CK2 associates with kinetochore protein Mif2/CENP-C. Loss of CK2 results in defects in chromosome segregation, short mitotic spindle, and activation of the spindle assembly checkpoint. CK2 phosphorylates Mif2 and Ndc10. CK2 phosphorylation plays antagonistic and synergistic roles with Aurora B phosphorylation of Mif2 and Ndc10, respectively.
Abstract
Protein kinase CK2 is one of the most conserved kinases in eukaryotic cells and plays essential roles in diverse processes. While we know that CK2 plays a role(s) in cell division, our understanding of how CK2 regulates cell cycle progression is limited. In this study, we revealed a regulatory role for CK2 in kinetochore function. The kinetochore is a multi-protein complex that assembles on the centromere of a chromosome and functions to attach chromosomes to spindle microtubules. To faithfully segregate chromosomes and maintain genomic integrity, the kinetochore is tightly regulated by multiple mechanisms, including phosphorylation by Aurora B kinase. We found that a loss of CK2 kinase activity inhibits anaphase spindle elongation and results in chromosome missegregation. Moreover, a lack of CK2 activates the spindle assembly checkpoint. We demonstrate that CK2 associates with Mif2, the Saccharomyces cerevisiae homologue of human CENP-C, which serves as an important link between the inner and outer kinetochore. Furthermore, we show Mif2 and the inner kinetochore protein Ndc10 are phosphorylated by CK2, and this phosphorylation plays antagonistic and synergistic roles with Aurora B phosphorylation of these targets, respectively.
INTRODUCTION
Protein kinase CK2 (formerly known as casein kinase 2) is a highly conserved serine/threonine kinase present in all eukaryotes (Litchfield, 2003). CK2 phosphorylates more than 300 substrates and has been linked to a wide variety of cellular processes, including gene expression, signal transduction, cell morphology/polarity, cell survival, and apoptosis (Meggio and Pinna, 2003). Abnormally high levels of CK2 have been observed in all cancers examined to date (Trembley et al., 2009).
CK2 has been functionally implicated in every stage of the cell cycle (Hanna et al., 1995; St-Denis et al., 2009). In vertebrate cells, CK2 has been shown to localize to the mitotic apparatus and to phosphorylate proteins required for mitosis (St-Denis and Litchfield, 2009), suggesting that it plays an important role in mitosis. Human CK2 has two catalytic subunit isoforms (α and α′) and one regulatory subunit (β), whereas budding yeast CK2 has two catalytic (Cka1 and Cka2) and two regulatory (Ckb1 and Ckb2) subunits. The two catalytic subunits exhibit both overlapping and independent cellular functions (Canton and Litchfield, 2006). In budding yeast, deletion of any of these subunits individually or both regulatory subunits together has no apparent phenotype. However, deletion of both catalytic subunits is lethal. Partial loss-of-function cka1 alleles display apolar morphology and disruption of the actin cytoskeleton (Rethinaswamy et al., 1998). By contrast, cka2 alleles arrest at G1 and G2/M phases (Hanna et al., 1995), suggesting that Cka1 primarily plays a role in cell polarity, while Cka2 functions in cell cycle progression.
Three lines of evidence suggest that CK2 might regulate kinetochore function. First, a systematic yeast two-hybrid screen identified interactions between Mif2 and two CK2 subunits, Cka2 and Ckb2 (Wong et al., 2007). Second, based on their amino acid sequences, Mif2 and Ndc10 (Cbf2) were predicted to be CK2 substrates (Glover, 1998). Third, when endogenous Mif2 was purified from yeast, its predicted CK2 site (Ser325) was found to be phosphorylated (Westermann et al., 2003).
Most kinetochore proteins are conserved across eukaryotes. Studies in Saccharomyces cerevisiae have contributed significantly to our understanding of kinetochore composition, function, and regulation. The budding yeast kinetochore is composed of more than 65 proteins that are divided into three groups (inner, central, and outer kinetochore) according to their relative positions between centromere DNA and microtubules (Westermann et al., 2007). Ndc10 is part of the inner kinetochore CBF3 complex (Ndc10, Cep3, Ctf13, and Skp1) that forms a structural foundation for assembling the entire kinetochore (Sorger et al., 1994). In addition to its centromere-binding activity, Ndc10 plays essential roles in spindle stability, spindle pole body (SPB) integrity, and cytokinesis (Rodrigo-Brenni et al., 2004; Bouck and Bloom, 2005; Widlund et al., 2006; Romao et al., 2008). Performance of Ndc10 functions is strongly correlated with cell cycle–dependent changes in its localization (Bouck and Bloom, 2005; Widlund et al., 2006). Mif2 is the budding yeast homologue of CENP-C (Meluh and Koshland, 1995) and serves as an important link between the inner and central kinetochore. Mif2 binds to the Cse4/CENP-A-containing nucleosome and to the Mtw1/MIND complex, a central kinetochore subcomplex (Meluh and Koshland, 1997; Westermann et al., 2003). The central kinetochore connects to the outer kinetochore, which in turn couples the force generated by dynamic microtubules to chromosomes for their movement during anaphase (Miranda et al., 2005; Westermann et al., 2005; Westermann et al., 2006; Gestaut et al., 2008; Grishchuk et al., 2008).
Phosphorylation plays an important role in the regulation of kinetochore functions, and multiple kinases have been found to regulate kinetochores (Cheeseman et al., 2002; Li and Elledge, 2003; Shimogawa et al., 2006; Kemmler et al., 2009). Among them, the yeast Aurora B kinase, Ipl1, is a key regulator of kinetochores. Ipl1 forms the chromosomal passenger complex (CPC) with Sli15 (INCENP), Bir1 (survivin), and Nbl1 (borealin; Nakajima et al., 2009). The CPC has a dynamic localization throughout the cell cycle, as it moves from centromeres to the spindle at anaphase and concentrates at the spindle midzone before cytokinesis (Ruchaud et al., 2007). At the kinetochore, Ipl1 phosphorylates targets and is required to detach mono-oriented or improperly attached chromosomes that do not generate tension between sister chromatids (Cheeseman et al., 2002). Both Mif2 and Ndc10 are phosphorylated by Ipl1. Mif2 purified from yeast cells is phosphorylated at a predicted Ipl1 site. Moreover, mutation of this phosphorylation site contributes to cell cycle arrest (Westermann et al., 2003). A C-terminal fragment of Ndc10 (residues 679–894) is phosphorylated by Ipl1 in vitro (Biggins et al., 1999). In addition, Ndc10 showed reduced microtubule-binding activity in a cell extract from a type 1 protein phosphatase mutant (glc7–10; Sassoon et al., 1999), suggesting that phosphorylation of Ndc10 plays a role in regulating Ndc10 binding to microtubules.
In this study, we established a regulatory role for CK2 in kinetochore function and examined how CK2 and Ipl1 cooperate to regulate kinetochore proteins Mif2/CENP-C and Ndc10.
