentiation media (10% FCS; full details of the components of different culture conditions can be found in the Materials and Methods section). All cell populations cultured under these conditions were evaluated for cell growth after 3 days in culture using MTS assay.
The results showed that the GBM NG2+ cells were more proliferative compared with the GBM NG2− cells under basic conditions (Fig. 2A), SF conditions (Fig. 2B), conditioned media (Fig. 2C), and differentiation conditions (Fig. 2D). The difference was statistically significant in all experiments in Fig. 2A–D (n = 3; P< .001).
Fig. 2.
The proliferative advantage of the glioblastoma multiforme (GBM) neuroglia (NG)-2+ cells is cell autonomous. MTS assay of NG2+ and NG2− fractions from 3 cell lines cultured under (A) basic culture conditions, (B) serum-free culture conditions, (C) Using conditioned media, and (D) Under differentiation (fetal calf serum) culture conditions. Higher absorbance values are observed in the GBM NG2+ cultures under all different culture conditions. Limiting dilution assay analysis of (E) GBM NG2+ and (F) GBM NG2− cells shows that the GBM NG2+ cells contain a higher number of clonogenic cells as indicated by u values. On the y axis r is the number of nonresponding wells and n is the total number of wells used. The dotted lines in E and F represent the 95% confidence intervals. All differences in A, B, C and D are statistically significant (Kruskal-Wallis ANOVA on ranks; P< .001).
These observations indicated that external cues were not the main determinant of the robust proliferative activity of the GBM NG2+ cells, suggesting that the proliferative advantage of the GBM NG2+ population is cell autonomous.
To test this point further, we carried out a clonal evaluation using limiting dilution assay, as described in Materials and Methods. The GBM NG2+ and GBM NG2− cells were seeded into ECM-coated 96-well plates in decreasing densities.
Data analysis confirmed that the GBM NG2+ cultures contained 3–6 times more single cells with colony-forming ability compared with the GBM NG2− cultures (Fig. 2E and F and Supplementary Material, Table S2).
The Proliferative Advantage of the GBM NG2+ Cells Can Be Observed in Clinical Samples
So far, we have provided in vitro evidence that the GBM NG2+ cells were more proliferative compared with the corresponding GBM NG2− cells isolated from the same GBM samples. We confirmed that the proliferative advantage of the GBM NG2+ cells could be observed under different culture conditions. Moreover, we suggested that the proliferative capacity of the GBM NG2+ cells is cell autonomous and showed a higher level of clonogenicity.
However, it is possible that these data represent an artifact of in vitro culture. To validate our observations in vivo we took 10 GBM tumors from 10 patients. Tumors were acutely dissociated into single cell suspensions before being fixed. We then stained these suspensions with antibodies against NG2 and Ki67 and quantified and analyzed the coexpression of these 2 markers using flow cytometry.
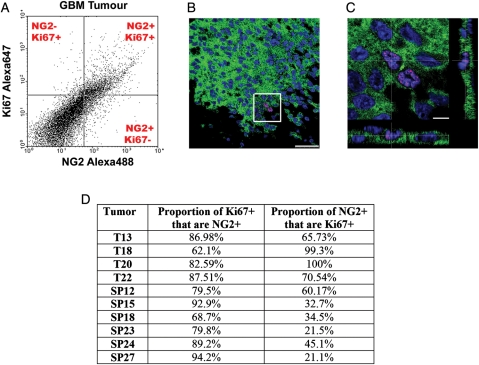
Similar to our in vitro findings, a large proportion of the GBM NG2+ cells in GBM tumors were Ki67+ (n = 10; mean = 55%; range = 22%–100%; Fig. 3A–D). Similarly, most GBM Ki67+ cells in GBM tumors were NG2+ (n = 10; mean = 83%; range = 62%–94%; Fig. 3A–D).
Fig. 3.
The proliferative pdvantage of the glioblastoma multiforme (GBM) neuroglia (NG)-2+ cells can be observed in clinical samples. (A) Flow cytometry analysis of the coexpression of Ki67 and NG2 in freshly dissociated and fixed GBM tumor samples. (B) Confocal micrograph of a GBM tumor showing cells coexpressing NG2 (green) and Ki67 (red). (C) Higher magnification of the highlighted area in panel B showing the coexpression of NG2 (green) and Ki67 (red) with Z-stack. (D) Table of quantitative data from 10 GBM tumor samples showing the proportions of coexpression of NG2 and the proliferative marker Ki67. Scale bars: 50 µm in B and 10 µm in C.
This indicates that the surface proteoglycan NG2 can be used to identify the proliferating compartment in GBM in real time on fresh clinical material.
GBM NG2+ Cells Exhibit Higher Tumorigenic Capability Compared with GBM NG2− Cells
Our data showed a significant difference in the proliferative and clonogenic capacities between the GBM NG2+ and GBM NG2− populations isolated from the same GBM samples.
In the normal adult brain, NG2+ progenitors play an important role in maintaining the brain tissue under normal and pathologic conditions. We therefore asked whether the GBM NG2+ cells would have a similar role in tumor maintenance. In addition, the involvement of the GBM NG2+ cells in GBM cell proliferation, which is one of the most fundamental hallmarks of cancer, identifies these cells as strong candidates to be involved in cancer growth and maintenance. This led us to evaluate and compare the in vivo tumorigenicity of the GBM NG2+ and GBM NG2− populations isolated from the same GBM sample.
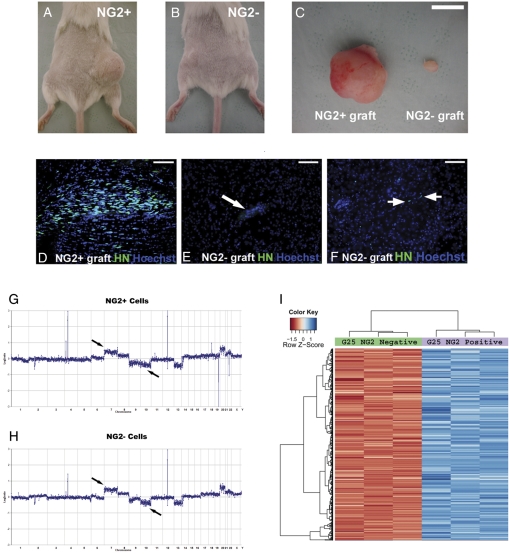
For initial screening purposes, we separated the GBM NG2+ and GBM NG2− populations using FACS from 3 GBM cell lines and immediately implanted the cells subcutaneously in 12 SCID mice. In all animals, we observed that the GBM NG2+ cells gave rise to subcutaneous tumor masses (Fig. 4A and C) that were clearly larger than tumor masses from the GBM NG2− cells at 3 months after implantation (Fig. 4B and C). These masses were formed by grafted human cells as shown by staining with anti–human nuclei (HN) antibodies (Supplementary Material, Fig. S1).
Fig. 4.
Glioblastoma multiforme (GBM) neuroglia (NG)-2+ cells exhibit higher tumorigenic capability compared with the GBM NG2− cells. (A) Subcutaneous mass developed in the right hindlimb after the implantation of GBM NG2+ cells. (B) Small subcutaneous mass developed in the left hindlimb after the implantation of the GBM NG2− cells. (C) Subcutaneous tumors excised from A and B. (D) Intracerebral tumor masses can be observed in mice after orthotopic implantation of the GBM NG2+ cells as shown by human nuclei staining. (E) Orthotopic implantations of the GBM NG2− cells do not form tumors. (F) Occasionally isolated HN+ cells can be observed in brains of animals implanted with the GBM NG2− cells. (G) Comparative genomic hybridization (CGH) of the GBM NG2+ cells. (H) CGH of the GBM NG2− cells. In G and H, black arrows indicate gain of chromosome 7 and loss of chromosome 10 (black arrows), which are typical cytogenetic abnormalities in GBM. Green arrows show abnormalities that are more pronounced in the GBM NG2+ cells, whereas red arrows indicate abnormalities that only exist in the GBM NG2+ cells. (I) Example of a heat map of genes differentially expressed by the GBM-NG2+ and GBM-NG2− cells. Scale bars: 100 µm in D, E, and F; 10 mm in C.
Next, we isolated the GBM NG2+ and GBM NG2− cells from 5 GBM cell lines and implanted them immediately orthotopically into the forebrains of a total of 28 SCID mice (Supplementary Material, Table S3). We found tumors generated by the implanted human GBM NG2+ cells in 12 of 14 animals. These were distinguished from host tissue by anti-HN antibodies (Fig. 4D). In contrast, only 2 of 14 tumors were found in mice grafted with human GBM NG2− cells (Supplementary Material, Table S3), with only narrow scar tissue seen at the site of implantation (Fig. 4E). In most cases of GBM NG2− implants, no human cells could be detected; however, occasionally a few scattered HN+ cells could be observed (Fig. 4F).
One possible reason for the low tumorigenic competency of the GBM NG2− cells is that they were normal cells contaminating our cell lines. Therefore, to exclude this possibility and confirm that both GBM NG2+ and GBM NG2− cells were tumor cells and not contaminating normal cells, we performed array comparative genomic hybridization (aCGH) on both populations. The data showed that both GBM NG2+ and GBM NG2− cells from 2 GBM cell lines tested had a molecular cytogenetic profile that was typical of GBM, including chromosome 7 gain and chromosome 10 deletion (Fig. 4G and H).
Interestingly, aCGH analysis showed subtle differences at the molecular cytogenetic profiles between the GBM NG2+ and GBM NG2− cells. The differences from one GBM sample are highlighted in Figure 4G and H, and a summary of the differences from 2 GBM samples are shown in Supplementary Material, Table S4. It is not possible based on our data to comment further on the significance of these cytogenetic variations owing to the natural heterogeneity of GBM.
Microarray Analysis Reveals a Distinct Molecular Signature of GBM NG2+ Cells Compared with GBM NG2− Cells from the Same Patient
To understand the molecular basis of our findings, expression microarray was performed on the GBM NG2+ and GBM NG2− populations derived from 2 GBM cell lines G25 and G30 (3 biological repeats for each GBM NG2+ and GBM NG2− population in each cell line; Fig. 4I).
Expression array data were analyzed according to the following criteria: detection rate >6.5, coefficient difference <−.5 or >.5, and adjusted FDR P< .05.
These data showed that 521 genes in G25 and 299 genes in G30 were overexpressed by the GBM NG2+ population compared with the GBM NG2− population (see Supplementary Microarray Data). When the 2 cell lines were matched together, the list of genes that were overexpressed in the GBM NG2+ cells compared with the GBM NG2− cells was shortened to include 134 genes. This in part reflects the high level of heterogeneity in GBM and highlights the possible wide range of potential mechanisms that can control the neoplastic process in different patients. Also, it led us to focus on comparing the GBM NG2+ and GBM NG2− populations in individuals rather than across several patients.
Interestingly, the genes overexpressed in the GBM NG2+ cells compared with the GBM NG2− cells include many members of the Mitosis and Cell Cycle Module (MCCM), which has previously been described.36–39 These include the Cell Division Cycle (CDC) family (CDC2, A4, A7, 25A, 28, 45), the Micro-Chromosome Maintenance (MCM) family (MCM2, 3, 4, 5, 6, 7, 8 and 10), and the Cyclin family (Cyclin D1, Cyclin E2, Cyclin F). In addition, several transcription factors were overexpressed in the GBM NG2+ cells, including Maternal Embryonic Leucine Zipper kinase (MELK), transcription factor AP-2 gamma (TFAP2C), and the E2F family (E2F7 and E2F2).
The genes overexpressed in the GBM NG2+ cells were further analyzed using GeneTrail Enrichment Tools providing a description of Kyoto Encyclopedia of Genes and Genomes (KEGG) pathways, Transfac, Transpath, and GeneOntology (see Supplementary Microarray Data).
Briefly, KEGG pathway studies showed that the GBM NG2+ population expressed genes responsible for DNA replication, cell cycle, and purine and pyrimidine metabolism pathways. Also, Transfac and Transpath studies demonstrated that the E2F transcription factor family was among the overexpressed genes.
Next, we studied the GeneOntology profiling, which gives an overview of the functional subcategories of the given sets of genes. We found that overexpressed genes in the GBM NG2+ population could be grouped into more than 200 subcategories. The most statistically significant among them were related to M phase, cell cycle, cell division (MCCM genes), and DNA modeling and repair (e.g., RAD51AP, POLE, TOP2A, CHEK1, TYMS; see Supplementary Microarray Data).
This distinct genetic signature is consistent with the observed characteristics and proliferative advantage of the GBM NG2+ cells and further supports the implication of the GBM NG2+ cells in cell proliferation in GBM.
Genes Associated With NG2+ Progenitors Are Widely Expressed in Astrocytic Tumors
In the normal brain, NG2+ progenitors coexpress NG2, Olig2, and PDGFRα. However, no quantitative data are available regarding the expression of all these markers in brain tumors. We therefore screened the prevalence of the expression of the mRNA of these genes in brain tumors.
We examined the expression of these markers in different World Health Organization (WHO) grades of astrocytic tumors by consulting data from an expression array dataset of 63 tumors (5 astrocytomas, WHO grade II; 19 anaplastic astrocytomas, WHO grade III; and 39 GBMs, WHO grade IV) previously published in the Gene Omnibus Database as dataset GSE1993 (GEO; www.ncbi.nlm.nih.gov/GEO40).
Data analysis indicated that NG2, Olig2, and PDGFRα genes were widely expressed in all grades of astrocytic tumors (Supplementary Material, Figure S3A–SC; Supplementary Material, Table S5). When we examined the GBM cohort in more detail, we noted that most tumors expressed Olig2 (87%), PDGFRα (72%), and NG2 (77%; Supplementary Material, Fig. S4A–S4C, Table S5). Two GBM tumors (GBM #2 and #14) lacked the expression of 2 markers, and only 1 GBM tumor (GBM #31) lacked the expression of all 3 markers (Supplementary Material, Fig. S3A–C).
These data were further supported by consulting random independent expression microarray datasets of 101 GBM samples published in Gene Expression Omnibus (Series Record: GSE8692, GSE3185, GSE9171, and GSE4290). All these independent datasets showed similar widespread expression of genes associated with NG2+ progenitors in astrocytomas, including GBM (Supplementary Material, Table S6).
The NG2 Proteoglycan Is Expressed by GBM Tumors
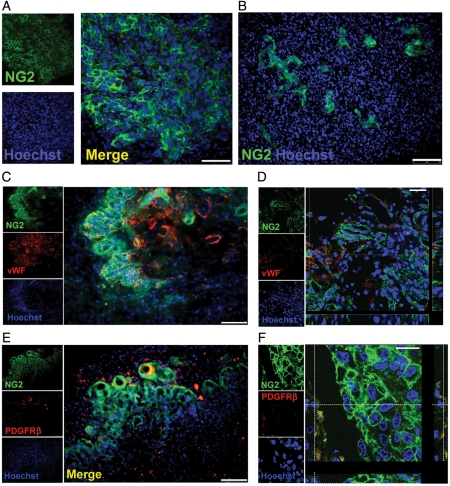
To confirm translation of the NG2 gene, we analyzed its expression in GBM tumors using immunohistochemistry. Our results confirmed expression of the proteoglycan NG2 in all tested GBM tumors (n = 17; selected example is shown in Fig. 5). The GBM NG2+ cells in the tumors were round-shaped (Fig. 5A) and lacked the characteristic processes described in normal process-bearing NG2+ progenitors in the normal adult brain.19,41
Fig. 5.