RESULTS
CK2 associates with kinetochores
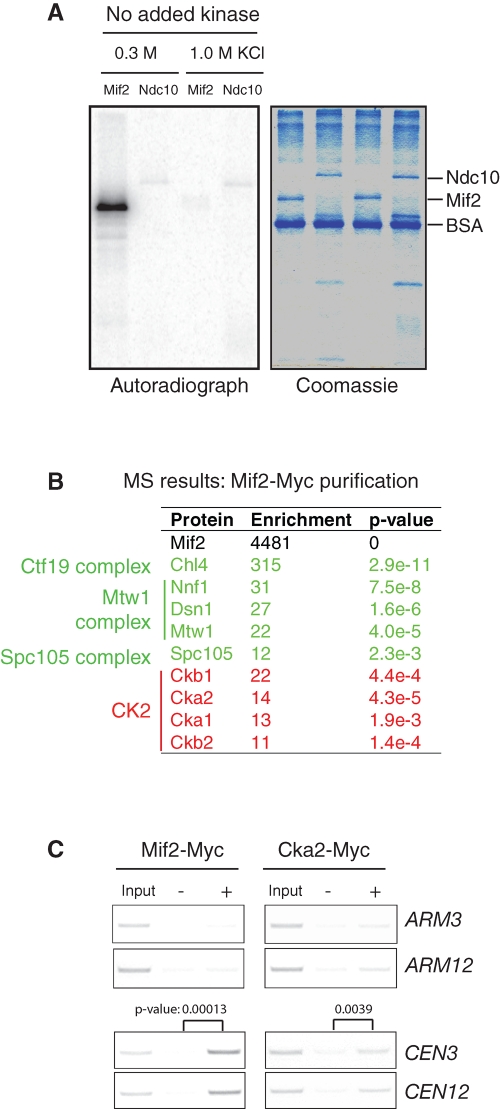
The kinetochore protein Mif2 interacts with two CK2 subunits, Cka2 and Ckb2, in a yeast two-hybrid screen (Wong et al., 2007), and its predicted CK2 site (Ser325) is phosphorylated in vivo (Westermann et al., 2003). In an attempt to determine whether Mif2 is a bona fide substrate of CK2, we overexpressed and purified full-length Mif2 from yeast cells. To our surprise, this purified Mif2 became phosphorylated in the presence of ATP without the addition of exogenous kinase (Figure 1A), indicating that a kinase activity copurified with Mif2. Moreover, this copurification appeared to be specific to Mif2 because under the same conditions, no kinase activity copurified with Ndc10, another kinetochore protein. This kinase activity disappeared when Mif2 was isolated using a more stringent wash buffer containing 1 M KCl. MS analysis revealed that CK2 copurifies with Mif2 (Figure 1B). A calculation based on peptide counts, an approximate measure of protein abundance, showed that all four subunits of CK2 were enriched 10- to 20-fold during the purification of Mif2. Additionally, kinetochore proteins known to interact with Mif2 were also enriched, validating our approach. These kinetochore proteins include Chl4 (Ctf19 complex), Nnf1, Dsn1, Mtw1 (Mtw1 complex), and Spc105 (Spc105 complex). Copurification of CK2 with Mif2 is consistent with the previously reported yeast two-hybrid interaction between Mif2 and CK2 (Wong et al., 2007) and may explain how Mif2, purified under our standard conditions, was phosphorylated in the absence of added kinase.
FIGURE 1:
CK2 associates with kinetochores. (A) A protein kinase activity copurifies with Mif2. Mif2-Myc and Ndc10-Myc were overexpressed using a GAL1 promoter in yeast and were affinity-purified using anti-Myc antibodies and a moderate (0.3 M KCl) or high (1 M KCl) salt concentration. The purified proteins were incubated with [γ32P]ATP for 30 min at 25°C. The samples were analyzed by autoradiography following SDS–PAGE. (B) MS identified all four subunits of CK2 as copurifying with Mif2. The peptide count for each protein was used to estimate its abundance. The enrichment of each protein is equal to a ratio of the peptide counts in the purified Mif2 sample to the total peptide counts (Peptide Atlas: www.peptideatlas.org). p Values for enrichment were calculated using Fisher's exact test. (C) Cka2, a catalytic subunit of CK2, associates with centromeric DNA. Cells expressing Mif2-Myc or Cka2-Myc at their endogenous loci were analyzed by ChIP. The centromere and arm of chromosome 3 and chromosome 12 were amplified by PCR from either the total input chromatin (Input), immunoprecipitated samples (+), or mock-treated (without antibody) controls (−). Three independent experiments were carried out, and one representative result is shown. For centromere binding, the results were quantified using ImageJ; p values were determined using Student's t test.
All four subunits of CK2 appeared to localize predominantly to the nucleus, but none of the subunits showed a distinctive kinetochore localization (Supplemental Figure S1). This observation is consistent with the fact that CK2 has multiple nuclear functions. To further test whether CK2 specifically associates with kinetochores/centromeres, we performed chromatin immunoprecipitations (ChIPs) using strains expressing Mif2-Myc or Cka2-Myc at their endogenous loci. Mif2 showed a strong centromere association but no association with chromosome arm regions. By comparison, Cka2 associated with both centromeres and chromosome arms (Figure 1C). We postulate that the weak association of CK2 with centromeric DNA could be due to the transient nature of enzyme–substrate interactions. Taken together, our results suggest that CK2 associates with centromeric DNA via its interaction with Mif2.
CK2 kinase activity is required for anaphase spindle elongation and chromosome segregation
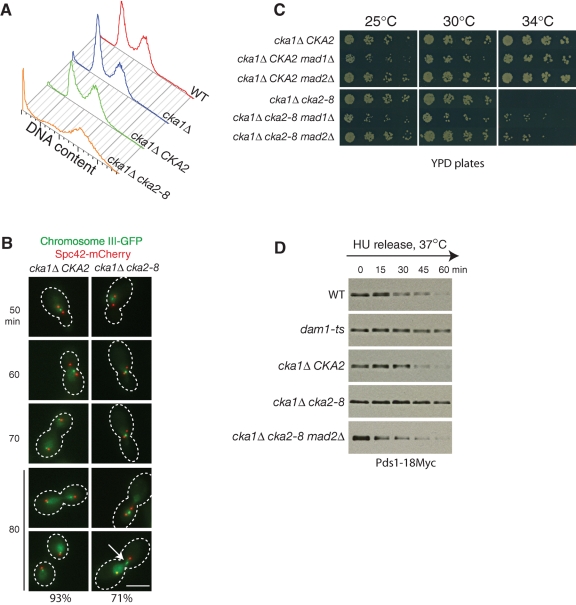
To investigate the role of CK2 in kinetochore function, we first examined the DNA content of a temperature-sensitive mutant, cka1Δ cka2–8. Mutations in the cka2–8 allele have been mapped to conserved residues within the kinase domain (Hanna et al., 1995). Wild-type and mutant haploid cells were synchronized in S phase with hydroxyurea and released into normal medium at the restrictive temperature (37°C). Deletion of CKA1 had no obvious effect on DNA content (Figure 2A). cka1Δ CKA2 cells had a DNA profile similar to wild-type cells at 37°C. By contrast, a cka1Δ cka2–8 strain demonstrated a massive accumulation of cells with 2C DNA content and cells with no DNA at 37°C (Figure 2A), suggesting a chromosome segregation defect.
FIGURE 2:
CK2 kinase activity is required for anaphase spindle elongation and chromosome segregation, and loss of CK2 kinase activates SAC. (A) Loss of CK2 kinase activity results in chromosome segregation defects. Wild-type, cka1Δ, cka1Δ CKA2, and cka1Δ cka2–8 cells were arrested by hydroxyurea at 25°C for 3 h, released into 37°C medium, and allowed to grow at 37°C for 2 h. Cellular DNA was stained by SYTOX green, and the DNA content was determined by flow cytometry. The complete genotypes of each strain can be found in Table S1. (B) Chromosomes missegregate in cka1Δ cka2–8 cells. Both cka1Δ cka2–8 and the isogenic wild-type strain cka1Δ CKA2 contain SPC42-mCherry (marking SPBs) and HIS3::PCu-LACI-GFP lacO::LEU2 (marking chromosome 3). Cells were arrested in G1 and released to a semirestrictive temperature, 34°C. Samples were collected and fixed every 10 min and subjected to fluorescence microscopy. At 80 min, 93% of cka1Δ CKA2 cells have segregated chromosomes. By contrast, 71% of cka1Δ cka2–8 cells failed to segregate chromosomes properly (n > 200). White arrow: a lagging chromosome. Scale bar: 5 μm. (C) Deletion of MAD2 and MAD1 partially rescues the growth of cka1Δ cka2–8. Serial dilutions of the indicated strains were spotted on YPD plates and grown at 25, 30, and 34°C for ∼36 h. (D) Loss of CK2 kinase activity activates the spindle assembly checkpoint. All strains express Pds1–18Myc. Cells were synchronized with hydroxyurea and released to 37°C medium. Equal numbers of cells were harvested every 15 min and subjected to Western blotting.