The neuroglia (NG)-2 proteoglycan is expressed by glioblastoma multiforme (GBM) tumors. (A) Round-shaped cells expressing NG2 are observed in GBM. (B) The GBM-NG2+ cells have a close relationship to the GBM blood vessels. (C) The majority of GBM NG2+ cells do not express the endothelial marker von Willebrand factor (vWF). (D) Confocal microscopy confirms that the majority of GBM NG2+ cells (green) are negative for vWF (red). (E) Few GBM NG2+ cells express the pericyte marker PDGFRβ. (F) Confocal micrograph showing that some cells coexpress NG2 (green) and PDGFRβ (red) but most GBM NG2+ cells are NG2+/PDGFRβ−. Scale bars: 50 µm in A and D; 100 µm in B, C, and E; and 25 µm in F.
Quantitative analysis showed that the proportion of GBM NG2+ cells varied among different samples (n = 10; mean = 30%; range = 6%–77% of total GBM cells).
Interestingly, we observed that the distribution of the GBM NG2+ cells in GBM tumors was not homogeneous and formed uneven, colony-like foci that were frequently located around and near blood vessels (Fig. 5A and B).
Since NG2 is also expressed by vascular cells in the nervous system,42 we wanted to determine whether the observed GBM NG2+ cells were of neural or vascular nature. We therefore tested the coexpression of NG2 and von Willebrand factor (vWF), a marker of endothelial cells,43,44 and PDGFRβ, a marker of pericytes in normal and pathologic vessels.42,45–47
Five different GBM tumors were used. We found negligible coexpression between NG2 and vWF (Fig. 5C and D), as very few vWF+ cells expressed NG2 at the luminal surface.
Similarly, few GBM NG2+ cells were positive for PDGFRβ (mean = 7%; range = 0:5%–29%; Fig. 5E and F), and only a small population of pericytic GBM NG2+/PDGFRβ+ cells could be observed within the total GBM cell population (mean = 2%; range = 0:25–8% of total GBM cells; Fig. 5F; Supplementary Material, Table S7).
Next, we screened immunohistochemically the GBM tumors for the expression of other markers associated with NG2+ progenitors. Similar to NG2, our data showed that Olig2 and PDGFRα were expressed in all tested GBM tumors (Supplementary Material, Fig. S5; negative controls are shown in Supplementary Material, Fig. S6).
The Phenotypic Identity of the GBM NG2+ Cells and Normal NG2+ Progenitors is Distinct
Normal NG2+ progenitors can be identified by the coexpression of 3 markers (NG2, Olig2, PDGFRα). Exploring the coexpression pattern of these 3 markers gives us the opportunity to interrogate the phenotypic identity of the GBM NG2+ cells and compare it with that of NG2+ progenitors.
We observed a small GBM NG2+/Olig2+ cell population that coexpressed NG2 and Olig2 (mean = 1%; range = 0:2%–5% of total GBM cells; Fig. 6A). Notably, most GBM Olig2+ cells expressed NG2 (mean = 81%; range = 18%–100%; Supplementary Material, Table S7), but only a small proportion of GBM NG2+ cells were Olig2+ (mean = 5%; range = 0.3%–15%; Supplementary Material, Table S7). Also observed were GBM NG2−/Olig2+ cells (mean = 1%; range = 0%–6% of total GBM cells; Fig. 6B) and GBM NG2+/Olig2− cells (mean = 47%; range = 16%–70% of total GBM cells; Fig. 6C).
Fig. 6.
The phenotypic identity of the glioblastoma multiforme (GBM) neuroglia (NG)-2+ cells is distinct from nonmalignant NG2+ progenitors. Patterns of coexpression are variable: (A) 1% of GBM cells coexpress NG2+ cells (green) and Olig2 (red), (B) 1% are NG2−/Olig2+, and (C) 47% of GBM cells are NG2+/Olig2−. Similarly, coexpression of NG2 (green) and platelet derived growth factor receptor alpha (PDGFRα; red) can be observed in GBM tumors (D). Also, GBM PDGFRα+ (red) cells can be negative for NG2 (green; E) and some GBM NG2+ cells (green) do not express PDGFRα (red; F). Scale bars: 25 µm in A, B, C, D, and E; 50 µm in F.
With respect to the coexpression of NG2 and PDGFRα (Fig. 6D), a large proportion of GBM NG2+ cells were consistent with PDGFRα+ (mean = 86%; range = 51%–100%; Supplementary Material, Table S7), whereas on average half the GBM PDGFRα+ cells were NG2+ (mean = 55%; range = 27%–75%; Supplementary Material, Table S7). Also found were GBM NG2−/PDGFRα+ cells (mean = 26%; range = 2%–54% of total GBM cells; Fig. 6E) and a small proportion of GBM NG2+/PDGFRα− cells (mean = 2%; range = 0%–5% of total GBM cells; Fig. 6F).
These data show that the coexpression patterns of NG2, Olig2, and PDGFRα in GBM samples were distinct from the pattern reported in NG2+ progenitors in the normal brain. Specifically, the marker expression was more variable and did not appear to adhere to any specific developmental pattern.
Discussion
GBM NG2+ Cells Exhibit Robust Proliferation Compared With GBM NG2− Cells from the Same Tumor
The expression of NG2 in GBM has been described previously33,48,49 and implicated in invasion, angiogenesis, and resistance to chemotherapy.50–52 However, a detailed description of the proliferative ability and tumorigenic potential of the GBM NG2+ population has not been described.
Our data reveal that GBM NG2+ cells show proliferative dominance compared with GBM NG2− cells from the same patient. This advantage is not related to external cues and appears to be a cell-autonomous process. Thus the GBM NG2+ cells exhibit robust proliferation in the absence of mitogens or under differentiation conditions. This cell population also exhibits a higher level of clonogenicity compared with the GBM NG2− population from the same patient. These in vitro data are validated in vivo using clinical samples confirming a similar proliferative advantage in the GBM NG2+ cells from GBM tumors. Our quantitative results indicate that NG2 expression delineates approximately 80% of actively cycling cells in multiple GBM tumors. This oncogenic advantage is maintained across all patients studied but with significant variability between individual subjects. These data raise the possibility that the growth of GBM could be significantly slowed by appropriate therapeutic targeting of the NG2 population in a subset of patients with GBM.
To further support our observations, we compared the genetic profile of the GBM NG2+ and GBM NG2− cells from the same tumor. Data from expression microarray reveal a distinct molecular signature of the GBM NG2+ cells characterized by the expression of genes associated with cell proliferation and cycling. It is well recognized that GBM tumors express MCCM genes that are involved in mitosis and cell cycling and are linked to higher-grade gliomas and clinically to poor survival.36–38,53 Our data indicate that these MCCM genes are overexpressed and enriched in the GBM NG2+ cells. The GBM NG2+ cells also overexpress a group of genes that can play significant roles in different aspects of the neoplastic process. These include transcription factors involved in proliferation, such as AP-2 gamma, MELK, E2F,54–57 and components of DNA replication, DNA repair, and nucleotide metabolism such as DNA polymerase y, thymidine kinase, and RAD51.58–60 These data show that enriching for GBM NG2+ cells identifies a population able to maintain the robust proliferation and growth of clinically symptomatic GBM.
The Role of NG2 in Cancer Remains to be Clarified
Recent data raise the possibility of a direct role for NG2 in cell proliferation in vitro61 and in an NG2 knockout model.62 The NG2 cytoplasmic domain can be phosphorylated by PKC (protein kinase C) and ERK (extracellular signal–regulated protein kinase), and, interestingly, the site of phosphorylated residues can determine the cell surface distribution of NG2 and its involvement in cell proliferation or invasion.63 In GBM NG2 may mediate cell proliferation indirectly by maintaining a progenitor state possibly by facilitating growth factors/receptor interaction.64 NG2 has been shown to bind platelet-derived growth factor–AA and facilitates its presentation to PDGFRα.65 Consistent with this are studies showing that normal activation of PDGFRα requires direct interaction with NG2.64,66,67 Similar observations have been reported regarding the NG2 interaction with other growth-promoting factors such as integrins, basic fibroblast growth factor, fibroblast growth factor receptors 1 and 3, epidermal growth factor receptor, and mitogen-activated protein kinase.62,63,65,68–71 The expression of NG2 has also been described in a variety of other cancers, including breast,72 melanoma,73 chondrosarcomas,74 and hematopoetic malignancies.75–79 Its expression is often associated with an aggressive clinical phenotype and poor prognosis. In GBM the expression of NG2 is associated with GBM migration, angiogenesis, and proliferation, which are recognized hallmarks of cancer.80 It remains to be determined whether the proteoglycan has a direct mechanistic role in these processes or serves an accessory function. Expression may also be associated with resistance to chemotherapy50 and radiotherapy (Dr. Checkenya; personal communication).
Coexpression of Markers Associated with the Oligodendroglial Lineage are Widespread in Astrocytoma
To date there has been no characterization of the pattern of coexpression of the glial progenitor markers NG2, Olig2, and PDGFRα in GBM, and it is not known whether the coexpression pattern of these markers reported in the normal CNS is recapitulated in GBM. NG2 expression is widespread in murine glioma81–83 but is more variable in human samples, being either widespread33,48 or absent84. Data on Olig2 expression in human GBM have also given conflicting results.85–95 Less controversial is the expression of PDGFRα in GBM, which has been shown to be widely expressed in gliomas of all grades, with frequent amplifications in GBM.96–99 Most of these reports addressed the expression of individual markers rather than patterns of coexpression. A small number of studies analyzed the expression of the NG2+ progenitor markers Olig2 and NG2 in an attempt to develop a diagnostic tool for oligodendrogliomas.84,100 Others used the screening studies of NG2, Olig2, or A2B5 expression to infer the histiogenesis of GBM and other gliomas.32,100
In our series, approximately 80% of GBM tumors expressed the 3 markers NG2, Olig2, and PDGFRα. This enabled us to characterize the coexpression of these markers and compare it with patterns expressed by glial progenitors in the normal brain.9,22–30,101 Our quantitative analysis confirms that the pattern of coexpression is distinct from the highly stereotyped pattern seen in the normal brain. Marker expression was more variable and did not appear to adhere to any specific developmental pattern. As a result, GBM NG2+ cells did not recapitulate the phenotypic characteristics seen in normal NG2+ progenitors in the normal brain.
We are not able to comment on any relationship between CD133 and NG2, since we are not proposing that NG2 is a cancer stem cell marker. In our samples, CD133+ and CD133–GBM cells proliferate equally (unpublished data) and expression is variable. For these reasons, together with a growing body of evidence suggesting that the importance of CD133 in GBM should be treated with caution,102 we have not pursued this further.
The expression of a specific developmental marker in cancer does not confirm its origin from normal cells that expressed the same marker.103 Hence the expression by GBM cells of markers associated with normal NG2+ progenitors does not constitute evidence that NG2+ progenitors are the cells of origin of GBM. It is plausible that these markers were expressed de novo in a stochastic manner as a result of environmental selective pressure.
Most NG2 Cells in Glioblastoma Are Not Pericytes
NG2 is expressed by pericytes and has also been implicated in normal blood vessel formation and pathologic angiogenesis in GBM.52,104,105 Detailed studies have shown a sparse distribution of pericytes in GBM vasculature compared with normal blood vessels,48,106,107 reflecting the dysregulation of normal pericyte-endothelial biology in GBM.
In our samples the expression of the pericytic marker PDGFRβ was sparsely distributed, and only a small number of PDGFRβ+ pericytes could be observed in the tumors. Nearly all PDGFRβ+ pericytes express NG2, but only a small number of GBM NG2+ cells express PDGFRβ. Similarly the coexpression of NG2 with the endothelial marker vWF was observed in only a small number of cells.
We show that cells expressing NG2 in GBM (GBM NG2+ cells) comprise a sub-population with a distinct proliferative advantage in clinically symptomatic patients. We also confirm that the GBM NG2+ cells can be interrogated directly from tumour samples avoiding a phase of cell culture. Targeting this proliferating compartment of GBM could attenuate disease progression and may even improve survival in a subset of patients.
Supplementary Material
Funding
This study was supported by The Evelyn Trust, Addenbrooke's Charitable Trust, the Royal College of Surgeons of Edinburgh, and the NIHR Cambridge Biomedical Research Centre.
Supplementary Material
Acknowledgments
We would like to thank Miss Roslin Russel, Dr Lawrence Petalidis, and Dr David Jones for help in microarray data analysis, Professor W Stallcup for the kind gift of PDGFRα/β antibodies, Mr Simon McCallum for technical help, and Dr Aviva M Tolkovsky and Dr Jessica Kwok for helpful comments on the manuscript.
The MRC UK, the National Institute for Health Research (NIHR), Addenbrookes Charitable Trust, the Royal College of Surgeons of Edinburgh (RCSEd), and the University of Aleppo Scholarships, Syria has supported this work.
Conflict of interest statement. None declared.
References
- 1.Stupp R, Mason WP, van den Bent MJ, et al. Radiotherapy plus concomitant and adjuvant temozolomide for glioblastoma. N Engl J Med. 2005;352(10):987–996. doi: 10.1056/NEJMoa043330. doi:10.1056/NEJMoa043330. [DOI] [PubMed] [Google Scholar]
- 2.Stupp R, van den Bent MJ, Hegi ME. Optimal role of temozolomide in the treatment of malignant gliomas. Curr Neurol Neurosci Rep. 2005;5(3):198–206. doi: 10.1007/s11910-005-0047-7. doi:10.1007/s11910-005-0047-7. [DOI] [PubMed] [Google Scholar]
- 3.Walker MD. The contemporary role of chemotherapy in the treatment of malignant brain tumor. Clin Neurosurg. 1978;25:388–396. doi: 10.1093/neurosurgery/25.cn_suppl_1.388. [DOI] [PubMed] [Google Scholar]
- 4.Walker MD, Alexander E, Jr., Hunt WE, et al. Evaluation of BCNU and/or radiotherapy in the treatment of anaplastic gliomas. A cooperative clinical trial. J Neurosurg. 1978;49(3):333–343. doi: 10.3171/jns.1978.49.3.0333. [DOI] [PubMed] [Google Scholar]
- 5.Hegi ME, Diserens AC, Gorlia T, et al. MGMT gene silencing and benefit from temozolomide in glioblastoma. N Engl J Med. 2005;352(10):997–1003. doi: 10.1056/NEJMoa043331. doi:10.1056/NEJMoa043331. [DOI] [PubMed] [Google Scholar]
- 6.Stallcup WB, Huang FJ. A role for the NG2 proteoglycan in glioma progression. Cell Adh Migr. 2008;2(3):192–201. doi: 10.4161/cam.2.3.6279. doi:10.4161/cam.2.3.6279. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Dawson MR, Polito A, Levine JM, Reynolds R. NG2-expressing glial progenitor cells: an abundant and widespread population of cycling cells in the adult rat CNS. Mol Cell Neurosci. 2003;24(2):476–488. doi: 10.1016/s1044-7431(03)00210-0. doi:10.1016/S1044-7431(03)00210-0. [DOI] [PubMed] [Google Scholar]