We then directly monitored chromosome segregation in cka1Δ cka2–8 cells. cka1Δ CKA2 and cka1Δ cka2–8 cells were synchronized with α-factor and released into medium at a semirestrictive temperature (34°C). In both strains, SPBs were identified by Spc42-mCherry and chromosome 3 was identified by the binding of LacI-GFP to lacO operator sites integrated 23 kb from the centromere of chromosome 3 (Straight et al., 1996). In both cka1Δ CKA2 and cka1Δ cka2–8 cells, SPBs duplicated and separated ∼50 min after α-factor release (Figure 2B). As the cell cycle progressed, mitotic spindles in cka1Δ CKA2 cells elongated, and by 70 min, the chromatids were separated in 93% of the cells. However, in cka1Δ cka2–8 cells, most of the spindles remained short (1–3 μm), and the sister chromatids failed to separate. At 80 min, in 71% of the mutant cells, the sister chromatids either failed to separate or separated with one chromatid lagging behind (Figure 2B). These results demonstrate that CK2 kinase activity is required for anaphase spindle elongation and normal sister-chromatid segregation.
Loss of CK2 kinase activity triggers activation of the spindle assembly checkpoint
The short spindle arrest seen in cka1Δ cka2–8 cells prompted us to determine whether loss of CK2 kinase activity activates the spindle assembly checkpoint (SAC). Deletion of the SAC component MAD2 (or MAD1) partially rescued cka1Δ cka2–8 growth at 34°C (Figure 2C). This was unexpected, because combining most mitotic mutants with checkpoint loss-of-function mutants results in synthetic sickness/lethality.
To directly test whether the SAC is activated in cka1Δ cka2–8 cells, we analyzed the levels of securin (Pds1 in yeast), which are an indicator of SAC activation. When active, the SAC inhibits activation of the anaphase-promoting complex (APC), a multi-subunit ubiquitin ligase required for Pds1 degradation. Wild-type, dam1ts, cka1Δ CKA2, cka1Δ cka2–8, and cka1Δ cka2–8 mad2Δ cells containing Pds1–18Myc were arrested with hydroxyurea and released at the restrictive temperature (37°C). As expected, Pds1 levels decreased in wild-type cells but were stabilized in kinetochore mutant dam1ts cells after release from hydroxyurea (Figure 2D). Pds1 levels in cka1Δ CKA2 cells decreased in a similar manner similar to wild-type cells. However, Pds1 was stabilized in cka1Δ cka2–8 cells, and this Pds1 stabilization was dependent on MAD2. This result indicates that loss of CK2 function activates the checkpoint. In other words, in wild-type cells, CK2 activity plays a role in silencing the checkpoint. Activation of the checkpoint in cka1Δ cka2–8 cells causes these cells to arrest with short spindles, as can be observed in Figure 2B.
CK2 phosphorylates Mif2 and Ndc10 in vitro
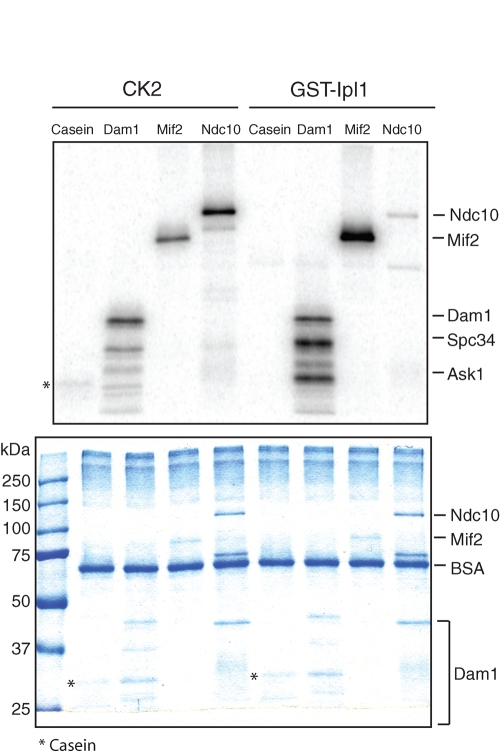
On the basis of our findings that CK2 interacts with the kinetochore protein Mif2 and that CK2 is required for chromosome segregation, we hypothesized that CK2 may regulate chromosome segregation by phosphorylating kinetochore proteins. To investigate this possibility, we first tested whether CK2 directly phosphorylates Mif2 and Ndc10, two kinetochore proteins that are predicted to be substrates of CK2 (Glover, 1998). When we purified Mif2 under stringent conditions using 1 M KCl, Mif2 was not phosphorylated in the absence of added kinases (Figure 1A). CK2 and Ipl1 were purified from yeast and Escherichia coli (Padmanabha and Glover, 1987; Kang et al., 2001), respectively; they phosphorylated their known substrates, casein and the Dam1 complex, respectively (Figure 3; Cheeseman et al., 2002). Consistent with previous studies (Biggins et al., 1999; Westermann et al., 2003), Ipl1 phosphorylates Mif2 and Ndc10. In addition, we observed that CK2 readily phosphorylates both Mif2 and Ndc10. The Dam1 complex was also phosphorylated by CK2, but the phosphorylation pattern differed from the pattern resulting from Ipl1 phosphorylation, highlighting the different specificities of the two kinases. We analyzed the in vitro phosphorylation sites of Mif2 and Ndc10 using MS. The predicted CK2 sites in Mif2 (Ser325) and Ndc10 (Thr104) were indeed phosphorylated (Figure S2). Several other residues were also phosphorylated in the in vitro kinase assays (Figure S2).
FIGURE 3:
CK2 directly phosphorylates Mif2 and Ndc10 in vitro. Mif2-Myc and Ndc10-Myc were overexpressed and affinity-purified as described previously. A stringent wash buffer containing 1 M KCl was used to remove Mif2-associated proteins prior to the kinase assay. The Dam1 complex purified from E. coli was used as a control. CK2 and GST-Ipl1 were purified from yeast and E. coli, respectively. Substrates (∼100 ng) were incubated with [γ32P]ATP in the presence of kinases (∼2 ng). Top, autoradiography; bottom, Coommassie Brilliant Blue staining.
Because both CK2 and Ipl1 phosphorylate Mif2 and Ndc10, it is possible that one phosphorylation event affects the other. We therefore carried out sequential kinase assays and found that phosphorylation by one kinase had no effect on phosphorylation by the other kinase for both substrates (Figure S3), indicating that these two phosphorylation events are independent in vitro.
CK2 phosphorylation of Mif2 in vivo antagonizes Aurora B phosphorylation in control of Mif2 stability
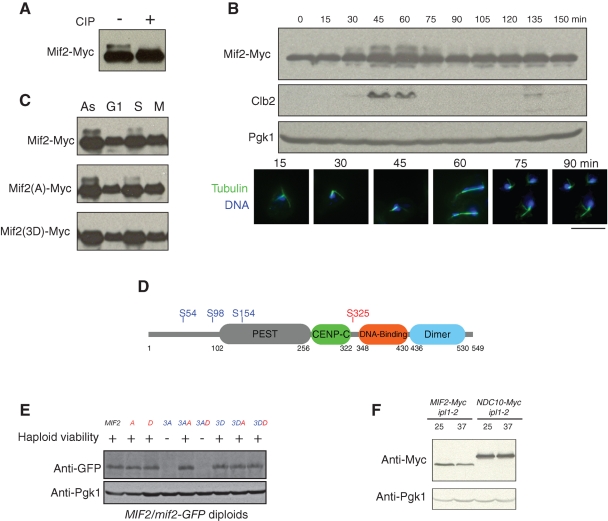
In budding yeast, Mif2 serves as an important link between the inner and outer kinetochore (Westermann et al., 2007). Similarly, in metazoan cells, CENP-C, Mif2's homologue, plays a bridging role by recognizing CENP-A nucleosomes and initiating kinetochore assembly (Milks et al., 2009; Carroll et al., 2010). A Mif2-Myc fusion protein migrates as two bands on SDS–PAGE, and phosphatase treatment showed that the slower-migrating band represents phosphorylated Mif2 (Figure 4A). To better understand the function of Mif2 phosphorylation, we first assessed the Mif2 phosphorylation state during the cell cycle. Mif2 phosphorylation peaks concomitant with mitotic spindle elongation and chromosome segregation (Figure 4B). This correlation suggests that the cell cycle–dependent phosphorylation is integral to Mif2 function. Since CK2 is believed to be constitutively active throughout the cell cycle (Niefind et al., 2009; Tripodi et al., 2010), cell cycle changes in Mif2 phosphorylation may be due to a different kinase. Consistently, we found that the slower-migrating band is a result of Ipl1 phosphorylation, since mutation of the Ipl1 sites, but not the CK2 site (see the following section), eliminated the slower-migrating band (Figure 4C).