- 8.Dimou L, Simon C, Kirchhoff F, Takebayashi H, Gotz M. Progeny of Olig2-expressing progenitors in the gray and white matter of the adult mouse cerebral cortex. J Neurosci. 2008;28(41):10434–10442. doi: 10.1523/JNEUROSCI.2831-08.2008. doi:10.1523/JNEUROSCI.2831-08.2008. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Geha S, Pallud J, Junier MP, et al. NG2+/Olig2+ cells are the major cycle-related cell population of the adult human normal brain. Brain Pathol. 2010;20(2):399–411. doi: 10.1111/j.1750-3639.2009.00295.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Gensert JM, Goldman JE. Heterogeneity of cycling glial progenitors in the adult mammalian cortex and white matter. J Neurobiol. 2001;48(2):75–86. [PubMed] [Google Scholar]
- 11.Goldman SA, Sim F. Stem Cells: Nuclear Reprogramming and Therapeutic Applications. Vol 265. New York, NY: John Wiley & Sons, Ltd; 2005. Neural progenitor cells of the adult brain. [Google Scholar]
- 12.Rivers LE, Young KM, Rizzi M, et al. PDGFRA/NG2 glia generate myelinating oligodendrocytes and piriform projection neurons in adult mice. Nat Neurosci. 2008;11(12):1392–1401. doi: 10.1038/nn.2220. doi:10.1038/nn.2220. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Tang DG, Tokumoto YM, Apperly JA, Lloyd AC, Raff MC. Lack of replicative senescence in cultured rat oligodendrocyte precursor cells. Science. 2001;291(5505):868–871. doi: 10.1126/science.1056780. doi:10.1126/science.1056780. [DOI] [PubMed] [Google Scholar]
- 14.Zhu X, Bergles DE, Nishiyama A. NG2 cells generate both oligodendrocytes and gray matter astrocytes. Development. 2008;135(1):145–157. doi: 10.1242/dev.004895. [DOI] [PubMed] [Google Scholar]
- 15.Buffo A, Vosko MR, Erturk D, et al. Expression pattern of the transcription factor Olig2 in response to brain injuries: implications for neuronal repair. Proc Natl Acad Sci U S A. 2005;102(50):18183–18188. doi: 10.1073/pnas.0506535102. doi:10.1073/pnas.0506535102. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Fancy SP, Zhao C, Franklin RJ. Increased expression of Nkx2.2 and Olig2 identifies reactive oligodendrocyte progenitor cells responding to demyelination in the adult CNS. Mol Cell Neurosci. 2004;27(3):247–254. doi: 10.1016/j.mcn.2004.06.015. [DOI] [PubMed] [Google Scholar]
- 17.Zawadzka M, Rivers LE, Fancy SP, et al. CNS-resident glial progenitor/stem cells produce Schwann cells as well as oligodendrocytes during repair of CNS demyelination. Cell Stem Cell. 2010;6(6):578–590. doi: 10.1016/j.stem.2010.04.002. doi:10.1016/j.stem.2010.04.002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Zhao JW, Raha-Chowdhury R, Fawcett JW, Watts C. Astrocytes and oligodendrocytes can be generated from NG2+ progenitors after acute brain injury: intracellular localization of oligodendrocyte transcription factor 2 is associated with their fate choice. Eur J Neurosci. 2009;29(9):1853–1869. doi: 10.1111/j.1460-9568.2009.06736.x. doi:10.1111/j.1460-9568.2009.06736.x. [DOI] [PubMed] [Google Scholar]
- 19.Berry M, Hubbard P, Butt AM. Cytology and lineage of NG2-positive glia. J Neurocytol. 2002;31(6–7):457–467. doi: 10.1023/a:1025735513560. doi:10.1023/A:1025735513560. [DOI] [PubMed] [Google Scholar]
- 20.Chandran S, Hunt D, Joannides A, Zhao C, Compston A, Franklin RJ. Myelin repair: the role of stem and precursor cells in multiple sclerosis. Philos Trans R Soc Lond B Biol Sci. 2008;363(1489):171–183. doi: 10.1098/rstb.2006.2019. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Nishiyama A, Komitova M, Suzuki R, Zhu X. Polydendrocytes (NG2 cells): multifunctional cells with lineage plasticity. Nat Rev Neurosci. 2009;10(1):9–22. doi: 10.1038/nrn2495. doi:10.1038/nrn2495. [DOI] [PubMed] [Google Scholar]
- 22.Levine JM, Stallcup WB. Plasticity of developing cerebellar cells in vitro studied with antibodies against the NG2 antigen. J Neurosci. 1987;7(9):2721–2731. doi: 10.1523/JNEUROSCI.07-09-02721.1987. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Marconi S, De Toni L, Lovato L, et al. Expression of gangliosides on glial and neuronal cells in normal and pathological adult human brain. J Neuroimmunol. 2005;170(1–2):115–121. doi: 10.1016/j.jneuroim.2005.03.025. doi:10.1016/j.jneuroim.2005.03.025. [DOI] [PubMed] [Google Scholar]
- 24.Pringle NP, Mudhar HS, Collarini EJ, Richardson WD. PDGF receptors in the rat CNS: during late neurogenesis, PDGF alpha-receptor expression appears to be restricted to glial cells of the oligodendrocyte lineage. Development. 1992;115(2):535–551. doi: 10.1242/dev.115.2.535. [DOI] [PubMed] [Google Scholar]
- 25.Raff MC, Miller RH, Noble M. A glial progenitor cell that develops in vitro into an astrocyte or an oligodendrocyte depending on culture medium. Nature. 1983;303(5916):390–396. doi: 10.1038/303390a0. doi:10.1038/303390a0. [DOI] [PubMed] [Google Scholar]
- 26.Raff MC, Miller RH, Noble M. Glial cell lineages in the rat optic nerve. Cold Spring Harb Symp Quant Biol. 1983;48(Pt 2):569–572. doi: 10.1101/sqb.1983.048.01.061. [DOI] [PubMed] [Google Scholar]
- 27.Chang A, Nishiyama A, Peterson J, Prineas J, Trapp BD. NG2-positive oligodendrocyte progenitor cells in adult human brain and multiple sclerosis lesions. J Neurosci. 2000;20(17):6404–6412. doi: 10.1523/JNEUROSCI.20-17-06404.2000. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Gogate N, Verma L, Zhou JM, et al. Plasticity in the adult human oligodendrocyte lineage. J Neurosci. 1994;14(8):4571–4587. doi: 10.1523/JNEUROSCI.14-08-04571.1994. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Kennedy PG, Fok-Seang J. Studies on the development, antigenic phenotype and function of human glial cells in tissue culture. Brain. 1986;109(Pt 6):1261–1277. doi: 10.1093/brain/109.6.1261. doi:10.1093/brain/109.6.1261. [DOI] [PubMed] [Google Scholar]
- 30.Nunes MC, Roy NS, Keyoung HM, et al. Identification and isolation of multipotential neural progenitor cells from the subcortical white matter of the adult human brain. Nat Med. 2003;9(4):439–447. doi: 10.1038/nm837. doi:10.1038/nm837. [DOI] [PubMed] [Google Scholar]
- 31.Mori T, Wakabayashi T, Takamori Y, Kitaya K, Yamada H. Phenotype analysis and quantification of proliferating cells in the cortical gray matter of the adult rat. Acta Histochem Cytochem. 2009;42(1):1–8. doi: 10.1267/ahc.08037. doi:10.1267/ahc.08037. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Colin C, Baeza N, Tong S, et al. In vitro identification and functional characterization of glial precursor cells in human gliomas. Neuropathol Appl Neurobiol. 2006;32(2):189–202. doi: 10.1111/j.1365-2990.2006.00740.x. doi:10.1111/j.1365-2990.2006.00740.x. [DOI] [PubMed] [Google Scholar]
- 33.Fael Al-Mayhani TM, Ball SL, Zhao JW, et al. An efficient method for derivation and propagation of glioblastoma cell lines that conserves the molecular profile of their original tumours. Journal of neuroscience methods. 2009;176(2):192–199. doi: 10.1016/j.jneumeth.2008.07.022. doi:10.1016/j.jneumeth.2008.07.022. [DOI] [PubMed] [Google Scholar]