FIGURE 4:
CK2 phosphorylation of Mif2 antagonizes Aurora B phosphorylation in control of Mif2 stability. (A) Mif2 is a phospho-protein in vivo. Cell extracts were incubated with calf intestinal alkaline phosphatase (CIP) for 1 h at 30°C and subjected to immunoblotting. (B) Phosphorylation of Mif2 is coordinated with chromosome segregation. Wild-type cells were synchronized in G1 and released. Cells were collected every 15 min and subjected to immunoblotting and immunofluorescence. Green: tubulin; blue: DNA. Scale bar: 5 μm. (C) Aurora B phosphorylation of Mif2 is cell cycle–dependent. Cells were untreated (As: asynchronous) or arrested in G1 phase with α-factor, in S phase with hydroxyurea, or in M phase with nocodazole. Cell extracts were analyzed by immunoblotting. Mif2(A) represents Mif2(S325A), in which the CK2 site is mutated; Mif2(3D) represents Mif2(S54D S98D S154D), in which the Ipl1 sites are mutated. (D) Mif2 contains one putative CK2 phosphorylation site (S235) and three putative Aurora B phosphorylation sites (S54, S98, S154). Mif2 contains a PEST sequence (gray), a conserved CENP-C motif (green), a DNA-binding domain (red), and a dimerization domain (blue). (E) CK2 phosphorylation antagonizes Ipl1 phosphorylation in control of Mif2 stability. Viability of the haploid mif2 phospho-mutants was indicated. +: viable; −: lethal. The mif2 mutant alleles in the heterozygous diploids (MIF2/mif2) were tagged with GFP, and the resultant GFP fusion protein levels were examined by immunoblotting using a GFP antibody (Roche). Red: the CK2 site; blue: Ipl1 sites. A = S325A; D = S325D; 3A = S54A S98A S154A; 3AA = S54A S98A S154A S325A; 3AD = S54A S98A S154A S325D; 3D = S54D S98D S154D; 3DA = S54D S98D S154D S325A; 3DD = S54D S98D S154D S325D. (F) Compromising Ipl1 kinase activity results in a decreased Mif2 protein levels. ipl1–2 cells expressing Mif2-Myc or Ndc10-Myc at their endogenous loci were incubated at the permissive temperature (25°C) or the restrictive temperature (37°C) for 3 h. Mif2-Myc and Ndc10-Myc protein levels were assessed by immunoblotting.
Mif2 consists of four functional domains: a PEST sequence, the CENP-C domain, a DNA-binding domain, and a dimerization domain (Figure 4D; Cohen et al., 2008). Domain deletion analysis indicated the CENP-C domain and the DNA-binding domain are essential, the PEST domain and the dimerization domain are required for mitotic progression, while no function could be associated with the N-terminal portion of Mif2 (Cohen et al., 2008). CK2 prefers to phosphorylate serine and threonine followed by at least three acidic amino residues (Meggio and Pinna, 2003). The consensus sequence for Ipl1 phosphorylation has been proposed as [RK]-X-[TS]-[ILV] (X represents any amino residue) based on analysis of known Ipl1 targets (Cheeseman et al., 2002). Based on these criteria, Mif2 contains one CK2 site (Glover, 1998) and three putative Ipl1 sites (Westermann et al., 2003). While the three Ipl1 sites cluster at the N-terminus of Mif2, the single CK2 site resides in the essential portion of Mif2 (Figure 4D). Substitution of the CK2 site with alanine resulted in reduced Mif2 phosphorylation by CK2 (Figure S4), confirming that Ser325 is a CK2 target site. The residual phosphorylation of Mif2 is likely due to the fact that the kinase is less specific under in vitro conditions or that other CK2 sites may exist in Mif2. However, substitution of Ser325 with alanine or aspartate had no obvious effect on cell growth, protein levels (Figure 4E), or localization (Figure S5).
We then tested whether CK2 phosphorylation and Ipl1 phosphorylation of Mif2 have any antagonistic or synergistic effects. When the three Ipl1 sites were mutated to alanine (mif2(3A)), the protein was no longer detectable and lethality resulted (Figure 4E). Intriguingly, mutation of the CK2 site to alanine restored the protein levels and reversed the lethality of mif2(3A), whereas mutation of the CK2 site to aspartate showed no rescue activity. Previously, the N-terminus of human CENP-C was shown to be essential for its destruction (Lanini and McKeon, 1995). Given the fact that all three Ipl1 sites in Mif2 are clustered at its N-terminus, our results suggest that Ipl1 phosphorylation at the N-terminus of Mif2 is critical for Mif2 stability. To further understand the role of Ipl1 in Mif2 stability, we compared Mif2 protein levels in ipl1–2 cells at the permissive and restrictive temperatures. We found that compromising Ipl1 kinase activity causes a decrease of Mif2 protein levels (Figure 4F), confirming that Ipl1 phosphorylation of Mif2 stabilizes Mif2. Additionally, this effect is specific for Mif2, because Ndc10 levels did not change under the same conditions. Ipl1 phosphorylation of Mif2 occurs concurrently with chromosome segregation and spindle elongation (Figure 4B), indicating that Mif2 phosphorylation by Ipl1 ensures Mif2 kinetochore function by maintaining its stability. Our results also showed that CK2 phosphorylation of Mif2 plays an antagonistic role with Aurora B in Mif2 stability.
Synergistic effects of CK2 and Aurora B in regulating Ndc10 localization
Ndc10 contains one putative CK2 site (Glover, 1998) and four putative Ipl1 sites (Figure 5A). A systematic mutation study showed that substitution of the CK2 and Ipl1 sites with alanine, separately or in combination, had no effect on cell viability. Furthermore, substitution of the CK2 site with aspartate had no growth phenotype. By contrast, substitution of Ipl1 sites alone or together with substitution of the CK2 site with aspartate resulted in lethality (Figure 5C).
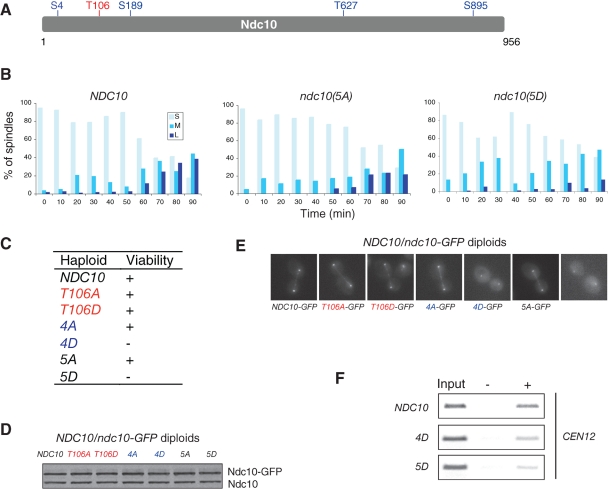
FIGURE 5:
CK2 phosphorylation of Ndc10 is synergistic with phosphorylation by Aurora B to control Ndc10 localization. (A) Ndc10 contains one putative CK2 phosphorylation site (red) and four putative Aurora B phosphorylation sites (blue). (B) Viability of the haploid ndc10 phospho-mutants. (C) Mitotic spindles fail to elongate in the ndc10(5D) mutant. ndc10Δ cells carrying indicated wild-type or mutant plasmid were synchronized by α-factor and released at 30°C. Cells were collected every 10 min and subjected to immunofluorescence to visualize mitotic spindles and DNA. Spindles were scored as short (S, <3 μm), medium (M, 3–6 μm) and long (L, > 6 μm). More than 100 cells were scored at each time point. The percentages of spindle sizes are presented. (D) CK2 and Ipl1 phosphorylation does not alter Ndc10 protein levels. The ndc10 mutant alleles in heterozygous diploids were tagged with GFP, and the resultant GFP fusion protein levels were examined by immunoblotting using an anti-Ndc10 antibody. (E) Anaphase localization of wild-type and ndc10 phospho-mutants in NDC10/ndc10-GFP heterozygous diploids. (F) Heterozygous diploid cells (NDC10/NDC10-GFP, NDC10/ndc10(4D)-GFP, and NDC10/ndc10(5D)-GFP) were analyzed by ChIP using a GFP antibody. The centromere of chromosome 12 was amplified by PCR from either the total input chromatin (Input), immunoprecipitated samples (+), or mock-treated (without antibody) controls (−).