- 34.Fiegler H, Carr P, Douglas EJ, et al. DNA microarrays for comparative genomic hybridization based on DOP-PCR amplification of BAC and PAC clones. Genes, Chromosomes & Cancer. 2003;36(4):361–374. doi: 10.1002/gcc.10155. doi:10.1002/gcc.10155. [DOI] [PubMed] [Google Scholar]
- 35.Cory AH, Owen TC, Barltrop JA, Cory JG. Use of an aqueous soluble tetrazolium/formazan assay for cell growth assays in culture. Cancer Commun. 1991;3(7):207–212. doi: 10.3727/095535491820873191. [DOI] [PubMed] [Google Scholar]
- 36.Hodgson JG, Yeh RF, Ray A, et al. Comparative analyses of gene copy number and mRNA expression in glioblastoma multiforme tumors and xenografts. Neuro Oncol. 2009;11(5):477–487. doi: 10.1215/15228517-2008-113. doi:10.1215/15228517-2008-113. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Horvath S, Zhang B, Carlson M, et al. Analysis of oncogenic signaling networks in glioblastoma identifies ASPM as a molecular target. Proc Natl Acad Sci U S A. 2006;103(46):17402–17407. doi: 10.1073/pnas.0608396103. doi:10.1073/pnas.0608396103. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Phillips HS, Kharbanda S, Chen R, et al. Molecular subclasses of high-grade glioma predict prognosis, delineate a pattern of disease progression, and resemble stages in neurogenesis. Cancer Cell. 2006;9(3):157–173. doi: 10.1016/j.ccr.2006.02.019. doi:10.1016/j.ccr.2006.02.019. [DOI] [PubMed] [Google Scholar]
- 39.Whitfield ML, Sherlock G, Saldanha AJ, et al. Identification of genes periodically expressed in the human cell cycle and their expression in tumors. Mol Biol Cell. 2002;13(6):1977–2000. doi: 10.1091/mbc.02-02-0030.. doi:10.1091/mbc.02-02-0030. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Petalidis LP, Oulas A, Backlund M, et al. Improved grading and survival prediction of human astrocytic brain tumors by artificial neural network analysis of gene expression microarray data. Mol Cancer Ther. 2008;7(5):1013–1024. doi: 10.1158/1535-7163.MCT-07-0177. doi:10.1158/1535-7163.MCT-07-0177. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Butt AM, Hamilton N, Hubbard P, Pugh M, Ibrahim M. Synantocytes: the fifth element. J Anat. 2005;207(6):695–706. doi: 10.1111/j.1469-7580.2005.00458.x. doi:10.1111/j.1469-7580.2005.00458.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Ozerdem U, Grako KA, Dahlin-Huppe K, Monosov E, Stallcup WB. NG2 proteoglycan is expressed exclusively by mural cells during vascular morphogenesis. Dev Dyn. 2001;222(2):218–227. doi: 10.1002/dvdy.1200. doi:10.1002/dvdy.1200. [DOI] [PubMed] [Google Scholar]
- 43.Jaffe EA, Nachman RL, Becker CG, Minick CR. Culture of human endothelial cells derived from umbilical veins. Identification by morphologic and immunologic criteria. The Journal of Clinical Investigation. 1973;52(11):2745–2756. doi: 10.1172/JCI107470. doi:10.1172/JCI107470. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Sporn LA, Marder VJ, Wagner DD. Inducible secretion of large, biologically potent von Willebrand factor multimers. Cell. 1986;46(2):185–190. doi: 10.1016/0092-8674(86)90735-x. doi:10.1016/0092-8674(86)90735-X. [DOI] [PubMed] [Google Scholar]
- 45.D'Amore PA, Smith SR. Growth factor effects on cells of the vascular wall: a survey. Growth Factors. 1993;8(1):61–75. doi: 10.3109/08977199309029135. doi:10.3109/08977199309029135. [DOI] [PubMed] [Google Scholar]
- 46.Lindahl P, Johansson BR, Leveen P, Betsholtz C. Pericyte loss and microaneurysm formation in PDGF-B-deficient mice. Science. 1997;277(5323):242–245. doi: 10.1126/science.277.5323.242. doi:10.1126/science.277.5323.242. [DOI] [PubMed] [Google Scholar]
- 47.Ozerdem U, Monosov E, Stallcup WB. NG2 proteoglycan expression by pericytes in pathological microvasculature. Microvasc Res. 2002;63(1):129–134. doi: 10.1006/mvre.2001.2376. doi:10.1006/mvre.2001.2376. [DOI] [PubMed] [Google Scholar]
- 48.Schrappe M, Klier FG, Spiro RC, Waltz TA, Reisfeld RA, Gladson CL. Correlation of chondroitin sulfate proteoglycan expression on proliferating brain capillary endothelial cells with the malignant phenotype of astroglial cells. Cancer Res. 1991;51(18):4986–4993. [PubMed] [Google Scholar]
- 49.He J, Liu Y, Xie X, et al. Identification of cell surface glycoprotein markers for glioblastoma-derived stem-like cells using a lectin microarray and LC-MS/MS approach. J Proteome Res. 2010;9(5):2565–2572. doi: 10.1021/pr100012p. doi:10.1021/pr100012p. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Chekenya M, Krakstad C, Svendsen A, et al. The progenitor cell marker NG2/MPG promotes chemoresistance by activation of integrin-dependent PI3K/Akt signaling. Oncogene. 2008;27(39):5182–5194. doi: 10.1038/onc.2008.157. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Wiranowska M, Ladd S, Smith SR, Gottschall PE. CD44 adhesion molecule and neuro-glial proteoglycan NG2 as invasive markers of glioma. Brain cell biology. 2006;35(2–3):159–172. doi: 10.1007/s11068-007-9009-0. [DOI] [PubMed] [Google Scholar]
- 52.Brekke C, Lundervold A, Enger PO, et al. NG2 expression regulates vascular morphology and function in human brain tumours. Neuroimage. 2006;29(3):965–976. doi: 10.1016/j.neuroimage.2005.08.026. doi:10.1016/j.neuroimage.2005.08.026. [DOI] [PubMed] [Google Scholar]
- 53.Facoetti A, Ranza E, Grecchi I, et al. Immunohistochemical evaluation of minichromosome maintenance protein 7 in astrocytoma grading. Anticancer Res. 2006;26(5A):3513–3516. [PubMed] [Google Scholar]
- 54.Alonso MM, Alemany R, Fueyo J, Gomez-Manzano C. E2F1 in gliomas: a paradigm of oncogene addiction. Cancer Lett. 2008;263(2):157–163. doi: 10.1016/j.canlet.2008.02.001. doi:10.1016/j.canlet.2008.02.001. [DOI] [PubMed] [Google Scholar]
- 55.Marie SK, Okamoto OK, Uno M, et al. Maternal embryonic leucine zipper kinase transcript abundance correlates with malignancy grade in human astrocytomas. Int J Cancer. 2008;122(4):807–815. doi: 10.1002/ijc.23189. doi:10.1002/ijc.23189. [DOI] [PubMed] [Google Scholar]
- 56.Nakano I, Masterman-Smith M, Saigusa K, et al. Maternal embryonic leucine zipper kinase is a key regulator of the proliferation of malignant brain tumors, including brain tumor stem cells. J Neurosci Res. 2008;86(1):48–60. doi: 10.1002/jnr.21471. doi:10.1002/jnr.21471. [DOI] [PubMed] [Google Scholar]
- 57.Cancer Genome Atlas Research Network. Comprehensive genomic characterization defines human glioblastoma genes and core pathways. Nature. 2008;455(7216):1061–1068. doi: 10.1038/nature07385. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58.Helleday T, Petermann E, Lundin C, Hodgson B, Sharma RA. DNA repair pathways as targets for cancer therapy. Nat Rev Cancer. 2008;8(3):193–204. doi: 10.1038/nrc2342. doi:10.1038/nrc2342. [DOI] [PubMed] [Google Scholar]
- 59.Kastan MB, Bartek J. Cell-cycle checkpoints and cancer. Nature. 2004;432(7015):316–323. doi: 10.1038/nature03097. doi:10.1038/nature03097. [DOI] [PubMed] [Google Scholar]