To further explore how ndc10(5D) affects spindle morphology, we created plasmid-based ndc10 phospho-mutants. Unlike the genomically integrated ndc10(5D) mutants used in the analysis described above, ndc10Δ cells harboring the ndc10(5D) plasmid were viable and exhibited a slow-growth phenotype (unpublished data). The viability observed when the mutant was on a plasmid is likely due to the fact that overexpression of the ndc10(5D) mutant sustains viability. We synchronized the ndc10Δ cells containing wild-type NDC10, ndc10(5A), or ndc10(5D) plasmids using α-factor arrest and release. Compared to the wild-type NDC10 cells, both ndc10(5A) and ndc10(5D) cells showed slower mitotic spindle elongation (Figure 5B). At 90 min after release, ∼40% of NDC10 cells displayed long, late anaphase–phase spindles (>6 μm), whereas only 20% of ndc10(5A) and 10% of ndc10(5D) cells had long anaphase spindles.
To understand mechanistically how phosphorylation regulates Ndc10 function, we tagged the ndc10(4D) mutant with green fluorescent protein (GFP) in a heterozygous diploid, in which one copy of the wild-type NDC10 allele maintained cell viability. Mutation of the Ndc10 phosphorylation sites did not appear to affect the stability of the protein (Figure 5D). However, unlike wild-type Ndc10-GFP, Ndc10(4D)-GFP appeared to localize to kinetochores but no longer localized along anaphase spindles (Figure 5E). Further mutation of the CK2 site to aspartate in the ndc10(4D) mutant, creating an ndc10(5D) mutant, reduced Ndc10 association with kinetochores. In an attempt to quantify signal intensity of ndc10 mutants relative to the kinetochore marker Mtw1-red fluorescent protein (RFP), we noticed that the fluorescence intensity of Mtw1-RFP was affected in ndc10(4D) and ndc10(5D) mutants (unpublished data). This is likely due to the fact that Ndc10 is a component of the CBF3 complex, whose binding to centromeres is required for assembling the entire kinetochore. Reduced association of the Ndc10 mutants to centromeres resulted in a weaker signal of Mtw1-RFP at kinetochores. Lacking a kinetochore marker that maintains constant levels in the wild-type and mutant cells, we decided to examine localization of wild-type Ndc10, Ndc10(4D), and Ndc10(5D) at kinetochores using the ChIP assay (Figure 5F). We found that Ndc10(4D) has weaker centromere binding. Moreover, additional mutation of the CK2 site, Ndc10(5D), led to a further decrease in centromere binding. These results indicate that phosphorylation by Ipl1 and CK2 regulate Ndc10 localization synergistically. Our results also suggest that Ipl1 phosphorylation of Ndc10 regulates Ndc10 localization on anaphase spindles, whereas CK2 phosphorylation of Ndc10 controls the targeting of Ndc10 to kinetochores.
DISCUSSION
Previously, CK2 was known to regulate cell division, but its specific role in cell division had not been established. In this study, we revealed a mitotic role for CK2 in the regulation of the kinetochore proteins Mif2 and Ndc10. Our results show that CK2 antagonizes SAC signaling. Loss of CK2 function activates SAC and results in a cell cycle arrest with short spindles. Notably, this defect is different from the defects observed in ipl1ts cells, which do not display a uniform arrest phenotype. In ipl1ts mutants, budding and cytokinesis are largely normal, while chromosomes fail to segregate properly (Biggins et al., 1999; Kim et al., 1999; Biggins and Murray, 2001). A recent study reported that, in fission yeast, CK2 is required for the SAC through the regulation of Mad2 stability (Shimada et al., 2009). When CK2 kinase activity was impaired in fission yeast, mitosis occurred despite a spindle defect, a phenotype that is different from what we observed in cka1Δ cka2–8 cells, where spindle elongation and chromosome segregation were blocked (Figure 2B). It is therefore possible that CK2 regulates mitosis through a different mechanism in fission yeast. Our data suggest that the absence of CK2 activity delays cell cycle progression in a manner dependent on SAC signaling.
We demonstrated that CK2 directly phosphorylates Mif2/CENP-C and Ndc10. CENP-C is a critical bridging component of kinetochores for both mitosis and meiosis. In budding yeast, Mif2 connects Cse4-containing nucleosomes and the Mtw1 complex (Westermann et al., 2003). During meiosis, Mif2 was found to interact with the monopolin complex, and this interaction was found to be essential for sister-chromatid coorientation in meiosis I (Corbett et al., 2010). In fission yeast, Cnp3/CENP-C functions as a scaffold for the recruitment of kinetochore proteins specific for mitosis and meiosis (Tanaka et al., 2009). In Xenopus egg extracts, CENP-C, together with CENP-N, binds directly to CENP-A nucleosomes and recruits other nonhistone CENPs (Carroll et al., 2010). More recently, the CENP-C N-terminal region was found to interact directly with the Mis12 complex in Drosophila and human cells. The high affinity between CENP-C and the Mis12 complex is sufficient to recruit the core outer kinetochore components and serves as the crucial link between the inner and outer kinetochore (Przewloka et al., 2011; Screpanti et al., 2011). Thus a number of CENP-C interactions may potentially be subjected to regulation.
Our study of Mif2 indicated that N-terminally clustered Ipl1 phosphorylation of Mif2 is required for Mif2 stability. In human cells, deletion of the N-terminus of CENP-C does not disrupt CENP-C binding to centromeres but instead results in massive accumulation of CENP-C diffusely in the nucleus. It has been hypothesized that the N-terminus of CENP-C serves as a destruction sequence that prevents the accumulation of CENP-C when it is mistargeted (Lanini and McKeon, 1995). However, how the N-terminus of CENP-C signals for degradation is not clear. We found that Mif2, when not phosphorylated by Ipl1, is quickly degraded. Intriguingly, Ipl1-regulated Mif2 degradation is counterregulated by CK2. Given the important role of Mif2 in kinetochore function, its control by dual regulatory inputs is not surprising. It is now important to test whether human CENP-C is regulated in a similar manner.
Our mutagenesis study dissected the roles of Aurora B and CK2 kinases in Ndc10 regulation. Phospho-mimicking Ndc10 mutations at Ipl1 sites abolished its targeting to the anaphase spindle, but not its localization to kinetochores, whereas the phospho-mimicking mutant of Ndc10 at the CK2 site combined with mutations of the Ipl1 sites additionally affected Ndc10 localization to the kinetochore. The former finding is consistent with two previous observations. First, Ndc10-GFP localizes normally in ipl1–321 cells (Bouck and Bloom, 2005). Similarly, in our study, substituting alanines for the Ndc10 residues phosphorylated by Ipl1 (ndc10(4A)) does not affect Ndc10 localization. Second, the Ndc10-containing CBF3 complex displayed a lower microtubule-binding activity in glc7–10 extracts (Sassoon et al., 1999). Glc7 is a phosphatase whose function counteracts that of Ipl1 kinase. Presumably, in glc7–10 cells, Ndc10 is hyperphosphorylated or constitutively phosphorylated, and therefore loses its ability to bind to microtubules. Our study showed that Ndc10(4D), a constitutively phosphorylated Ndc10, consistently failed to target to anaphase spindles. Localization of Ndc10 to the spindle depends on Ndc10 binding to Bir1, an essential component of the CPC, and the Ndc10-Bir1 interaction is subject to regulation of Cdk1 phosphorylation (Widlund et al., 2006). In addition to regulation of Ndc10 targeting to spindles by Ipl1, Ndc10 is also sumoylated, and its sumoylation is required for its binding to Bir1 and, hence, Ndc10 spindle localization (Montpetit et al., 2006). Whether Ipl1 phosphorylation of Ndc10 regulates its sumoylation remains to be tested. It appears that Ndc10 is finely tuned by multiple mechanisms to ensure its proper localization to spindles. In this study, we established the existence of another regulatory mechanism for Ndc10 by CK2. Ndc10 is a CK2 substrate, and CK2 phosphorylation of Ndc10, together with its phosphorylation by Aurora B, regulates Ndc10 localization to kinetochores. In total, our data establish an important, previously unappreciated role for CK2 in mitotic regulation, mediated at least in part by direct phosphorylation of the kinetochore proteins Mif2 and Ndc10.