- 60.Short SC, Martindale C, Bourne S, Brand G, Woodcock M, Johnston P. DNA repair after irradiation in glioma cells and normal human astrocytes. Neuro Oncol. 2007;9(4):404–411. doi: 10.1215/15228517-2007-030. doi:10.1215/15228517-2007-030. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 61.Xiong J, Wang Y, Zhu Z, et al. NG2 proteoglycan increases mesangial cell proliferation and extracellular matrix production. Biochem Biophys Res Commun. 2007;361(4):960–967. doi: 10.1016/j.bbrc.2007.07.113. doi:10.1016/j.bbrc.2007.07.113. [DOI] [PubMed] [Google Scholar]
- 62.Kucharova K, Stallcup WB. The NG2 proteoglycan promotes oligodendrocyte progenitor proliferation and developmental myelination. Neuroscience. 2010;166(1):185–194. doi: 10.1016/j.neuroscience.2009.12.014. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 63.Makagiansar IT, Williams S, Mustelin T, Stallcup WB. Differential phosphorylation of NG2 proteoglycan by ERK and PKCalpha helps balance cell proliferation and migration. J Cell Biol. 2007;178(1):155–165. doi: 10.1083/jcb.200612084. doi:10.1083/jcb.200612084. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 64.Grako KA, Ochiya T, Barritt D, Nishiyama A, Stallcup WB. PDGF (alpha)-receptor is unresponsive to PDGF-AA in aortic smooth muscle cells from the NG2 knockout mouse. J Cell Sci. 1999;112(Pt 6):905–915. doi: 10.1242/jcs.112.6.905. [DOI] [PubMed] [Google Scholar]
- 65.Goretzki L, Burg MA, Grako KA, Stallcup WB. High-affinity binding of basic fibroblast growth factor and platelet-derived growth factor-AA to the core protein of the NG2 proteoglycan. J Biol Chem. 1999;274(24):16831–16837. doi: 10.1074/jbc.274.24.16831. doi:10.1074/jbc.274.24.16831. [DOI] [PubMed] [Google Scholar]
- 66.Nishiyama A, Lin XH, Giese N, Heldin CH, Stallcup WB. Co-localization of NG2 proteoglycan and PDGF alpha-receptor on O2A progenitor cells in the developing rat brain. J Neurosci Res. 1996;43(3):299–314. doi: 10.1002/(SICI)1097-4547(19960201)43:3<299::AID-JNR5>3.0.CO;2-E. doi:10.1002/(SICI)1097-4547(19960201)43:3<299::AID-JNR5>3.0.CO;2-E. [DOI] [PubMed] [Google Scholar]
- 67.Nishiyama A, Lin XH, Giese N, Heldin CH, Stallcup WB. Interaction between NG2 proteoglycan and PDGF alpha-receptor on O2A progenitor cells is required for optimal response to PDGF. J Neurosci Res. 1996;43(3):315–330. doi: 10.1002/(SICI)1097-4547(19960201)43:3<315::AID-JNR6>3.0.CO;2-M. doi:10.1002/(SICI)1097-4547(19960201)43:3<315::AID-JNR6>3.0.CO;2-M. [DOI] [PubMed] [Google Scholar]
- 68.Chatterjee N, Stegmuller J, Schatzle P, et al. Interaction of syntenin-1 and the NG2 proteoglycan in migratory oligodendrocyte precursor cells. J Biol Chem. 2008;283(13):8310–8317. doi: 10.1074/jbc.M706074200. doi:10.1074/jbc.M706074200. [DOI] [PubMed] [Google Scholar]
- 69.Hayakawa-Yano Y, Nishida K, Fukami S, et al. Epidermal growth factor signaling mediated by grb2 associated binder1 is required for the spatiotemporally regulated proliferation of olig2-expressing progenitors in the embryonic spinal cord. Stem Cells. 2007;25(6):1410–1422. doi: 10.1634/stemcells.2006-0584. doi:10.1634/stemcells.2006-0584. [DOI] [PubMed] [Google Scholar]
- 70.Makagiansar IT, Williams S, Dahlin-Huppe K, Fukushi J, Mustelin T, Stallcup WB. Phosphorylation of NG2 proteoglycan by protein kinase C-alpha regulates polarized membrane distribution and cell motility. J Biol Chem. 2004;279(53):55262–55270. doi: 10.1074/jbc.M411045200. doi:10.1074/jbc.M411045200. [DOI] [PubMed] [Google Scholar]
- 71.Yang J, Price MA, Neudauer CL, et al. Melanoma chondroitin sulfate proteoglycan enhances FAK and ERK activation by distinct mechanisms. J Cell Biol. 2004;165(6):881–891. doi: 10.1083/jcb.200403174. doi:10.1083/jcb.200403174. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 72.Wang X, Osada T, Wang Y, et al. CSPG4 protein as a new target for the antibody-based immunotherapy of triple-negative breast cancer. J Natl Cancer Inst. 2010;102(19):1496–1512. doi: 10.1093/jnci/djq343. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 73.Burg MA, Grako KA, Stallcup WB. Expression of the NG2 proteoglycan enhances the growth and metastatic properties of melanoma cells. J Cell Physiol. 1998;177(2):299–312. doi: 10.1002/(SICI)1097-4652(199811)177:2<299::AID-JCP12>3.0.CO;2-5. [DOI] [PubMed] [Google Scholar]
- 74.Leger O, Johnson-Leger C, Jackson E, Coles B, Dean C. The chondroitin sulfate proteoglycan NG2 is a tumour-specific antigen on the chemically induced rat chondrosarcoma HSN. Int J Cancer. 1994;58(5):700–705. doi: 10.1002/ijc.2910580514. doi:10.1002/ijc.2910580514. [DOI] [PubMed] [Google Scholar]
- 75.Behm FG, Smith FO, Raimondi SC, Pui CH, Bernstein ID. Human homologue of the rat chondroitin sulfate proteoglycan, NG2, detected by monoclonal antibody 7.1, identifies childhood acute lymphoblastic leukemias with t(4;11)(q21;q23) or t(11;19)(q23;p13) and MLL gene rearrangements. Blood. 1996;87(3):1134–1139. [PubMed] [Google Scholar]
- 76.Hilden JM, Smith FO, Frestedt JL, et al. MLL gene rearrangement, cytogenetic 11q23 abnormalities, and expression of the NG2 molecule in infant acute myeloid leukemia. Blood. 1997;89(10):3801–3805. [PubMed] [Google Scholar]
- 77.Mauvieux L, Delabesse E, Bourquelot P, et al. NG2 expression in MLL rearranged acute myeloid leukaemia is restricted to monoblastic cases. Br J Haematol. 1999;107(3):674–676. doi: 10.1046/j.1365-2141.1999.01730.x. doi:10.1046/j.1365-2141.1999.01730.x. [DOI] [PubMed] [Google Scholar]
- 78.Smith FO, Rauch C, Williams DE, et al. The human homologue of rat NG2, a chondroitin sulfate proteoglycan, is not expressed on the cell surface of normal hematopoietic cells but is expressed by acute myeloid leukemia blasts from poor-prognosis patients with abnormalities of chromosome band 11q23. Blood. 1996;87(3):1123–1133. [PubMed] [Google Scholar]
- 79.Zangrando A, Intini F, te Kronnie G, Basso G. Validation of NG2 antigen in identifying BP-ALL patients with MLL rearrangements using qualitative and quantitative flow cytometry: a prospective study. Leukemia. 2008;22(4):858–861. doi: 10.1038/sj.leu.2404952. doi:10.1038/sj.leu.2404952. [DOI] [PubMed] [Google Scholar]
- 80.Hanahan D, Weinberg RA. The hallmarks of cancer. Cell. 2000;100(1):57–70. doi: 10.1016/s0092-8674(00)81683-9. doi:10.1016/S0092-8674(00)81683-9. [DOI] [PubMed] [Google Scholar]
- 81.Bulloch K, Stallcup WB, Cohn M. The derivation and characterization of neuronal cell lines from rat and mouse brain. Brain research. 1977;135(1):25–36. doi: 10.1016/0006-8993(77)91049-6. doi:10.1016/0006-8993(77)91049-6. [DOI] [PubMed] [Google Scholar]