MATERIALS AND METHODS
Yeast strains and plasmids
The yeast strains used in this study are listed in Table S1. Cells were grown in yeast–peptone–dextrose (YPD) or selective medium at 25°C unless specified.
Protein purification and identification
Mif2 and Ndc10 were overexpressed in yeast and purified as described previously (Rodal et al., 2003) with the following modifications: 1) The wash buffer contained either 300 mM or 1 M KCl. 2) After TEV cleavage, the protease containing a His tag was removed by nickel nitrilotriacetic acid (Ni-NTA) resin. Proteins that copurified with Mif2 were identified by MS as described previously (Nakajima et al., 2009).
In vitro kinase assays
Mif2 and Ndc10 were expressed and purified as described in the previous section. CK2 (kindly provided by Claiborne Glover III, University of Georgia, Athens, GA) was purified from yeast (Padmanabha and Glover, 1987). GST-Ipl1 was purified from E. coli. To determine the concentrations of the kinases and substrates, both substrates and kinases were subjected to SDS–PAGE along with bovine serum albumin (BSA) standards and stained with GelCode Blue (Thermo Scientific, Lafayette, CO). The protein concentrations were determined by measuring the intensities of the protein bands using Odyssey Imaging System (LI-COR Biosciences, Lincoln, NE). Substrate (100 ng) was mixed with kinase (2 ng), 10× buffer (200 mM HEPES-KOH, pH7.5, 100 mM MgCl2, 100 mM MnCl2, 10 mM dithiothreitol [DTT]), 2 μCi of [γ32P]ATP, and cold ATP (final concentration 30 μM). Reactions were incubated at room temperature for 30 min and stopped with 4× SDS loading buffer.
Chromatin immunoprecipitation
Chromatin immunoprecipitation was performed as described previously (Kang et al., 2001).
Flow cytometry
Yeast cell samples were prepared as described previously (Haase, 2004). DNA content was analyzed using a Coulter FC-500 Analyzer (Beckman Coulter, Brea, CA).
Fluorescence microscopy
Indirect immunofluorescence microscopy was performed as described previously (Cheeseman et al., 2002). For imaging GFP or mCherry fusion proteins in synchronized cells, the cells were collected at each time point and fixed with paraformaldehyde as described previously (Biggins et al., 1999). Fixed and live-cell microscopy were performed as described previously (Kaksonen et al., 2003). All images were obtained using an Olympus IX71 microscope equipped with a 100× numerical aperture 1.4 objective and an Orca-ER camera (Hamamatsu, Bridgewater, NJ) and processed using ImageJ (http://rsbweb.nih.gov/ij).
Mass spectrometry analysis
Samples were prepared as described in the Supplemental Materials. Data-dependent tandem MS (MS/MS) analysis was performed with an LTQ-Orbitrap mass spectrometer (ThermoFisher, San Jose, CA). Peptides eluted from the LC column were directly electrosprayed into the mass spectrometer with the application of a distal 2.5-kV spray voltage. A cycle of one full-scan MS spectrum (m/z 300–2000) was acquired, followed by eight MS/MS events, sequentially generated on the first to the eighth most intense ions selected from the full MS spectrum at a 35% normalized collision energy. This number of microscans was done for both MS and MS/MS scans, and the maximum ion injection time was 50 and 100 ms, respectively. The dynamic exclusion settings used were as follows: repeat count, 1; repeat duration, 30 s; exclusion list size, 100; and exclusion duration, 180 s. MS scan functions and high-performance liquid chromatography solvent gradients were controlled by the Xcalibur data system (ThermoFisher).
Full MS and tandem mass spectra were extracted from raw files, and the tandem mass spectra were searched against a S. cerevisiae protein database (http://downloads.yeastgenome.org/sequence/). To accurately estimate peptide probabilities and false discovery rates, we used a reverse-decoy database containing the reversed sequences of all the proteins appended to the target database (Peng et al., 2003). Tandem mass spectra were matched to sequences using the ProLuCID (Xu et al., 2006) algorithm.
ProLuCID searches were done on an Intel Xeon 80-processor cluster running under the Linux operating system. The peptide mass search tolerance was set to 3 Da for spectra acquired on the LTQ-Orbitrap instrument. The mass of the amino acid cysteine was statistically modified by +57.02146 Da, to take into account the carboxyamidomethylation of the sample, and serine, threonine, and tyrosine were treated as differentially modified by +79.9663 Da for phosphorylation. No enzymatic cleavage conditions were imposed on the database search, so the search space included all candidate peptides whose theoretical mass fell within the mass tolerance window, regardless of their tryptic status (Lu et al., 2009).
The validity of peptide/spectrum matches (PSMs) was assessed in DTASelect (Tabb et al., 2002; Cociorva et al., 2007), using two SEQUEST-defined (Eng et al., 1994) parameters: the cross-correlation score (XCorr) and normalized difference in cross-correlation scores (DeltaCN). The search results were grouped by charge state (+1, +2, +3, and > +3) and tryptic status (fully tryptic, half-tryptic, and nontryptic), resulting in 12 distinct subgroups. In each one of these subgroups, the distribution of XCorr, DeltaCN, and DeltaMass (the mass difference between theoretical and experimental precursor mass in ppm) values for 1) direct and 2) decoy database PSMs was obtained, then the direct and decoy subsets were separated by discriminant analysis. Full separation of the direct and decoy PSM subsets is not generally possible; therefore peptide match probabilities were calculated based on a nonparametric fit of the direct and decoy score distributions. A peptide probability of 90% was set as the minimum threshold. The false discovery rate was calculated as the percentage of reverse-decoy PSMs among all the PSMs that passed the 90% probability threshold. In addition, we required that every protein be supported by at least a unique peptide with probability greater than 99%. After this last filtering step, we estimate that both the protein and peptide false discovery rates were reduced to between 0.0% and 0.5%. The phosphorylation sites of peptides were evaluated using the software DeBunker (Lu et al., 2007) and the in-house algorithm Pinpointer, and further confirmed by manual inspection of all tandem mass spectra.
Supplementary Material
Acknowledgments
We are grateful to Claiborne Glover III for sharing cka2ts mutant plasmids and CK2 purified from yeast. We thank Philip Hieter for providing the anti-Ndc10 antibody. We thank Yidi Sun, Alphée Michelot, Adrianne Pigula, and Jonathan Wong for experimental help and suggestions. This research was supported by National Institutes of Health grants GM47842 to G.B. and P41 RR011823 to J.Y. III.
Abbreviations used:
- ALP
alkaline phosphatase
- APC
anaphase-promoting complex
- ChIP
chromatin immunoprecipitations
- CPC
chromosomal passenger complex
- GFP
green fluorescent protein
- MS
mass spectrometry
- Ni-NTA
nickel nitrilotriacetic acid
- PSMs
peptide/spectrum matches
- RFP
red fluorescent protein
- SAC
spindle assembly checkpoint
- SPB
spindle pole body
- YPD
yeast–peptone–dextrose.
Footnotes
This article was published online ahead of print in MBoC in Press (http://www.molbiolcell.org/cgi/doi/10.1091/mbc.E10-11-0915) on June 1, 2011.
REFERENCES
- Biggins S, Murray AW. The budding yeast protein kinase Ipl1/Aurora allows the absence of tension to activate the spindle checkpoint. Genes Dev. 2001;15:3118–3129. doi: 10.1101/gad.934801. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Biggins S, Severin FF, Bhalla N, Sassoon I, Hyman AA, Murray AW. The conserved protein kinase Ipl1 regulates microtubule binding to kinetochores in budding yeast. Genes Dev. 1999;13:532–544. doi: 10.1101/gad.13.5.532. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bouck DC, Bloom KS. The kinetochore protein Ndc10p is required for spindle stability and cytokinesis in yeast. Proc Natl Acad Sci USA. 2005;102:5408–5413. doi: 10.1073/pnas.0405925102. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Canton DA, Litchfield DW. The shape of things to come: an emerging role for protein kinase CK2 in the regulation of cell morphology and the cytoskeleton. Cell Signal. 2006;18:267–275. doi: 10.1016/j.cellsig.2005.07.008. [DOI] [PubMed] [Google Scholar]
- Carroll CW, Milks KJ, Straight AF. Dual recognition of CENP-A nucleosomes is required for centromere assembly. J Cell Biol. 2010;189:1143–1155. doi: 10.1083/jcb.201001013. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cheeseman IM, Anderson S, Jwa M, Green EM, Kang J, Yates JR, III, Chan CS, Drubin DG, Barnes G. Phospho-regulation of kinetochore-microtubule attachments by the Aurora kinase Ipl1p. Cell. 2002;111:163–172. doi: 10.1016/s0092-8674(02)00973-x. [DOI] [PubMed] [Google Scholar]
- Cociorva D, Tabb DL, Yates JR., III Validation of tandem mass spectrometry database search results using DTASelect. Current Protoc Bioinformatics. 2007;Chap 13 (Unit 13.4) doi: 10.1002/0471250953.bi1304s16. [DOI] [PubMed] [Google Scholar]
- Cohen RL, Espelin CW, De Wulf P, Sorger PK, Harrison SC, Simons KT. Structural and functional dissection of Mif2p, a conserved DNA-binding kinetochore protein. Mol Biol Cell. 2008;19:4480–4491. doi: 10.1091/mbc.E08-03-0297. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Corbett KD, Yip CK, Ee LS, Walz T, Amon A, Harrison SC. The monopolin complex crosslinks kinetochore components to regulate chromosome-microtubule attachments. Cell. 2010;142:556–567. doi: 10.1016/j.cell.2010.07.017. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Eng JK, McCormack AL, Yates JR., III An approach to correlate tandem mass spectral data of peptides with amino acid sequences in a protein database. J Am Soc Mass Spectrom. 1994;5:976–989. doi: 10.1016/1044-0305(94)80016-2. [DOI] [PubMed] [Google Scholar]
- Gestaut DR, Graczyk B, Cooper J, Widlund PO, Zelter A, Wordeman L, Asbury CL, Davis TN. Phosphoregulation and depolymerization-driven movement of the Dam1 complex do not require ring formation. Nat Cell Biol. 2008;10:407–414. doi: 10.1038/ncb1702. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Glover CV III. On the physiological role of casein kinase II in Saccharomyces cerevisiae. Prog Nucleic Acid Res Mol Biol. 1998;59:95–133. doi: 10.1016/s0079-6603(08)61030-2. [DOI] [PubMed] [Google Scholar]
- Grishchuk EL, Spiridonov IS, Volkov VA, Efremov A, Westermann S, Drubin D, Barnes G, Ataullakhanov FI, McIntosh JR. Different assemblies of the DAM1 complex follow shortening microtubules by distinct mechanisms. Proc Natl Acad Sci USA. 2008;105:6918–6923. doi: 10.1073/pnas.0801811105. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Haase SB. Cell cycle analysis of budding yeast using SYTOX Green. Curr Protoc Cytom. 2004 doi: 10.1002/0471142956.cy0723s26. Chap 7, Unit 7.23. [DOI] [PubMed] [Google Scholar]
- Hanna DE, Rethinaswamy A, Glover CV. Casein kinase II is required for cell cycle progression during G1 and G2/M in Saccharomyces cerevisiae. J Biol Chem. 1995;270:25905–25914. doi: 10.1074/jbc.270.43.25905. [DOI] [PubMed] [Google Scholar]
- Kaksonen M, Sun Y, Drubin DG. A pathway for association of receptors, adaptors, and actin during endocytic internalization. Cell. 2003;115:475–487. doi: 10.1016/s0092-8674(03)00883-3. [DOI] [PubMed] [Google Scholar]
- Kang J, Cheeseman IM, Kallstrom G, Velmurugan S, Barnes G, Chan CS. Functional cooperation of Dam1, Ipl1, and the inner centromere protein (INCENP)-related protein Sli15 during chromosome segregation. J Cell Biol. 2001;155:763–774. doi: 10.1083/jcb.200105029. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kemmler S, Stach M, Knapp M, Ortiz J, Pfannstiel J, Ruppert T, Lechner J. Mimicking Ndc80 phosphorylation triggers spindle assembly checkpoint signalling. EMBO J. 2009;28:1099–1110. doi: 10.1038/emboj.2009.62. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kim JH, Kang JS, Chan CS. Sli15 associates with the ipl1 protein kinase to promote proper chromosome segregation in Saccharomyces cerevisiae. J Cell Biol. 1999;145:1381–1394. doi: 10.1083/jcb.145.7.1381. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lanini L, McKeon F. Domains required for CENP-C assembly at the kinetochore. Mol Biol Cell. 1995;6:1049–1059. doi: 10.1091/mbc.6.8.1049. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Li Y, Elledge SJ. The DASH complex component Ask1 is a cell cycle-regulated Cdk substrate in Saccharomyces cerevisiae. Cell Cycle. 2003;2:143–148. [PubMed] [Google Scholar]
- Litchfield DW. Protein kinase CK2: structure, regulation and role in cellular decisions of life and death. Biochem J. 2003;369:1–15. doi: 10.1042/BJ20021469. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lu B, Ruse C, Xu T, Park SK, Yates J III. Automatic validation of phosphopeptide identifications from tandem mass spectra. Anal Chem. 2007;79:1301–1310. doi: 10.1021/ac061334v. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lu B, Xu T, Park SK, McClatchy DB, Liao L, Yates JR., III Shotgun protein identification and quantification by mass spectrometry in neuroproteomics. Methods Mol Biol. 2009;566:229–259. doi: 10.1007/978-1-59745-562-6_16. [DOI] [PubMed] [Google Scholar]
- Meggio F, Pinna LA. One-thousand-and-one substrates of protein kinase CK2? FASEB J. 2003;17:349–368. doi: 10.1096/fj.02-0473rev. [DOI] [PubMed] [Google Scholar]
- Meluh PB, Koshland D. Evidence that the MIF2 gene of Saccharomyces cerevisiae encodes a centromere protein with homology to the mammalian centromere protein CENP-C. Mol Biol Cell. 1995;6:793–807. doi: 10.1091/mbc.6.7.793. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Meluh PB, Koshland D. Budding yeast centromere composition and assembly as revealed by in vivo cross-linking. Genes Dev. 1997;11:3401–3412. doi: 10.1101/gad.11.24.3401. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Milks KJ, Moree B, Straight AF. Dissection of CENP-C-directed centromere and kinetochore assembly. Mol Biol Cell. 2009;20:4246–4255. doi: 10.1091/mbc.E09-05-0378. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Miranda JJ, De Wulf P, Sorger PK, Harrison SC. The yeast DASH complex forms closed rings on microtubules. Nat Struct Mol Biol. 2005;12:138–143. doi: 10.1038/nsmb896. [DOI] [PubMed] [Google Scholar]