- 82.Johansson FK, Goransson H, Westermark B. Expression analysis of genes involved in brain tumor progression driven by retroviral insertional mutagenesis in mice. Oncogene. 2005;24(24):3896–3905. doi: 10.1038/sj.onc.1208553. doi:10.1038/sj.onc.1208553. [DOI] [PubMed] [Google Scholar]
- 83.Wilson SS, Baetge EE, Stallcup WB. Antisera specific for cell lines with mixed neuronal and glial properties. Dev Biol. 1981;83(1):146–153. doi: 10.1016/s0012-1606(81)80017-6. doi:10.1016/S0012-1606(81)80017-6. [DOI] [PubMed] [Google Scholar]
- 84.Shoshan Y, Nishiyama A, Chang A, et al. Expression of oligodendrocyte progenitor cell antigens by gliomas: implications for the histogenesis of brain tumors. Proc Natl Acad Sci U S A. 1999;96(18):10361–10366. doi: 10.1073/pnas.96.18.10361. doi:10.1073/pnas.96.18.10361. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 85.Aguirre-Cruz L, Mokhtari K, Hoang-Xuan K, et al. Analysis of the bHLH transcription factors Olig1 and Olig2 in brain tumors. J Neurooncol. 2004;67(3):265–271. doi: 10.1023/b:neon.0000024190.56750.81. doi:10.1023/B:NEON.0000024190.56750.81. [DOI] [PubMed] [Google Scholar]
- 86.Marie Y, Sanson M, Mokhtari K, et al. OLIG2 as a specific marker of oligodendroglial tumour cells. Lancet. 2001;358(9278):298–300. doi: 10.1016/S0140-6736(01)05499-X. doi:10.1016/S0140-6736(01)05499-X. [DOI] [PubMed] [Google Scholar]
- 87.Lu QR, Park JK, Noll E, et al. Oligodendrocyte lineage genes (OLIG) as molecular markers for human glial brain tumors. Proc Natl Acad Sci U S A. 2001;98(19):10851–10856. doi: 10.1073/pnas.181340798. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 88.Mokhtari K, Paris S, Aguirre-Cruz L, et al. Olig2 expression, GFAP, p53 and 1p loss analysis contribute to glioma subclassification. Neuropathol Appl Neurobiol. 2005;31(1):62–69. doi: 10.1111/j.1365-2990.2004.00612.x. doi:10.1111/j.1365-2990.2004.00612.x. [DOI] [PubMed] [Google Scholar]
- 89.Bannykh SI, Stolt CC, Kim J, Perry A, Wegner M. Oligodendroglial-specific transcriptional factor SOX10 is ubiquitously expressed in human gliomas. J Neurooncol. 2006;76(2):115–127. doi: 10.1007/s11060-005-5533-x. doi:10.1007/s11060-005-5533-x. [DOI] [PubMed] [Google Scholar]
- 90.Bouvier C, Bartoli C, Aguirre-Cruz L, et al. Shared oligodendrocyte lineage gene expression in gliomas and oligodendrocyte progenitor cells. J Neurosurg. 2003;99(2):344–350. doi: 10.3171/jns.2003.99.2.0344. [DOI] [PubMed] [Google Scholar]
- 91.Ikota H, Kinjo S, Yokoo H, Nakazato Y. Systematic immunohistochemical profiling of 378 brain tumors with 37 antibodies using tissue microarray technology. Acta Neuropathol. 2006;111(5):475–482. doi: 10.1007/s00401-006-0060-1. doi:10.1007/s00401-006-0060-1. [DOI] [PubMed] [Google Scholar]
- 92.Ligon KL, Alberta JA, Kho AT, et al. The oligodendroglial lineage marker OLIG2 is universally expressed in diffuse gliomas. J Neuropathol Exp Neurol. 2004;63(5):499–509. doi: 10.1093/jnen/63.5.499. [DOI] [PubMed] [Google Scholar]
- 93.Ohnishi A, Sawa H, Tsuda M, et al. Expression of the oligodendroglial lineage-associated markers Olig1 and Olig2 in different types of human gliomas. J Neuropathol Exp Neurol. 2003;62(10):1052–1059. doi: 10.1093/jnen/62.10.1052. [DOI] [PubMed] [Google Scholar]
- 94.Riemenschneider MJ, Koy TH, Reifenberger G. Expression of oligodendrocyte lineage genes in oligodendroglial and astrocytic gliomas. Acta Neuropathol. 2004;107(3):277–282. doi: 10.1007/s00401-003-0809-8. doi:10.1007/s00401-003-0809-8. [DOI] [PubMed] [Google Scholar]
- 95.Yokoo H, Nobusawa S, Takebayashi H, et al. Anti-human Olig2 antibody as a useful immunohistochemical marker of normal oligodendrocytes and gliomas. Am J Pathol. 2004;164(5):1717–1725. doi: 10.1016/S0002-9440(10)63730-3. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 96.Guha A, Feldkamp MM, Lau N, Boss G, Pawson A. Proliferation of human malignant astrocytomas is dependent on Ras activation. Oncogene. 1997;15(23):2755–2765. doi: 10.1038/sj.onc.1201455. doi:10.1038/sj.onc.1201455. [DOI] [PubMed] [Google Scholar]
- 97.Hermanson M, Funa K, Hartman M, et al. Platelet-derived growth factor and its receptors in human glioma tissue: expression of messenger RNA and protein suggests the presence of autocrine and paracrine loops. Cancer Res. 1992;52(11):3213–3219. [PubMed] [Google Scholar]
- 98.Nister M, Claesson-Welsh L, Eriksson A, Heldin CH, Westermark B. Differential expression of platelet-derived growth factor receptors in human malignant glioma cell lines. J Biol Chem. 1991;266(25):16755–16763. [PubMed] [Google Scholar]
- 99.Nister M, Libermann TA, Betsholtz C, et al. Expression of messenger RNAs for platelet-derived growth factor and transforming growth factor-alpha and their receptors in human malignant glioma cell lines. Cancer Res. 1988;48(14):3910–3918. [PubMed] [Google Scholar]
- 100.Colin C, Virard I, Baeza N, et al. Relevance of combinatorial profiles of intermediate filaments and transcription factors for glioma histogenesis. Neuropathol Appl Neurobiol. 2007;33(4):431–439. doi: 10.1111/j.1365-2990.2007.00829.x. doi:10.1111/j.1365-2990.2007.00829.x. [DOI] [PubMed] [Google Scholar]
- 101.Rowitch DH, Kriegstein AR. Developmental genetics of vertebrate glial-cell specification. Nature. 2010;468(7321):214–222. doi: 10.1038/nature09611. [DOI] [PubMed] [Google Scholar]
- 102.Bidlingmaier S, Zhu X, Liu B. The utility and limitations of glycosylated human CD133 epitopes in defining cancer stem cells. J Mol Med. 2008;86(9):1025–1032. doi: 10.1007/s00109-008-0357-8. doi:10.1007/s00109-008-0357-8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 103.Goldstein AS, Huang J, Guo C, Garraway IP, Witte ON. Identification of a cell of origin for human prostate cancer. Science. 2010;329(5991):568–571. doi: 10.1126/science.1189992. doi:10.1126/science.1189992. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 104.Chekenya M, Hjelstuen M, Enger PO, et al. NG2 proteoglycan promotes angiogenesis-dependent tumor growth in CNS by sequestering angiostatin. FASEB J. 2002;16(6):586–588. doi: 10.1096/fj.01-0632fje. [DOI] [PubMed] [Google Scholar]
- 105.Ozerdem U, Stallcup WB. Pathological angiogenesis is reduced by targeting pericytes via the NG2 proteoglycan. Angiogenesis. 2004;7(3):269–276. doi: 10.1007/s10456-004-4182-6. doi:10.1007/s10456-004-4182-6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 106.Long DM. Capillary ultrastructure and the blood-brain barrier in human malignant brain tumors. J Neurosurg. 1970;32(2):127–144. doi: 10.3171/jns.1970.32.2.0127. [DOI] [PubMed] [Google Scholar]
- 107.Rojiani AM, Dorovini-Zis K. Glomeruloid vascular structures in glioblastoma multiforme: an immunohistochemical and ultrastructural study. J Neurosurg. 1996;85(6):1078–1084. doi: 10.3171/jns.1996.85.6.1078. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.