- Montpetit B, Hazbun TR, Fields S, Hieter P. Sumoylation of the budding yeast kinetochore protein Ndc10 is required for Ndc10 spindle localization and regulation of anaphase spindle elongation. J Cell Biol. 2006;174:653–663. doi: 10.1083/jcb.200605019. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nakajima Y, Tyers RG, Wong CC, Yates JR, III, Drubin DG, Barnes G. Nbl1p: a Borealin/Dasra/CSC-1-like protein essential for Aurora/Ipl1 complex function and integrity in Saccharomyces cerevisiae. Mol Biol Cell. 2009;20:1772–1784. doi: 10.1091/mbc.E08-10-1011. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Niefind K, Raaf J, Issinger OG. Protein kinase CK2 in health and disease: protein kinase CK2: from structures to insights. Cell Mol Life Sci. 2009;66:1800–1816. doi: 10.1007/s00018-009-9149-8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Padmanabha R, Glover CV. Casein kinase II of yeast contains two distinct alpha polypeptides and an unusually large beta subunit. J Biol Chem. 1987;262:1829–1835. [PubMed] [Google Scholar]
- Peng J, Elias JE, Thoreen CC, Licklider LJ, Gygi SP. Evaluation of multidimensional chromatography coupled with tandem mass spectrometry (LC/LC-MS/MS) for large-scale protein analysis: the yeast proteome. J Proteome Res. 2003;2:43–50. doi: 10.1021/pr025556v. [DOI] [PubMed] [Google Scholar]
- Przewloka MR, Venkei Z, Bolanos-Garcia VM, Debski J, Dadlez M, Glover DM. CENP-C is a structural platform for kinetochore assembly. Curr Biol. 2011;21:399–405. doi: 10.1016/j.cub.2011.02.005. [DOI] [PubMed] [Google Scholar]
- Rethinaswamy A, Birnbaum MJ, Glover CV. Temperature-sensitive mutations of the CKA1 gene reveal a role for casein kinase II in maintenance of cell polarity in Saccharomyces cerevisiae. J Biol Chem. 1998;273:5869–5877. doi: 10.1074/jbc.273.10.5869. [DOI] [PubMed] [Google Scholar]
- Rodal AA, Manning AL, Goode BL, Drubin DG. Negative regulation of yeast WASp by two SH3 domain-containing proteins. Curr Biol. 2003;13:1000–1008. doi: 10.1016/s0960-9822(03)00383-x. [DOI] [PubMed] [Google Scholar]
- Rodrigo-Brenni MC, Thomas S, Bouck DC, Kaplan KB. Sgt1p and Skp1p modulate the assembly and turnover of CBF3 complexes required for proper kinetochore function. Mol Biol Cell. 2004;15:3366–3378. doi: 10.1091/mbc.E03-12-0887. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Romao M, Tanaka K, Sibarita JB, Ly-Hartig NT, Tanaka TU, Antony C. Three-dimensional electron microscopy analysis of ndc10–1 mutant reveals an aberrant organization of the mitotic spindle and spindle pole body defects in Saccharomyces cerevisiae. J Struct Biol. 2008;163:18–28. doi: 10.1016/j.jsb.2008.03.015. [DOI] [PubMed] [Google Scholar]
- Ruchaud S, Carmena M, Earnshaw WC. The chromosomal passenger complex: one for all and all for one. Cell. 2007;131:230–231. doi: 10.1016/j.cell.2007.10.002. [DOI] [PubMed] [Google Scholar]
- Sassoon I, Severin FF, Andrews PD, Taba MR, Kaplan KB, Ashford AJ, Stark MJ, Sorger PK, Hyman AA. Regulation of Saccharomyces cerevisiae kinetochores by the type 1 phosphatase Glc7p. Genes Dev. 1999;13:545–555. doi: 10.1101/gad.13.5.545. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Screpanti E, De Antoni A, Alushin GM, Petrovic A, Melis T, Nogales E, Musacchio A. Direct binding of Cenp-C to the Mis12 complex joins the inner and outer kinetochore. Curr Biol. 2011;21:391–398. doi: 10.1016/j.cub.2010.12.039. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Shimada M, Yamamoto A, Murakami-Tonami Y, Nakanishi M, Yoshida T, Aiba H, Murakami H. Casein kinase II is required for the spindle assembly checkpoint by regulating Mad2p in fission yeast. Biochem Biophys Res Commun. 2009;388:529–532. doi: 10.1016/j.bbrc.2009.08.030. [DOI] [PubMed] [Google Scholar]
- Shimogawa MM, et al. Mps1 phosphorylation of Dam1 couples kinetochores to microtubule plus ends at metaphase. Curr Biol. 2006;16:1489–1501. doi: 10.1016/j.cub.2006.06.063. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sorger PK, Severin FF, Hyman AA. Factors required for the binding of reassembled yeast kinetochores to microtubules in vitro. J Cell Biol. 1994;127:995–1008. doi: 10.1083/jcb.127.4.995. [DOI] [PMC free article] [PubMed] [Google Scholar]
- St-Denis NA, Derksen DR, Litchfield DW. Evidence for regulation of mitotic progression through temporal phosphorylation and dephosphorylation of CK2α. Mol Cell Biol. 2009;29:2068–2081. doi: 10.1128/MCB.01563-08. [DOI] [PMC free article] [PubMed] [Google Scholar]
- St-Denis NA, Litchfield DW. Protein kinase CK2 in health and disease: from birth to death: the role of protein kinase CK2 in the regulation of cell proliferation and survival. Cell Mol Life Sci. 2009;66:1817–1829. doi: 10.1007/s00018-009-9150-2. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Straight AF, Belmont AS, Robinett CC, Murray AW. GFP tagging of budding yeast chromosomes reveals that protein-protein interactions can mediate sister chromatid cohesion. Curr Biol. 1996;6:1599–1608. doi: 10.1016/s0960-9822(02)70783-5. [DOI] [PubMed] [Google Scholar]
- Tabb DL, McDonald WH, Yates JR., III DTASelect and Contrast: tools for assembling and comparing protein identifications from shotgun proteomics. J Proteome Res. 2002;1:21–26. doi: 10.1021/pr015504q. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tanaka K, Chang HL, Kagami A, Watanabe Y. CENP-C functions as a scaffold for effectors with essential kinetochore functions in mitosis and meiosis. Dev Cell. 2009;17:334–343. doi: 10.1016/j.devcel.2009.08.004. [DOI] [PubMed] [Google Scholar]
- Trembley JH, Wang G, Unger G, Slaton J, Ahmed K. Protein kinase CK2 in health and disease: CK2: a key player in cancer biology. Cell Mol Life Sci. 2009;66:1858–1867. doi: 10.1007/s00018-009-9154-y. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tripodi F, Cirulli C, Reghellin V, Marin O, Brambilla L, Schiappelli MP, Porro D, Vanoni M, Alberghina L, Coccetti P. CK2 activity is modulated by growth rate in Saccharomyces cerevisiae. Biochem Biophys Res Commun. 2010;398:44–50. doi: 10.1016/j.bbrc.2010.06.028. [DOI] [PubMed] [Google Scholar]
- Westermann S, Avila-Sakar A, Wang HW, Niederstrasser H, Wong J, Drubin DG, Nogales E, Barnes G. Formation of a dynamic kinetochore-microtubule interface through assembly of the Dam1 ring complex. Mol Cell. 2005;17:277–290. doi: 10.1016/j.molcel.2004.12.019. [DOI] [PubMed] [Google Scholar]
- Westermann S, Cheeseman IM, Anderson S, Yates JR, III, Drubin DG, Barnes G. Architecture of the budding yeast kinetochore reveals a conserved molecular core. J Cell Biol. 2003;163:215–222. doi: 10.1083/jcb.200305100. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Westermann S, Drubin DG, Barnes G. Structures and functions of yeast kinetochore complexes. Annu Rev Biochem. 2007;76:563–591. doi: 10.1146/annurev.biochem.76.052705.160607. [DOI] [PubMed] [Google Scholar]
- Westermann S, Wang HW, Avila-Sakar A, Drubin DG, Nogales E, Barnes G. The Dam1 kinetochore ring complex moves processively on depolymerizing microtubule ends. Nature. 2006;440:565–569. doi: 10.1038/nature04409. [DOI] [PubMed] [Google Scholar]
- Widlund PO, Lyssand JS, Anderson S, Niessen S, Yates JR, III, Davis TN. Phosphorylation of the chromosomal passenger protein Bir1 is required for localization of Ndc10 to the spindle during anaphase and full spindle elongation. Mol Biol Cell. 2006;17:1065–1074. doi: 10.1091/mbc.E05-07-0640. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wong J, et al. A protein interaction map of the mitotic spindle. Mol Biol Cell. 2007;18:3800–3809. doi: 10.1091/mbc.E07-06-0536. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Xu T, Venable JD, Park SK, Cociorva D, Lu B, Liao L, Wohlschlegel J, Hewel J, Yates JR., III ProLuCID, a fast and sensitive tandem mass spectra-based protein identification program. Mol Cell Proteomics. 2006;5:S174. [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.